Abstract
It is proposed that the precellular stage of biological evolution unraveled within networks of inorganic compartments that harbored a diverse mix of virus‐like genetic elements. This stage of evolution might makes up the Last Universal Cellular Ancestor (LUCA) that more appropriately could be denoted Last Universal Cellular Ancestral State (LUCAS). Such a scenario recapitulates the ideas of J. B. S. Haldane sketched in his classic 1928 essay. However, unlike in Haldane's day, considerable support for this scenario exits today: lack of homology between core DNA replication system components in archaea and bacteria, distinct membrane chemistries and enzymes of lipid biosynthesis in archaea and bacteria, spread of several viral hallmark genes among diverse groups of viruses, and the extant archaeal and bacterial chromosomes appear to be shaped by accretion of diverse, smaller replicons. Under the viral model of precellular evolution, the key components of cells originated as components of virus‐like entities. The two surviving types of cellular life forms, archaea and bacteria, might have emerged from the LUCAS independently, along with, probably, numerous forms now extinct.
Keywords: comparative genomics, evolution of cells, evolution of viruses, origin of membranes, viral hallmark genes
Comparative Genomics, Ancestral Gene Repertoires, and Last Universal Cellular Ancestor
As numerous complete genomes from diverse walks of life become available, comparative genomics turns into a truly powerful methodology. 1 , 2 , 3 , 4 It has the ability not only to determine which genes are conserved and which are not, but also to reconstruct the gene composition of ancestral life forms including the hypothetical Last Universal Common (Cellular) Ancestor (LUCA)—under certain assumptions, of course. 5 , 6 , 7 , 8 , 9 The key assumption is that genes shared by many diverse extant species are most likely to be inherited from the common ancestor of these species—in particular, genes that are present in all modern cellular life forms hark back to LUCA. The number of such ubiquitous genes is very small, fewer than 60, and nearly all of them encode proteins involved in translation and the core transcription machinery. 5 , 6 , 7 This limited repertoire of genes obviously could not provide for a viable life form, so a considerable number of genes that must have been present in LUCA were lost or displaced in some lines of descent during the subsequent evolution.
Consequently, reconstruction approaches have to be applied in order to delineate the likely gene complement of LUCA. The simplest reconstruction methods are based on the principle of evolutionary parsimony, that is, attempt to derive the evolutionary scenario that includes the smallest number of elementary events (the most parsimonious scenario). 10 , 11 , 12 The set of relevant events is small: (i) gene “birth,” that is, emergence of a new gene, typically, via gene duplication followed by radical divergence, (ii) gene acquisition via horizontal gene transfer (HGT), and (iii) gene loss.
Counting these events for different scenarios and choosing the one with the minimum number of events seems to be a straightforward task. However, realization of this goal meets with hurdles at several levels. First, in order to derive the patterns of presence‐absence of a gene in a set of lineages (phyletic pattern), which are used as the input for the reconstruction methods, it is necessary to robustly identify orthologous genes, that is, genes that evolved from a single ancestor gene in the common ancestor of the compared species. 13 , 14 Identification of orthologs is a nontrivial task for relatively fast‐evolving genes from distant species and, especially, for any genes with a history of multiple duplications and losses. Second, and more fundamentally, reliable reconstruction of the course of evolution and of the ancestral gene sets is hampered by the uncertainty associated with the relative probabilities or rates of different events, in particular, gene loss versus horizontal gene transfer. Third, even phyletic patterns based on reliably delineated sets of orthologs hardly contain all the information that is required for the evolutionary reconstruction. In principle, even a gene that is found in all modern cellular life forms might not be inherited from LUCA: its ubiquity could instead result from an HGT sweep. Fourth, reconstruction methods based on parsimony are inherently limited as they have no capability to identify ancestral genes that have been lost in all or all but one of the extant lineages. Thus, the estimates of the gene content of ancestral forms are conservative, and the extent of underestimate is uncertain. Finally, to generate evolutionary scenarios, the parsimony reconstructions rely on a particular topology of the “tree of life.” Even apart from the major uncertainties that are inherent in deep phylogenetic trees, any such tree at best reflects the history of a small fraction of highly conserved genes: figuratively speaking, it is “a tree of one percent.” 15 Worse yet, the very adequacy of the “tree of life” concept is questionable considering the extensive HGT that is part and parcel of the evolution of prokaryotes. 16 , 17 A more adequate probabilistic framework, such as that provided by maximum likelihood models, is required to produce more realistic estimates, but such models can be prohibitively complex, and the approach to parameter estimation is unclear. Neither is it clear how the reconstruction can be performed in a tree‐independent fashion.
All the difficulties and uncertainties of evolutionary reconstructions notwithstanding, parsimony analyses combined with less formal attempts on the reconstruction of the deep past of particular functional systems leave no serious doubts that LUCA already possessed at least several hundred genes. This diverse gene complement consists of genes encoding proteins of information processing systems including not only the core structural components (e.g., a minimal set of ribosomal proteins) but also some “accessory” proteins, for example, a considerable variety of RNA modification enzymes; numerous metabolic pathways including the central energy metabolism and the biosynthesis of amino acids, nucleotides, and some coenzymes; and some crucial membrane proteins, such as the subunits of the signal recognition particle (SRP) and the H+‐ATPase. 11 , 18 , 19 In addition, a considerable number of RNA species such as three rRNAs, tRNA of all specificities, and the SRP 7S RNA are confidently traced back to LUCA.
However, there are also gaping holes in the reconstructed gene repertoire of LUCA. The two most important ones are (i) the absence of the central parts of the DNA replication machinery, namely, the polymerases that are responsible for the initiation (primases) and elongation of DNA replication, and for gap‐filling after primer removal, and the principal DNA helicases, and (ii) the absence of most enzymes of lipid biosynthesis. These proteins fail to make it into the reconstructed gene repertoire of LUCA because the respective processes in bacteria, on the one hand, and archaea on the other hand are catalyzed by distinct, unrelated enzymes and, in the case of membrane phospholipids, yield chemically distinct membranes (the archaeal membrane phospholipids are isoprenoid ethers of glycerol 1‐phosphate whereas bacterial lipids fatty acid esthers of glycerol 3‐phosphate, i.e., the lipids in the two domains differ not only in their chemical composition but also in chirality). 20 , 21 , 22 , 23 , 24 Thus, the reconstructed gene set of LUCA seems to display a remarkable nonuniformity in that some functional systems seem to reach elaborate complexity almost indistinguishable from that in modern organisms whereas others are rudimentary or missing. This strange picture is remarkably similar to Woese's general concept of nonsimultaneous “crystallization” of different cellular systems at the early stages of evolution 25 and prompts one to step back and take a more general view at the LUCA problem.
Why There Must Have Been a LUCA and What Do We Know About It for Certain?
The year 2009 is the Darwin year when the world celebrates his 200th birthday and the 150th anniversary of On the Origin of Species. 26 It also happens to be the 150th jubilee of the idea of LUCA that, to my knowledge, was clearly proposed by Darwin for the first time (the acronym itself, of course, is much younger: it was coined in 1996 at a special meeting on the last common ancestor of modern life forms 27 ). In the famous final passage of the Origin, Darwin wrote: “There is grandeur in this view of life, with its several powers, having been originally breathed into a few forms or into one; and that, whilst this planet has gone cycling on according to the fixed law of gravity, from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved.” 26 In Darwin's day, this was an incredibly bold conjecture considering that the only empirical support came from phenotypic similarities between diverse organisms (paradoxically, Darwin's prescience might have been helped by the obscurity of microbes at the time so that he, effectively, considered multicellular organisms).
The advances of molecular biology and, later, comparative genomics forcefully vindicated Darwin's insight. The (near) universality of the genetic code complemented by the universal conservation of ∼50 proteins involved in the core translation functions, ∼30 structural RNAs, and the three core subunits of the DNA‐dependent RNA polymerase 5 , 6 , 7 comprise strong evidence in support of the existence of some form of LUCA. Importantly, most of these molecules show a clear‐cut pattern of phylogenetic relationships, with the three domains of life (bacteria, archaea, and eukaryota) being well separated in phylogenetic trees, and the archaeal and eukaryotic sequences showing greater similarity to each other, which suggest rooting the tree between the archaeo‐eukaryotic and bacterial branches. 6 , 28 This rooting was supported by the phylogenetic analysis of ancient paralogous genes, namely, translation factors and membrane ATPase subunits, that are thought to derive from gene duplications antedating LUCA. 29 , 30
Although it has been suggested that this tree topology is a long‐branch attraction artifact and so the root position has been challenged, 31 , 32 , 33 it appears clear that there is a substantial, even if numerically relatively small, set of genes that are not only common to all cellular life forms but also share a (largely) common history. The existence of this evolutionarily coherent gene set that is, in all likelihood, ancestral to all extant cellular life appears to, effectively, prove the existence of an ancestral state that can be reasonably denoted LUCA. The real issue, then, is not whether or not a LUCA existed but rather what it was like, that is, which features of this entity we can infer with confidence and which (so far) remain uncertain.
It seems to make sense to think of LUCA in two distinct contexts:
-
(i)
complexity that can be expressed as the number of distinct genes; and
-
(ii)
the degree of organizational and biological similarity to modern cells—for brevity and convenience, this property can be denoted “cellularity.”
These two characteristics are likely to correlate but are not necessarily tightly coupled let alone deterministically linked. In principle, it is not inconceivable that LUCA was a cellular entity that was substantially simpler than any modern cell (at least, a free‐living one) in terms of its genetic content or, conversely, that considerable genetic complexity evolved prior to the emergence of cellular organization (Fig. 1).
Figure 1.
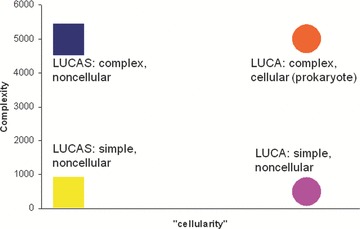
Genetic complexity and “cellularity” of LUCA(S): the space of logical possibilities. “Cellularity” is the degree of similarity to the organization of modern cells. The notion of “cellularity” is qualitative, there are no specific units. The complexity scale also could be considered arbitrary, but the units of complexity can be assumed to (roughly) represent the number of genes. The primordial virus world model delineated in this article implies the complex, noncellular LUCAS.
All the uncertainties involved notwithstanding, it seems to be extremely likely that LUCA was fairly complex, that is, had at least about as many genes as the simplest of the modern free‐living prokaryotes, namely, on the order of a 1000 genes or more. Figures in this range have been inferred by all algorithmic methods for ancestral gene set reconstruction. 5 , 11 , 12 , 19 However, given the uncertainty associated with these approaches (see above), the more compelling argument for a complex LUCA is the complexity of the modern translation machinery that comprises indisputable LUCA heritage. The functioning of such an advanced translation system is predicated on commensurate metabolic capabilities, including not only the pathways for the synthesis of all nucleotides and (nearly) all amino acids, but also those for at least some coenzymes, for example, S‐adenosylmethionine, the cofactor of the numerous RNA methylases several of which can be traced back to LUCA with a high confidence. 18 , 34 Furthermore, the evolutionary relationships of some translation system components imply that these proteins are products of preceding complex evolution. A case in point are the aminoacyl‐tRNA synthetases (aaRS), the 20 enzymes (one for each amino acid) that are essential for translation and of which, at least, 18 are confidently traced back to LUCA. 35 , 36 The core catalytic domains of the aaRS represent two distinct classes that possess unrelated structural folds and cover 10 amino acid specificities each. Analysis of the evolutionary history of the catalytic domains of Class I aaRS indicates that they all make up one cluster of terminal branches in the elaborate tree of the “Rossmann‐like” protein domains. 37 , 38 Thus, the diversification of the aaRS, that was already (nearly) complete in LUCA, was preceded by complex protein evolution including the divergence of many families of enzymes. The same argument applies to translation factors, RNA methylases, and other groups of proteins involved in translation. 18 Logically, these observations clinch the case for a LUCA whose genetic complexity was, in the least, not much lower than that of simple modern prokaryotes.
However, it is far from being obvious that LUCA resembled modern prokaryotes in terms of cellular organization as well. The “uniformitarian assumption,” namely, that LUCA was a more or less regular, modern type is often accepted, effectively, by default in the discussions of early evolution, even if rarely discussed explicitly. 39 , 40 , 41 However, any reconstruction of LUCA must account for the evolution of the features that are not immediately traceable back to the common ancestor of archaea and bacteria, the two main ones being DNA replication and membrane biogenesis (and chemistry). The uniformitarian hypotheses of LUCA would explain the lack of conservation of these key systems in one of two ways:
-
(i)
LUCA somehow combined both versions of these systems, with subsequent differential loss in the archaeal and bacterial lineages; and
-
(ii)
LUCA had a particular version of each of these systems, with subsequent nonorthologous displacement in archaea or bacteria.
Specifically, with respect to membrane biogenesis, it has been proposed that LUCA had a mixed, heterochiral membrane, with the two versions with opposite chiralities emerging as a result of subsequent specialization in archaea and bacteria. 24 With regard to the DNA replication, a hypothesis has been developed under which one of the modern replication systems is ancestral whereas the other system evolved in viruses and subsequently displaced the original one in either the archaeal or the bacterial lineage. 42
By contrast, radical proposals on LUCA's nature take a “what you see is what you get” approach by postulating that LUCA lacked those key features that are not homologous in extant archaea and bacteria, at least, in their modern form. The possibility that LUCA was radically different from any known cells has been brought up, originally, in the concept of “progenote,” a hypothetical, primitive entity in which the link between the genotype and the phenotype was not yet firmly established. 43 In its original form, the progenote idea involves primitive, imprecise translation, a notion that is not viable given the extensive diversification of proteins prior to LUCA that is demonstrated beyond doubt by the analysis of diverse protein superfamilies (see above). More realistically, it can be proposed that the emergence of the major features of cells was substantially asynchronous 25 so that LUCA closely resembled modern cells in some ways but was distinctly “primitive” in others. The results of comparative genomics provide clues for distinguishing advanced and primitive features of LUCA. Thus, focusing on the major areas of nonhomology between archaea and bacteria, it has been hypothesized that LUCA:
- (i)
- (ii)
Figure 2.
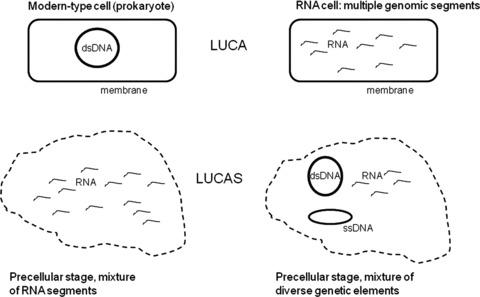
Distinct possible organizations of LUCA(S). The dashed line schematically denotes an unspecified form of compartmentalization in the precellular LUCAS.
With respect to the DNA genome and replication, the conundrum to explain was the combination of nonhomologous and conserved components of the DNA replication machinery as well as the universal conservation of the core transcription machinery. To account for this mixed pattern of conservation and diversity, it has been suggested that LUCA had a retrovirus‐like replication cycle, with the conserved transcription machinery involved in the transcription of provirus‐like dsDNA molecules and the conserved components of the DNA replication system playing accessory roles in this process. 22 This speculative scheme combined, in the same hypothetical replication cycle, the conserved proteins that are involved in transcription and replication with proteins, such as reverse transcriptase (RT) that, among the extant life forms, are seen, primarily or exclusively, in viruses and other selfish genetic elements. The proposal formally accounts for the universal conservation of these proteins but has no direct analogy in extant genetic systems.
The other major area of nonhomology between archaea and bacteria, lipid biosynthesis (along with lipid chemistry) prompted the notion of a noncellular, although compartmentalized LUCA. Specifically, it has been proposed that LUCA might have been a diverse population of expressed genetic elements that dwelled in networks of inorganic compartments. 23 A major hurdle for the models of non‐membrane‐bounded LUCA is that several membrane proteins and even molecular complexes, such as the proton ATPase and the signal recognition particle (SRP), are nearly universal among modern cellular life forms and, in all likelihood, were present in LUCA. 45
A more careful consideration of the “genomic” (lack of homology of the core components of the DNA replication systems in archaea and bacteria) and the “membrane” (radical difference in between the phospholipids and the enzymes of lipid biosynthesis between archaea and bacteria) challenges to LUCA suggests that the two are tightly linked. A complex LUCA without a large DNA genome similar to modern bacterial and archaeal genomes could only have a genome consisting of several hundred segments of RNA (or provirus‐like DNA), each several kilobases in size. This limitation is dictated by the dramatically lower stability of RNA molecules compared to DNA and is empirically supported by the fact that the largest known RNA genomes (those of coronaviruses) are ∼30 kb in size. 46 It has been proposed that LUCA represented a bona fide RNA cell that subsequently radiated into three major RNA cell lineages (the ancestors of bacteria, archaea, and eukarya) in which the genome was independently replaced by DNA as a result of acquisition of the DNA replication machinery from distinct viruses. 44 However, the necessity to possess hundreds of genomic RNA segments seems to raise an insurmountable obstacle for a RNA cell because a reasonable accuracy of genome partitioning into daughter cells during cell division would require elaborate mechanisms of genome segregation of a kind not found in modern prokaryotes. Otherwise, the change in the gene complement brought about by each cell division would, effectively, prevent reproduction. Those segregation mechanisms that do operate in modern bacteria (and, probably, archaea) involve pumping of dsDNA into daughter cells with the help of a specific ATPase and, probably, coevolved with large dsDNA genomes. 47 , 48 , 49 , 50 Thus, if LUCA indeed lacked a large dsDNA genome and instead had a “collective” genome comprising numerous RNA segments, it must have been a life form distinct from modern cells, perhaps, actually, a noncellular one.
Another broadly discussed aspect of early life forms, including LUCA, is the rampant HGT that is often considered a prerequisite for the evolution of complex life. 51 , 52 Indeed, HGT is the route of rapid innovation, and innovation was bound to be rapid at the earliest stages of life's evolution. Moreover, it has been recently suggested and illustrated by mathematical modeling that the very universality of the genetic code might be linked to the critical role of HGT at the early phase of evolution: in the presence of extensive HGT, a single version of the code would necessarily sweep the population of ancestral life forms, whereas any organisms with deviant code would be unable to benefit from HGT and, being isolated from other organisms, would be eliminated by selection. 53 , 54 Analogies with the history of human civilization are obvious and, perhaps, illuminating: the existence of a lingua franca greatly accelerates progress, and conversely, isolated communities are stalled in their development and doomed to eventual extinction. Constant, extensive HGT is an intrinsic feature of the models of noncellular, compartmentalized LUCA 45 but certainly cannot be taken for granted within the framework of the cellular LUCA models. An updated version of the non‐cellular LUCA model is presented below.
A Noncellular but Compartmentalized LUCA(S): A Community of Diverse Replicators and the Playground of Early Evolution
Russell and coworkers proposed that networks of microcompartments that exist at both extant and ancient hydrothermal vents, and consist, primarily, of iron sulfide could be ideal habitats for early life. These inorganic compartment networks provide gradients of temperature and pH that could fuel primordial energetics, and versatile catalytic surfaces for primitive biochemistry. 55 , 56 These might have been the sites of prebiological and precellular biological evolution, from mixtures of organic molecules to the putative, primordial RNA world to the independent escapes of archaeal and bacterial cells. 23 , 45 These compartments are envisaged being inhabited by diverse populations of genetic elements, initially, segments of RNA, subsequently, larger and more complex RNA molecules encompassing one or a few protein‐coding genes, and later yet, also DNA segments of gradually increasing size (Fig. 3). Notably, a computer simulation study has shown that, in the presence of thermal gradient that inevitably exists at a hydrothermal vent, extremely high concentrations of small molecules and polymers can be reached, 57 a condition that would substantially facilitate a variety of reactions including RNA ligation. 58
Figure 3.

The primordial virus world model of precellular evolution. Some major stages in precellular evolution are denoted to the right. Thin, waved lines indicate RNA elements, and circles and ovals indicate DNA elements. Hexagons denote virus‐like particles, and those enclosed in rounded rectangles denote membrane‐containing virions. DNA segments replicated by an archaeal‐type replication machinery are shown in red, and those replicated by the bacterial‐type replication system are shown in blue; the two types of membranes are similarly color coded. The archaeal‐type replication system might have antedated the bacterial systems during precellular evolution as argued elsewhere. 106 Arrows between compartments indicate horizontal transfer of the contents.
Thus, early life forms, likely including LUCA, are perceived as complex ensembles of genetic elements that inhabited networks of inorganic compartments. 45 , 59 A key feature of this model is that genetic elements with different replication and expression strategies (including replicating DNA segments) encoding distinct replication machineries would coexist within a network or even within the same compartment. Thus, the earlier, somewhat artificial scheme, in which the universally conserved components of the DNA replication machinery were implicated in a primordial, retrovirus‐like replication cycle, 22 might be superfluous. The model of the compartmentalized primordial gene pool implies evolution of the retrovirus‐like replication cycle within the RNA‐protein world and subsequent evolution of diverse DNA replication systems (Fig. 3) but does not necessarily require the components of these distinct genetic systems to function together within the same replication cycle.
This model explains the lack of homology between the membranes, membrane biogenesis systems, and the DNA replication machineries of archaea and bacteria by inferring a LUCA that did not have a single, large DNA genome and was not a membrane‐bounded cell. However, under this model, the primordial, precellular life forms are envisaged as “laboratories” in which various strategies of genome replication‐expression as well as rudimentary forms of biogenic compartmentalization were “invented” and tried out (Fig. 3 and see below).
The central point of this scenario of life's early evolution is the virus‐like nature of the perceived precellular life forms. The idea that viruses could be related to the first life forms is almost as old as virology itself. Apparently, it was first proposed by Felix d’Herelle, the discoverer of bacteriophages 60 and was incorporated and developed by J. B. S. Haldane in his classic 1928 essay on the origin of life. 61 Haldane came up with the striking speculation that the first self‐reproducing agents were viruses or virus‐like agents and that a virus stage in life's evolution preceded the emergence of cells. Subsequently, the concept of the primordial origin of viruses was, largely, abandoned as it became obvious that viruses were obligate intracellular parasites that depend on the host cells for most of their functions; instead, the scenarios of cell degeneration or escaped cellular genes became dominant in the thinking on the origins of viruses. 62 , 63 , 64
Very recently, the study of fundamental aspects of virus evolution experienced a true renaissance that led to the proliferation of hypotheses and models that revolve around the concept that viruses were important contributors to the origin and evolution of cells. 42 , 44 , 59 , 65 , 66 , 67 , 68 , 69 , 70 In particular, Forterre proposed the hypothesis of “three DNA cells and thee DNA viruses” according to which modern‐type DNA‐based cells evolved when three distinct DNA viruses displaced the original RNA genomes in three cellular lineages (progenitors of bacteria, archaea, and eukaryotes, respectively); the DNA viruses themselves are thought to have evolved as parasites of these primordial RNA cells. 44 However, as discussed above, RNA cells do not appear to be a viable proposition. Therefore, the alternative scenario that seems to reconcile the results of comparative genomics and the general logic of precellular evolution revives Haldane's idea at a new level and involves evolution of diverse virus‐like elements and even virus‐like particles prior to the advent of modern‐type cells. 59
The emergence of cells is the epitome of the problems encountered by all explanations of the evolution of complex biological structures, the crucial conundrum of biology that was first recognized and explored by Darwin in his famous discussion of the evolution of the animal eyes. 26 Darwin's solution, with some embellishments, has since become the standard scenario for the origin of complex systems: the intermediates might not be fit to perform the function of the final, complex structure but they are good enough for either a simplified version of that function or, perhaps, a distinct function that is not always easy to deduce from the present one. For the latter case, Gould coined the succinct term exaptation, that is, recruitment of a structure for a new function. 71 The virus‐like early stage in life's early evolution belongs to the same family of solutions and might be the most plausible if not the only way to avoid the ultimate “irreducible complexity” trap associated with the origin of cellular organization itself.
Like all biological evolution, precellular evolution was undoubtedly driven, in large part, by natural selection. Selection enters the scene with the appearance of replicating entities, initially, it is currently presumed, RNA molecules replicated by ribozymes, and subsequently, after the emergence of translation, RNA molecules replicating with the aid of proteins. 72 , 73 These earliest stages of evolution are beyond the scope of this discussion. It is important to note, however, that one of the central aspects of the model of a virus‐like, compartmentalized, precellular stage of evolution is a gradual transition from selection at the level of individual genetic elements to group selection for ensembles of such elements encoding both enzymes directly involved in replication and proteins responsible for accessory functions, such as translation and nucleic acid precursor synthesis. 45 , 74
Ensembles of “selfish cooperators” could potentially evolve by two routes: (i) physical joining of genetics elements and (ii) compartmentalization. 45 The former route is considered to be the onset of the evolution of operons including the ribosomal‐RNA polymerase superoperon, the only substantially conserved feature of the genome organization between archaea and bacteria. 75 , 76 The compartmentalization route would depend on the evolution of virus‐like particles that could harbor (relatively) stable sets of genomic segments resembling the extant RNA viruses with multipartite genomes. Unlike cells, the virions of viruses with small genomes, particularly, the nearly ubiquitous icosahedral (spherical) capsids, are simple, symmetrical structures that, in many cases, are formed by self‐assembly of a single capsid protein. 77 , 78 , 79 , 80 Thus, it is attractive to speculate that simple virus‐like particles were the first form of genuine, biological compartmentalization that were important at the precellular stage of evolution. In addition to the benefit of compartmentalization, virus‐like particles would protect genetic elements (especially, RNA) from degradation and could be vehicles for gene transfer within and between networks of inorganic compartments.
Most of the spherical viruses with relatively complex genomes possess molecular motors for DNA or RNA packaging within the capsid; 79 , 81 , 82 , 83 , 84 at least in some cases, these machines also mediate extrusion of mRNA transcripts from the capsid. 85 , 86 The viral packaging and extrusion machines contain motor ATPases of at least three distinct families that seem to share a common architecture, forming hexameric channels through which DNA or RNA is actively translocated. 86 , 87 Notably, one of the groups of viral packaging ATPases is a branch of the FtsK‐HerA superfamily that also includes prokaryotic ATPases responsible for DNA pumping into daughter cells during cell division 50 whereas another family is homologous to bacterial twitching mobility ATPases (Ref. 86 and EVK, unpublished observations). In membrane‐containing virions of many viruses, the packaging motors translocate the DNA or RNA both across the capsid and the lipid membrane of the virion. It is tempting to hypothesize that viral packaging machines were evolutionary precursors of the cellular pumping and motility ATPases. Moreover, the H+‐ATPase/ATP synthase, the key, universal membrane enzyme and the centerpiece of modern cellular energetics, also forms a similar hexameric channel 88 and might have started out as part of the packaging/extrusion machinery in a still uncharacterized (possibly, extinct) class of virus‐like agents. Indeed, a recent comparative‐genomic analysis has suggested that that the common ancestor of the two major branches of membrane ATPases, F‐ATPases typically found in bacteria and V‐ATPases characteristic of archaea and eukaryotes, evolved from a common ancestor that functioned as a protein or RNA translocase. 89 More generally, it seems an attractive possibility that primordial viral membranes were intermediate steps in the evolution of membranes that antedated the emergence evolution of the first cellular membranes, a major challenge in terms of evolution of complexity. Just as genome replication of virus‐like agents can be viewed as the original test ground for replication strategies, 42 two of which have been subsequently recruited for the two major lineages of cellular life forms, evolving virus particles might have been the “laboratory” for testing molecular devices that were later incorporated into the membranes of emerging cells (Fig. 3).
From the selection for gene ensembles, there is a direct path to selection for compartment contents such that compartments sustaining rapid replication of genetic elements would “infect” adjacent compartment and, effectively, propagate their “genomes” 45 : primordial virus‐like particles would have been important for this process. The precellular equivalent of HGT, that is, transfer of the genetic content between compartments, is part and parcel of this model, in agreement with the general concept that rampant HGT was an essential feature of the early stages of life's evolution. 51 , 53 , 54 After a substantial degree of complexity has been reached through the evolution of selfish cooperators within the networks of inorganic compartments, repeated escapes of cell‐like entities that combined (relatively) large DNA genomes and membranes containing transport and translocation devices (originally evolved in virus‐like agents, under this model) became possible. There is no telling how many such attempts have failed quickly and how many might have been initially successful but the fact is that only two, archaea and bacteria (assuming a symbiotic scenario for the origin of eukaryotes 90 ), or three, archaea, bacteria, and eukarya (assuming the so‐called archezoan scenario of eukaryotic origin 91 ) survived for extended time intervals (the scenario for the origin of eukaryotes is peripheral in this context and is outside the scope of this article). The first successful escapes of cellular life forms from the hypothetical precellular pool would correspond to the “Darwinian threshold” for cellular life postulated by Woese, 51 that is, the threshold beyond which HGT would be substantially curtailed, and evolution of distinct lineages (species) of cellular organisms could take off.
Like other models of the early stages of evolution of biological complexity, and perhaps, even more explicitly, the “primordial virus world” scenario outlined here faces the problem of takeover by selfish elements. 74 , 92 , 93 If the primordial parasites became too aggressive, they would kill off their hosts within a compartment and could survive only by infecting a new compartment (where they could be dangerous again). Devastating “pandemics” sweeping through entire networks and eventually wiping out their entire content are imaginable, and indeed, this would be the likely fate of many, if not most, primordial “organisms.” The conditions for the survival of precellular life forms were, first, emergence of temperate virus‐like agents that do not kill the host, and second, early invention of defense mechanisms, likely, based on RNA interference (RNAi). The ubiquity of both temperate selfish elements and RNAi‐based defense systems in all major branches of cellular life 94 , 95 suggests that these phenomena evolved at a very early, quite possibly, precellular stage of evolution.
The primordial virus world model of precellular evolution sketched here seems to offer plausible, even if, to a large extent, speculative solutions to many puzzles associated with the origin of cells. Comparative genomics of viruses and other selfish elements seems to provide substantial empirical support for this model. Considering that, under the primordial virus world scenario, the first cells emerged from a noncellular ancestral state in multiple, independent escapes, it seems sensible to replace the acronym LUCA with LUCAS, for Last Common Ancestral State.
Viral Hallmark Genes: The Heritage of the Precellular Virus World
Viruses and other selfish replicons show remarkable diversity in terms of both replication‐expression strategy and genomic complexity. 62 , 69 , 70 , 96 , 97 , 98 The selfish replicons constituting the virus world span, roughly, the same range of genome sizes, about four orders of magnitude (from ∼102 nucleotides in the smallest viroid genome to >106 nucleotides in the giant mimivirus) as genomes of cellular life forms (from ∼2 × 105 nucleotides in the smallest bacterial genome to ∼3 × 109 nucleotides in mammals, some extremely large plant and animal genomes excluded). Predictably, within such a huge span of genome size, viruses show a tremendous variety of gene repertoires. In viruses with large genomes, such as poxviruses, the mimivirus or T‐even bacteriophages, there are many genes with readily recognizable homologs in cellular life forms that, clearly, have been transferred from the host at a relatively late stage of viral evolution. 99 , 100 , 101 The origins of many other viral genes remain obscure as they are present in one or more lineages of viruses but not in any sequenced genomes of cellular life forms. Conceivably, such genes are products of rapid evolution at the base of the respective viral lineages so that the traces of their origin have been obliterated.
In addition, however, a distinct class of viral genes shows a truly remarkable distribution. These “viral hallmark genes” are shared by many groups of viruses with extremely diverse replication‐expression strategies, genome sizes, and host ranges (Table 1). 59 No single hallmark gene is found in all groups of viruses' but, together, the partially overlapping distribution ranges of the hallmark genes cover almost the entirety of the virus world. There are only very distant homologs of the viral hallmark genes in cellular organisms, and all viral members of the respective gene families appear to have a common origin. All hallmark genes encode proteins with central, essential roles in the replication, expression, and virion morphogenesis of the respective viruses (Table 1). The relative contribution of the hallmark genes to the gene complement of a virus strongly depends on the genome size. Viruses with small genomes, such as most of the RNA viruses, often have only a few genes, so that the hallmark genes make up the majority. 102 By contrast, in viruses with large genomes, the hallmark genes account only for a small fraction of the gene complement. Considering the broad range of genome sizes and gene contents, and the even more dramatic, qualitative difference between the replication‐expression strategies (e.g., positive‐strand RNA viruses contrasted to dsDNA viruses) of viruses sharing some of the hallmark genes, it is striking and certainly calls for an explanation that the life cycles of these diverse viruses center around homologous genes (such as those for the jelly‐roll capsid protein or the superfamily 3 helicase involved in genome replication).
Table 1.
The Viral Hallmark Genes and Proteins they Encodea
| Protein | Function | Virus groups | Homologs in cellular life forms | Comments |
|---|---|---|---|---|
| Jelly‐roll capsid protein (JRC) | Main capsid subunit of icosahedral virions | Picornaviruses, comoviruses, carmoviruses, dsRNA phage, NCLDV, herpesviruses, adenoviruses, papovaviruses, parvoviruses, icosahedral DNA phages, and archaeal viruses | Distinct jelly‐roll domains are present in eukaryotic nucleoplasmins and in protein‐protein interaction domains of certain enzymes | Certain icosahedral viruses, such as ssRNA phages and alphaviruses, have unrelated capsid proteins. In poxviruses, the JRC is not a virion protein but forms intermediate structures during virion morphogenesis |
| Superfamily 3 helicase (S3H) | Initiation and elongation of genome replication | Picornaviruses, comoviruses, eukaryotic RCR viruses, NCLDV, baculoviruses, some phages (e.g., P4), plasmids, particularly archaeal ones | S3H is a distinct, deep‐branching family of the AAA+ ATPase class | Fusion with primase in DNA viruses and plasmids |
| Archaeo‐eukaryotic DNA primase | Initiation of genome replication | NCLDV, herpesviruses, baculoviruses, some phages | All viral primases appear to form a clade within the archaeo‐eukaryotic primase family | Fusion with S3H in most NCLDV, some phages, and archaeal plasmids |
| UL9‐like superfamily 2 helicase | Initiation and elongation of genome replication | Herpesviruses, some NCLDV, some phages | Viral UL9‐like helicases form a distinct branch in the vast superfamily of DNA and RNA helicases | Fusion with primase in asfarviruses, mimiviruses |
| Rolling‐circle replication initiation endonuclease (RCRE)/origin‐binding protein | Initiation of genome replication | Small eukaryotic DNA viruses (parvo, gemini, circo, papova), phages, plasmids, and eukaryotic helitron transposons | No cellular RCRE or papovavirus‐type origin‐binding protein; however, these proteins have a derived form of the palm domain that is found in the majority of cellular DNA polymerases | Papovaviruses have an inactivated form of RCRE that functions as origin‐binding protein |
| Packaging ATPase of the FtsK family | DNA packaging into the virion | NCLDV, adenoviruses, polydnaviruses, some phages (e.g., P9, M13), nematode transposons | A distinct clade in the FtsK/HerA superfamily of P‐loop NTPases that includes DNA‐pumping ATPases of bacteria and archaea | |
| ATPase subunit of terminase | DNA packaging into the virion | Herpesviruses, tailed phages | The terminases comprise a derived family of P‐loop NTPases that is distantly related to Superfamily I/II helicases and AAA+ ATPases | |
| RNA‐dependent RNA polymerase (RdRp)/reverse transcriptase (RT) | Replication of RNA genomes | Positive‐strand RNA viruses and virus‐like elements, dsRNA viruses and virus‐like elements, retroid viruses/elements, possibly, negative‐strand RNA viruses | Another major group of palm‐fingers domains that are distinct from those in DNA polymerases; eukaryotic telomerase appears to be a RT derivative | The RdRps of dsRNA viruses are homologs of positive‐strand RNA virus polymerases. The provenance of negative‐strand RNA virus RdRp remains uncertain although sequence motif and, especially, structural analysis suggests their derivation from RdRps of positive‐strand RNA viruses. |
| B‐family DNA polymerase | Replication of large dsDNA genomes | Diverse bacteriophages with dsDNA genomes, NCLDV, adenoviruses, herpesviruses, baculoviruses, fungal dsDNA plasmids | A distinct family of palm‐finger domain polymerases | The hallmark status of this gene is somewhat uncertain as it is hard to demonstrate the monophyly of the viral polymerases; however, the polymerases of the viruses with genome‐linked terminal proteins do appear to be monophyletic. |
| Genome‐linked terminal protein | Adenoviruses, fungal dsDNA plasmids, several groups of bacteriophages | Protein involved in a distinct mechanism of DNA replication initiation |
aThe table is from Ref. 59, with modifications; see Ref. 59 for further details and references. The list of hallmark genes is not necessarily complete and is likely to grow with further sequencing of genomes from new groups of viruses, determination of structures of viral proteins, and comparative analysis.
Abbreviations: NCLDV, nucleo‐cytoplasmic large DNA viruses (of eukaryotes); RCR, rolling circle replication.
Various evolutionary scenarios accounting for the highly unusual phyletic spread of the viral hallmark genes have been examined in detail elsewhere. 59 In brief, the simplest explanation for the fact that the hallmark proteins involved in viral replication and virion formation are present in a broad variety of viruses but not in any cellular life forms seems to be that the latter actually never possessed these genes. Rather, the hallmark genes, probably, antedate cells and descend directly from the primordial pool of virus‐like genetic elements. Given the spread of the hallmark genes among numerous groups of extremely diverse viruses, a major corollary is that, at least, several lineages of viruses and other selfish elements with distinct genome structures and replication‐expression strategies derive from the precellular stage of evolution (although the current distribution of the hallmark genes, certainly, was affected by later HGT).
Conclusions
The concept of a precellular stage of biological evolution outlined here posits that the precellular stage of life's evolution took place within networks of inorganic compartments that hosted a diverse mix of virus‐like genetic elements. 45 , 59 It is further proposed that these ensembles of genetic elements were the ancestral state from which cells emerged, probably, in multiple, independent escapes only two or three of which (the ancestors of bacteria and archaea, and possibly, eukarya) yielded stable cellular lineages that enjoyed a long‐term evolutionary success. Considering this hypothetical consortial state of primordial life forms that eventually gave rise to cells, it seems reasonable to replace the acronym LUCA with LUCAS, for the Last Universal Common Ancestral State.
The viral model of cellular origin recapitulates, at a quite different stage in the development of biology, the early ideas of Haldane. 61 Since 1928, when Haldane's essay was published, the status of the model has radically changed. At this time, the support and, indeed, the incentives for this model derive from four lines of substantive comparative‐genomic evidence:
-
(i)
the lack of homology between the core components of the DNA replication systems in the two primary lines of descent of cellular life forms, archaea and bacteria;
-
(ii)
the similar lack of homology between the enzymes of membrane lipid biosynthesis in conjunction with distinct membrane chemistries in archaea and bacteria;
-
(iii)
the spread of viral hallmark genes among numerous and extremely diverse groups of viruses, in contrast to their absence in cellular life forms;
-
(iv)
the highly dynamic character of the extant prokaryotic world which is shaped by the interaction of the bacterial chromosomes and the mobilome, that is, the sum total of viruses, plasmids, and other selfish elements. 103 , 104
Although bacterial and archaeal chromosomes are large dsDNA molecules and are relatively stable over the short scale of evolution, these genomes of cellular life forms are in an equilibrium with the mobilome, and over the longer time scale, were shaped by accretion of diverse, smaller replicons. 104 , 105 Thus, there seems to be a continuity between the hypothetical, primordial virus stage of life's evolution and the dynamic prokaryotic world, the principal distinction being the additional compartmentalization that is brought about by the cellular organization and provides for the persistence of large genomes.
In addition to being compatible with multiple lines of empirical evidence, the viral model of early evolution seems to offer at least a tentative solution to the classic Darwinian challenge of the evolution of complex structures that can function only as a whole, in this case, the cell itself. This solution comes along the lines first outlined by Darwin himself, 26 that is, gradual evolution of the complex organization via intermediates whose functions are different from, even if mechanistically similar to, those of the fully developed structure. Under this model, primordial functions are envisaged to evolve as parts of the life cycles of virus‐like genetic elements. Within this context, the model addresses the most daunting challenges to the hypothesis of a precellular LUCA(S), namely, the universal conservation of some essential membrane proteins and complexes: the ancestors of these membrane devices might function within emerging membranes of virus‐like particles.
The primordial virus world model is, at least in parts, refutable and, potentially, testable. A discovery of an organism with an archaeal replication system but a bacterial membrane (or vice versa) would come close to a refutation. Further study of the diversity of viruses might reveal new membrane translocation devices, for instance, packaging machines homologous to the H+‐ATPases of cellular organisms. Such evidence would provide support for a role of viruses in the evolution of cellular membranes. Direct biochemical experiments on early evolution are inherently hard. However, this model might make them easier by splitting the gargantuan feat of evolving a cell into more manageable steps of evolution of virus‐like agents.
Conflicts of Interest
The author declares no conflicts of interest.
Acknowledgments
Valerian Dolja, Bill Martin, Tania Senkevich, and Yuri Wolf contributed to the development of various aspects of this model. I also thank the participants of the meeting on the LUCA at Fondacion Les Treilles (France), in September, 2007, and specifically, the organizers of the meeting, Patrick Forterre, Celine Brochier‐Armanet, and Simonetta Gribaldo, for most helpful discussions during which the acronym LUCAS was coined collectively. This work was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
References
- 1. Koonin, E.V. , Aravind L. & Kondrashov A.S.. 2000. The impact of comparative genomics on our understanding of evolution. Cell 101: 573–576. [DOI] [PubMed] [Google Scholar]
- 2. Wolfe, K.H. & Li W.H.. 2003. Molecular evolution meets the genomics revolution. Nat. Genet. 33(Suppl): 255–265. [DOI] [PubMed] [Google Scholar]
- 3. Doolittle, R.F. 2005. Evolutionary aspects of whole‐genome biology. Curr. Opin. Struct. Biol. 15: 248–253. [DOI] [PubMed] [Google Scholar]
- 4. Delsuc, F. , Brinkmann H. & Philippe H.. 2005. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6: 361–375. [DOI] [PubMed] [Google Scholar]
- 5. Koonin, E.V. 2003. Comparative genomics, minimal gene‐sets and the last universal common ancestor. Nat. Rev. Microbiol. 1: 127–136. [DOI] [PubMed] [Google Scholar]
- 6. Harris, J.K. et al 2003. The genetic core of the universal ancestor. Genome Res. 13: 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charlebois, R.L. & Doolittle W.F.. 2004. Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res. 14: 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mushegian, A. 2008. Gene content of LUCA, the last universal common ancestor. Front. Biosci. 13: 4657–4666. [DOI] [PubMed] [Google Scholar]
- 9. Glansdorff, N. , Xu Y. & Labedan B.. 2008. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct. 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snel, B. , Bork P. & Huynen M.A.. 2002. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 12: 17–25. [DOI] [PubMed] [Google Scholar]
- 11. Mirkin, B.G. et al 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunin, V. & Ouzounis C.A.. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13: 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fitch, W.M. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–106. [PubMed] [Google Scholar]
- 14. Koonin, E.V. 2005. Orthologs, paralogs and evolutionary genomics. Annu. Rev. Genet. 39: 309–338. [DOI] [PubMed] [Google Scholar]
- 15. Dagan, T. & Martin W.. 2006. The tree of one percent. Genome Biol. 7: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doolittle, W.F. & Bapteste E.. 2007. Pattern pluralism and the tree of life hypothesis. Proc. Natl. Acad. Sci. USA 104: 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koonin, E.V. 2007. The biological big bang model for the major transitions in evolution. Biol. Direct. 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anantharaman, V. , Koonin E.V. & Aravind L.. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30: 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ouzounis, C.A. et al 2006. A minimal estimate for the gene content of the last universal common ancestor—exobiology from a terrestrial perspective. Res. Microbiol. 157: 57–68. [DOI] [PubMed] [Google Scholar]
- 20. Mushegian, A.R. & Koonin E.V.. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93: 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgell, D.R. & Doolittle W.F.. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89: 995–998. [DOI] [PubMed] [Google Scholar]
- 22. Leipe, D.D. , Aravind L. & Koonin E.V.. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27: 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin, W. & Russell M.J.. 2003. On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 59–83; discussion 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pereto, J. , Lopez‐Garcia P. & Moreira D.. 2004. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 29: 469–477. [DOI] [PubMed] [Google Scholar]
- 25. Woese, C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. USA 95: 6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darwin, C. 1859. On the Origin of Species. Murray. London . [Google Scholar]
- 27. Lazcano, A. & Forterre P.. 1999. The molecular search for the last common ancestor. J. Mol. Evol. 49: 411–412. [PubMed] [Google Scholar]
- 28. Brown, J.R. & Doolittle W.F.. 1997. Archaea and the prokaryote‐to‐eukaryote transition. Microbiol. Mol. Biol. Rev. 61: 456–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iwabe, N. et al 1989. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA 86: 9355–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gogarten, J.P. et al 1989. Evolution of the vacuolar H+‐ATPase: Implications for the origin of eukaryotes. Proc. Natl. Acad. Sci. USA 86: 6661–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Forterre, P. & Philippe H.. 1999. Where is the root of the universal tree of life? Bioessays 21: 871–879. [DOI] [PubMed] [Google Scholar]
- 32. Lopez, P. , Forterre P. & Philippe H.. 1999. The root of the tree of life in the light of the covarion model. J. Mol. Evol. 49: 496–508. [DOI] [PubMed] [Google Scholar]
- 33. Philippe, H. & Forterre P.. 1999. The rooting of the universal tree of life is not reliable. J. Mol. Evol. 49: 509–523. [DOI] [PubMed] [Google Scholar]
- 34. Kozbial, P.Z. & Mushegian A.R.. 2005. Natural history of S‐adenosylmethionine‐binding proteins. BMC Struct. Biol. 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolf, Y.I. et al 1999. Evolution of aminoacyl‐tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9: 689–710. [PubMed] [Google Scholar]
- 36. Woese, C.R. et al 2000. Aminoacyl‐tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64: 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aravind, L. , Anantharaman V. & Koonin E.V.. 2002. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP‐ATPase nucleotide‐binding domains: Implications for protein evolution in the RNA. Proteins 48: 1–14. [DOI] [PubMed] [Google Scholar]
- 38. Aravind, L. et al 2002. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 12: 392–399. [DOI] [PubMed] [Google Scholar]
- 39. Forterre, P. et al 1992. The nature of the last universal ancestor and the root of the tree of life, still open questions. Biosystems 28: 15–32. [DOI] [PubMed] [Google Scholar]
- 40. Forterre, P. & Philippe H.. 1999. The last universal common ancestor (LUCA), simple or complex? Biol. Bull. 196: 373–375; discussion 375–377. [DOI] [PubMed] [Google Scholar]
- 41. Forterre, P. , Gribaldo S. & Brochier C.. 2005. [Luca: the last universal common ancestor]. Med. Sci. (Paris) 21: 860–865. [DOI] [PubMed] [Google Scholar]
- 42. Forterre, P. 1999. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 33: 457–465. [DOI] [PubMed] [Google Scholar]
- 43. Woese, C.R. & Fox G.E.. 1977. The concept of cellular evolution. J. Mol. Evol. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- 44. Forterre, P. 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. USA 103: 3669–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koonin, E.V. & Martin W.. 2005. On the origin of genomes and cells within inorganic compartments. Trends Genet. 21: 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gorbalenya, A.E. , et al. 2006. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 117: 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donachie, W.D. 2002. FtsK: Maxwell's demon? Mol. Cell. 9: 206–207. [DOI] [PubMed] [Google Scholar]
- 48. Errington, J. , Daniel R.A. & Scheffers D.J.. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67: 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiss, D.S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54: 588–597. [DOI] [PubMed] [Google Scholar]
- 50. Iyer, L.M. et al 2004. Comparative genomics of the FtsK‐HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32: 5260–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Woese, C.R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. USA 99: 8742–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koonin, E.V. & Galperin M.Y.. 2002. Sequence ‐ Evolution ‐ Function. Computational Approaches in Comparative Genomics. Kluwer Acad. Publ. New York . [PubMed] [Google Scholar]
- 53. Vetsigian, K. , Woese C. & Goldenfeld N.. 2006. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. USA 103: 10696–10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goldenfeld, N. & Woese C.. 2007. Biology's next revolution. Nature 445: 369. [DOI] [PubMed] [Google Scholar]
- 55. Russell, M.J. et al 1994. A hydrothermally precipitated catalytic iron sulphide membrane as a first step toward life. J. Mol. Evol. 39: 231–243. [Google Scholar]
- 56. Russell, M.J. & Hall A.J.. 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 154: 377–402. [DOI] [PubMed] [Google Scholar]
- 57. Baaske, P. et al 2007. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc. Natl. Acad. Sci. USA 104: 9346–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koonin, E.V. 2007. An RNA‐making reactor for the origin of life. Proc. Natl. Acad. Sci. USA 104: 9105–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koonin, E.V. , Senkevich T.G. & Dolja V.V.. 2006. The ancient virus world and evolution of cells. Biol. Direct. 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. D’Herelle, F. 1922. The Bacteriophage Its Role in Immunity. Williams and Wilkins. Baltimore . [Google Scholar]
- 61. Haldane, J.B.S. 1928. The origin of life. Rationalist Annual. 148: 3–10. [Google Scholar]
- 62. Agol, V.I. 1976. An aspect of the origin and evolution of viruses. Orig. Life. 7: 119–132. [DOI] [PubMed] [Google Scholar]
- 63. Luria, S.E. & Darnell J.. 1967. General Virology. John Wiley. New York . [Google Scholar]
- 64. Matthews, R.E. 1983. The origin of viruses from cells. Int. Rev. Cytol. Suppl. 15: 245–280. [DOI] [PubMed] [Google Scholar]
- 65. Forterre, P. 2002. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 5: 525–532. [DOI] [PubMed] [Google Scholar]
- 66. Forterre, P. 2003. The great virus comeback—from an evolutionary perspective. Res. Microbiol. 154: 223–225. [DOI] [PubMed] [Google Scholar]
- 67. Forterre, P. 2005. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 87: 793–803. [DOI] [PubMed] [Google Scholar]
- 68. Forterre, P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117: 5–16. [DOI] [PubMed] [Google Scholar]
- 69. Claverie, J.M. 2006. Viruses take center stage in cellular evolution. Genome Biol. 7: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Koonin, E.V. & Dolja V.V.. 2006. Evolution of complexity in the viral world: The dawn of a new vision. Virus Res. 117: 1–4. [DOI] [PubMed] [Google Scholar]
- 71. Gould, S.J. 1997. The exaptive excellence of spandrels as a term and prototype. Proc. Natl. Acad. Sci. USA 94: 10750–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Eigen, M. 1971. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58: 465–523. [DOI] [PubMed] [Google Scholar]
- 73. Wolf, Y.I. & Koonin E.V.. 2007. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol. Direct. 2: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Szathmary, E. & Demeter L.. 1987. Group selection of early replicators and the origin of life. J. Theor. Biol. 128: 463–486. [DOI] [PubMed] [Google Scholar]
- 75. Lathe, W.C. 3rd , Snel B. & Bork P.. 2000. Gene context conservation of a higher order than operons. Trends Biochem. Sci. 25: 474–479. [DOI] [PubMed] [Google Scholar]
- 76. Wolf, Y.I. et al 2001. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 11: 356–372. [DOI] [PubMed] [Google Scholar]
- 77. Klug, A. & Caspar D.L.. 1960. The structure of small viruses. Adv. Virus Res. 7: 225–325. [DOI] [PubMed] [Google Scholar]
- 78. Morgan, G.J. 2003. Historical review: Viruses, crystals and geodesic domes. Trends Biochem. Sci. 28: 86–90. [DOI] [PubMed] [Google Scholar]
- 79. Poranen, M.M. , Tuma R. & Bamford D.H.. 2005. Assembly of double‐stranded RNA bacteriophages. Adv. Virus Res. 64: 15–43. [DOI] [PubMed] [Google Scholar]
- 80. Hagan, M.F. & Chandler D.. 2006. Dynamic pathways for viral capsid assembly. Biophys. J. 91: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Catalano, C.E. 2000. The terminase enzyme from bacteriophage lambda: A DNA‐packaging machine. Cell Mol. Life Sci. 57: 128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grimes, S. , Jardine P.J. & Anderson D.. 2002. Bacteriophage phi 29 DNA packaging. Adv. Virus Res. 58: 255–294. [DOI] [PubMed] [Google Scholar]
- 83. Mindich, L. 2004. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 101: 83–92. [DOI] [PubMed] [Google Scholar]
- 84. Condit, R.C. , Moussatche N. & Traktman P.. 2006. In a nutshell: Structure and assembly of the vaccinia virion. Adv. Virus Res. 66: 31–124. [DOI] [PubMed] [Google Scholar]
- 85. Pirttimaa, M.J. et al 2002. Nonspecific nucleoside triphosphatase P4 of double‐stranded RNA bacteriophage phi6 is required for single‐stranded RNA packaging and transcription. J. Virol. 76: 10122–10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kainov, D.E. , Tuma R. & Mancini E.J.. 2006. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell Mol. Life Sci. 63: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Simpson, A.A. et al 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nakamoto, R.K. et al 2000. Molecular mechanisms of rotational catalysis in the F(0)F(1) ATP synthase. Biochim. Biophys. Acta 1458: 289–299. [DOI] [PubMed] [Google Scholar]
- 89. Mulkidjanian, A.Y. et al 2007. Inventing the dynamo machine: The evolution of the F‐type and V‐type ATPases. Nat. Rev. Microbiol. 5: 892–899. [DOI] [PubMed] [Google Scholar]
- 90. Embley, T.M. & Martin W.. 2006. Eukaryotic evolution, changes and challenges. Nature 440: 623–630. [DOI] [PubMed] [Google Scholar]
- 91. Poole, A. & Penny D.. 2007. Eukaryote evolution: engulfed by speculation. Nature 447: 913. [DOI] [PubMed] [Google Scholar]
- 92. Eigen, M. & Schuster P.. 1977. The hypercycle. A principle of natural self‐organization. Part A: Emergence of the hypercycle. Naturwissenschaften 64: 541–565. [DOI] [PubMed] [Google Scholar]
- 93. Zintzaras, E. , Santos M. & Szathmary E.. 2002. “Living” under the challenge of information decay: The stochastic corrector model vs. hypercycles. J. Theor. Biol. 217: 167–181. [DOI] [PubMed] [Google Scholar]
- 94. Zamore, P.D. & Haley B.. 2005. Ribo‐gnome: The big world of small RNAs. Science 309: 1519–1524. [DOI] [PubMed] [Google Scholar]
- 95. Sorek, R. , Kunin V. & Hugenholtz P.. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6: 181–186. [DOI] [PubMed] [Google Scholar]
- 96. Baltimore, D. 1971. Viral genetic systems. Trans. N. Y. Acad. Sci. 33: 327–332. [DOI] [PubMed] [Google Scholar]
- 97. Koonin, E.V. 2005. Virology: Gulliver among the Lilliputians. Curr. Biol. 15: R167–169. [DOI] [PubMed] [Google Scholar]
- 98. Claverie, J.M. et al 2006. Mimivirus and the emerging concept of “giant” virus. Virus Res. 117: 133–144. [DOI] [PubMed] [Google Scholar]
- 99. Senkevich, T.G. et al 1997. The genome of molluscum contagiosum virus: Analysis and comparison with other poxviruses. Virology 233: 19–42. [DOI] [PubMed] [Google Scholar]
- 100. Bugert, J.J. & Darai G.. 2000. Poxvirus homologues of cellular genes. Virus Genes 21: 111–133. [PubMed] [Google Scholar]
- 101. Iyer, L.M. et al 2006. Evolutionary genomics of nucleo‐cytoplasmic large DNA viruses. Virus Res. 117: 156–184. [DOI] [PubMed] [Google Scholar]
- 102. Koonin, E.V. et al 2008. The big bang of picorna‐like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6: 925–939. [DOI] [PubMed] [Google Scholar]
- 103. Frost, L.S. et al 2005. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 3: 722–732. [DOI] [PubMed] [Google Scholar]
- 104. Koonin, E.V. & Wolf Y.I.. 2008. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res., 36: 6688–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. McGeoch, A.T. & Bell S.D.. 2008. Extra‐chromosomal elements and the evolution of cellular DNA replication machineries. Nat. Rev. Mol. Cell Biol. 9: 569–574. [DOI] [PubMed] [Google Scholar]
- 106. Koonin, E.V. 2006. Temporal order of evolution of distinct DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct. 1: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]


