Abstract
Extensive horizontal gene transfer (HGT) among prokaryotes seems to undermine the tree of life (TOL) concept. However, the possibility remains that the TOL can be salvaged as a statistical central trend in the phylogenetic “forest of life” (FOL). A comprehensive comparative analysis of 6901 phylogenetic trees for prokaryotic genes revealed a signal of vertical inheritance that was particularly strong among the 102 nearly universal trees (NUTs), despite the high topological inconsistency among the trees in the FOL, most likely, caused by HGT. The topologies of the NUTs are similar to the topologies of numerous other trees in the FOL; although the NUTs cannot represent the FOL completely, they reflect a significant central trend. Thus, the original TOL concept becomes obsolete but the idea of a “weak” TOL as the dominant trend in the FOL merits further investigation. The totality of gene trees comprising the FOL appears to be a natural representation of the history of life given the inherent tree-like character of the replication process.
THE TREE OF LIFE CONCEPT IN THE AGE OF GENOMICS
The concept of the TOL introduced by Darwin, captured in the famous single illustration of the “Origin of species” (Darwin 1859), and used by Haeckel as the grand scheme of the history of the actual life-forms is the cornerstone of evolutionary biology and, arguably, of biology in general. For nearly 140 years after the publication of the Origin, phylogenetic trees, which were initially constructed using phenotypic characters but, following the seminal work of Zuckerkandl and Pauling (1962, 1965), increasingly relied on molecular sequence comparison, were viewed as a (more or less accurate) depiction of the evolution of the respective organisms. In other words, a tree built for a specific character or a gene was routinely equated with a “species tree.” The use of rRNA as the molecule of choice for phylogenetic reconstruction culminated in the now textbook three-domain TOL of Woese and coworkers (Pace et al. 1986; Woese 1987) and was the brilliant culmination of the heroic period of phylogenetics that brought hopes that the detailed, definitive topology of the TOL could be within reach.
Trouble for the TOL concept, however, started even before the advent of genomics as it became clear that common and essential genes of prokaryotes experienced multiple HGTs. So the idea of a “net of life” as a potential replacement for the TOL was proposed (Hilario and Gogarten 1993; Gogarten 1995). Generally, however, in the pregenomic era, HGT was viewed as a minor process of evolution, crucial in some areas such as the spread of antibiotic resistance but secondary for the general scheme of evolution. In the late 1990s, comparative genomics of prokaryotes dramatically changed this picture by showing that the patterns of gene distribution across genomes were typically patchy, whereas the topologies of gene-specific phylogenetic trees were often incongruent. These findings indicated that HGT was extremely common among prokaryotes (bacteria and archaea) (Doolittle 1999a,b, 2000; Martin 1999; Koonin et al. 2001; Gogarten et al. 2002; Koonin and Aravind 2002; Lawrence and Hendrickson 2003; Gogarten and Townsend 2005; Dagan et al. 2008) and could have been important also in the evolution of eukaryotes, especially, as a consequence of endosymbiotic events (Doolittle 1998; Martin and Herrmann 1998; Doolittle et al. 2003; Embley and Martin 2006). Thus, a perfect TOL turned out to be a chimera because extensive HGT prevents any single gene tree from being an accurate representation of the evolution of entire genomes. The realization that HGT among prokaryotes is the dominant rather than an exceptional mode of evolution led to the idea of “uprooting” the TOL, a development that is often interpreted as a paradigm shift in evolutionary biology (Pennisi 1999; Doolittle 2000; O'Malley and Boucher 2005).
Of course, the incongruence of gene phylogenies caused by HGT or other processes cannot alter the fact that all cellular life-forms are linked by a tree of cell divisions (Omnis cellula e cellula, according to the famous motto of Rudolf Virchow [1858]) that goes back to the earliest stages of evolution, with the exception of endosymbiotic events that were key to the evolution of eukaryotes but not prokaryotes (Lane and Archibald 2008). The problems with the TOL concept in the era of comparative genomics concern the TOL as it can be derived by the phylogenetic (phylogenomic) analysis of genes and genomes. Thus, the claim that HGT uproots the TOL more accurately means that extensive HGT has the potential to result in complete decoupling of molecular phylogenies from the actual tree of cells. Phylogenetic trees of genes also reflect the evolution of the respective molecular functions, so the phylogenomic analysis has straightforward biological connotations. Thus, the phylogenomic approach and not the abstract tree of cells reveals the actual history of the genetic content of organisms. Accordingly, we examine here the current status of the “phylogenomic TOL.”
The views of evolutionary biologists on the status of the TOL in the face of the ubiquitous HGT (O'Malley and Boucher 2005) span the entire range from (1) continued denial of the major role of HGT in the evolution of life (Kurland 2000; Kurland et al. 2003) to (2) “moderate” revision of the TOL concept (Wolf et al. 2002; Zhaxybayeva et al. 2004; Beiko et al. 2005; Ge et al. 2005; Kunin et al. 2005; Galtier and Daubin 2008) to (3) radical uprooting whereby the representation of the evolution of organisms (or genomes) as a TOL is declared meaningless (Bapteste et al. 2005; Doolittle and Bapteste 2007; Koonin 2007). The moderate approach maintains that all of the differences among individual gene trees notwithstanding, the TOL concept remains valid as a central trend that, at least in principle, can be revealed through a comprehensive comparison of gene tree topologies. The radical view counters that the massive HGT obliterates the very distinction between the vertical and horizontal routes of genetic information transmission, so the TOL concept should be abandoned in favor of a (broadly defined) network representation of evolution (Dagan et al. 2008). The TOL conundrum is fittingly emphasized in the recent debate on the “highly resolved tree of life” that was generated from a concatenation of alignments of 31 highly conserved proteins (Ciccarelli et al. 2006), only to be dismissed as a “tree of one percent” (of the genes in any given genome) that does not actually reflect the history of genomes (Dagan and Martin 2006).
We discuss here our recent effort of a comprehensive comparison of the phylogenetic trees for individual genes of prokaryotes. We refer to this set of ~7000 trees as the FOL. We show that there is indeed a central trend in the FOL but the deep splits in this topology cannot be unambiguously resolved, probably, owing to both extensive HGT and methodological problems of tree reconstruction. Nevertheless, computer simulations show that the observed pattern of evolution of archaea and bacteria is better compatible with a compressed cladogenesis (CC) model (Rokas et al. 2005; Rokas and Carroll 2006) than with a “big bang” model that includes non-tree-like phases of evolution (Koonin 2007). These findings are, in principle, compatible with the “TOL as a central trend” concept. However, we argue on more general grounds that the TOL is not a fundamentally necessary concept, and the entire FOL with its different trends could be the most adequate representation of the history of life. We now have the adequate computational methods and tools to identify and analyze these trends.
THE FOREST OF LIFE AND THE NEARLY UNIVERSAL TREES
We analyzed 6901 maximum likelihood phylogenetic trees that were built using clusters of orthologous gene (COG) databases that included a selected representative set of 100 prokaryotes (41 archaea and 59 bacteria) (Tatusov et al. 1997, 2003; Jensen et al. 2008). The majority of these trees include only a small number of species (<20); only 2040 trees included more than 20 species, and only a small set of NUTs included >90% of the analyzed prokaryotes. We sought to identify patterns in the FOL and, in particular, to determine whether there exists a central trend among the trees and whether the topologies of the NUTs reflect such a trend should it exist. To this end, we analyzed the complete, all-against-all matrix of the topological distances between the trees (Puigbò et al. 2007, 2009). This matrix was represented as a network of trees and was subject to classical multidimensional scaling (CMDS) analysis to detect potentially existing distinct clusters of trees. In addition, we introduced a new measure, the inconsistency score (IS), that determines how representative the topology of the given tree is of the entire FOL (IS is the fraction of the times the splits from a given tree are found in all trees of the FOL). Using the IS, we objectively examine trends in the FOL, without relying on the topology of a preselected “species tree” such as a supertree used in the most comprehensive previous study of HGT (Beiko et al. 2005) or a tree of concatenated highly conserved proteins or rRNAs (Mirkin et al. 2003; Ciccarelli et al. 2006; Dagan et al. 2008).
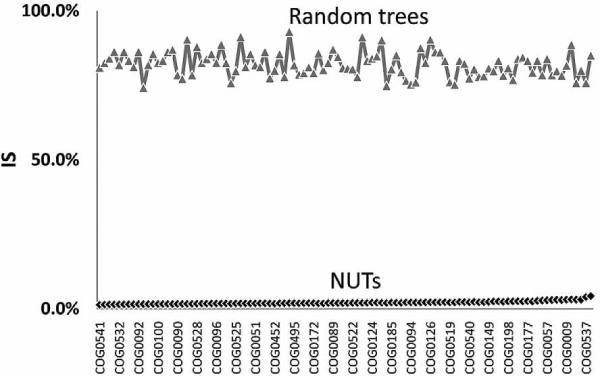
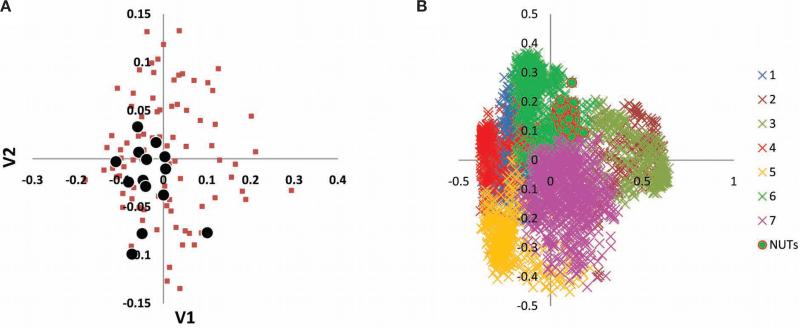
We began the systematic exploration of the FOL from the grove of 102 NUTs, most, although not all, of which, as expected, correspond to genes encoding components of information transmission systems, particularly, translation. The topologies of the NUTs were, in general, highly coherent. Indeed, the inconsistency among the NUTs ranged from 1.4% to 4.3%, whereas the mean value of inconsistency for an equal-sized set (102) of randomly generated trees with the same number of species was ~80% (Fig. 1). In 56% of the NUTs, archaeal and bacterial branches were perfectly separated, whereas the remaining 44% showed indications of HGT between archaea and bacteria (13% from archaea to bacteria, 23% from bacteria to archaea, and 8% in both directions). In the rest of the NUTs, interdomain gene transfer was not detected, but there were many probable HGT events within one or both domains (data not shown). We further analyzed the relationships among the 102 NUTs by embedding them into a 30-dimensional tree space using the CMDS procedure and the gap statistics analysis, which revealed a lack of significant clustering among the NUTs in the tree space: All of the NUTs seem to belong to a single unstructured cloud of points scattered around a single centroid (Fig. 2a). This organization of the tree space is best compatible with individual trees randomly deviating from a single dominant topology (“the TOL”), apparently as a result of HGT (but also, possibly, due to random errors of the tree construction procedure).
Figure 1.
The 102 NUTs have largely consistent topologies. (Black) Inconsistency scores of the 102 NUTs, (gray) IS values for the random trees produced by shuffling the branches in each of the NUTs are represented in black and ordered by increasing IS values. The IS of each NUT was calculated using as the reference set all 102 NUTs, and the IS of each random tree was similarly calculated using as the reference set of all 102 random trees. (Modified from Puigbò et al. 2009.)
Figure 2.
Clustering of the NUTs and the FOL using the classical multidimensional scaling method. (A) Best two-dimensional projection of the clustering of the 102 NUTs in a 30-dimensional space. (B) Best two-dimensional projection of the clustering of the 3789 COG trees in a 669-dimensional space. The seven clusters are color coded and the NUTs are shown by circles. (Modified from Puigbò et al. 2009.)
The overall conclusion on the evolutionary trends among the NUTs is that although the topologies of the NUTs were, for the most part, not identical, so that the NUTs could be separated by their degree of inconsistency (a proxy for the amount of HGT), the overall high consistency level indicated that the NUTs are scattered in the close vicinity of a consensus tree, with the HGT events distributed approximately randomly. These findings are compatible with previous reports on the apparently random distribution of HGT events in the history of highly conserved genes, in particular, those encoding proteins involved in translation (Brochier et al. 2002; Ge et al. 2005).
We further analyzed the structure of the FOL by embedding the 3789 COG trees (the subset of the FOL that included most trees with a large number of organisms) into a 669-dimensional space using the CMDS procedure and found that the optimal partitioning of this set yielded seven clusters of trees; notably, all of the NUTs formed a compact subset of cluster 6 (Fig. 2b). The trees that belonged to different clusters showed considerable differences in the distribution of the trees by the number of species, the partitioning of archaea-only and bacteria-only trees, and the functional classification of the respective COGs (Puigbò et al. 2009). The results of the CMDS clustering support the existence of several distinct “attractors” in the FOL, although trivial separation of the trees by size could substantially contribute to this finding. The key observation is that all of the NUTs occupy a compact and contiguous region of the tree space and, unlike the complete set of the trees, are not partitioned into distinct clusters (compare Fig. 2a,b).
As could be expected, the trees in the FOL show a strong signal of numerous HGT events including interdo-main gene transfers. Among the 1473 trees that include at least five archaeal species and at least five bacterial species, perfect separation of archaea and bacteria was seen only in 13%. This is the low bound for the fraction of trees that are free of interdomain HGT because, even for trees with a perfect separation of archaea and bacteria, HGT cannot be ruled out, for instance, in cases when a small compact archaeal branch is embedded within a bacterial lineage (or vice versa).
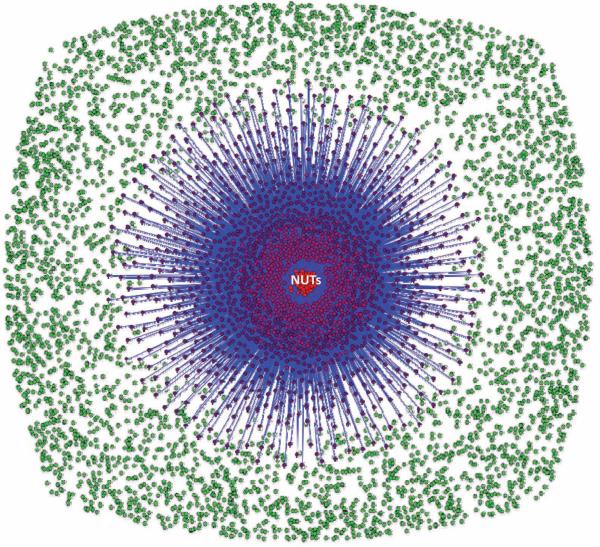
We constructed a network of all 6901 trees in the FOL and examined the position and the connectivity of the 102 NUTs in this network. At the 50% similarity cutoff and a p value <0.05, the 102 NUTs were connected to 2615 trees (38% of the FOL) (Fig. 3), and the mean similarity of the trees in the FOL to the NUTs was ~50%, with similar distributions of strongly, moderately, and weakly similar trees seen for most of the NUTs (Puigbò et al. 2009). In a sharp contrast, using the same similarity cutoff, 102 randomized NUTs were connected to only 33 trees (~0.5% of the trees in the FOL) and the mean similarity was ~28% to the trees in the FOL. These findings reveal the high and nonrandom topological similarity between the NUTs and a large part of the FOL and show that this similarity is not an artifact of the large number of species in the NUTs.
Figure 3.
Network of the 6901 trees comprising the FOL. (Red circles) 102 NUTs, (green circles) remainder of the trees. The NUTs are connected to trees with similar topologies (>80% blue, >90% violet, 100% red). (Modified from Puigbò et al. 2009.)
DEPENDENCE OF TREE INCONSISTENCY ON PHYLOGENETIC DEPTH: BIG BANG OR COMPRESSED CLADOGENESIS?
It is well known from many phylogenetic studies and was supported by the examination of a supernetwork of the NUTs (Puigbò et al. 2009) that deep internal nodes in phylogenetic trees tend to be poorly resolved compared to external nodes. Whether there actually is a discernible phylogenetic signal in the deepest nodes of the trees bears on the question of whether there is a central trend in the FOL that potentially could be approximated by the NUTs.
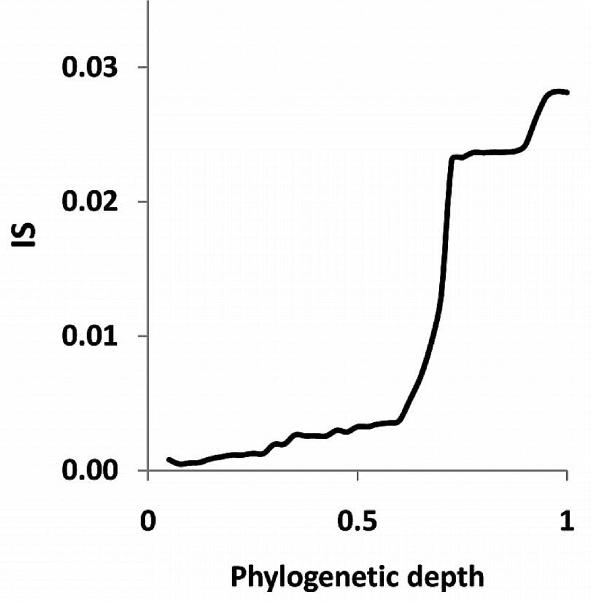
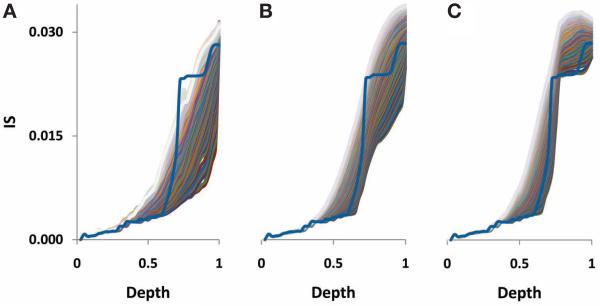
To explore the dependence of the inconsistency between trees on phylogenetic depth quantitatively, we used an ultra-metric tree that was produced from the supertree of the 102 NUTs and in which the phylogenetic distances were scaled from 0 to 1 (Puigbò et al. 2009). We found that the inconsistency of the FOL sharply increased, in a phase-transition-like fashion, between the depths of 0.7 and 0.8 (Fig. 4), suggesting that the evolutionary processes which were responsible for the formation of this part of the FOL could be qualitatively distinct from affected lesser phylogenetic depths. We considered two models of early evolution, at the level of archaeal and bacteria phyla: (1) Compressed clado-genesis (CC), under which there is a tree structure even at the deepest levels but the internal branches are extremely short (Rokas and Carroll 2006), and (2) biological big bang (BBB) model, where the early phase of evolution involved horizontal gene exchange so intensive that there is no signal of vertical inheritance in principle (Koonin 2007).
Figure 4.
Inconsistency versus phylogenetic depth plot for the 6901 trees in the FOL. The distances (on a 0–1 scale) are from the ultrametric tree that was produced from the supertree of the 102 NUTs. (Modified from Puigbò et al. 2009.)
The evolution of the FOL was simulated under each of the two models. We attempted to fit the observed IS-depth dependence (Fig. 4) with the respective curves obtained by simulating the BBB at different phylogenetic depths by randomly shuffling the tree branches at the given depth and modeling the subsequent evolution as a tree-like process with different rates of HGT. The clear-cut result is that only by simulating the BBB at the depth of 0.8, i.e., before the divergence of the major bacterial and archaeal phyla, could a good fit with the empirical data be reached (Fig. 5). In contrast, simulation of the BBB at the critical depth of 0.7 or above, which erases the phylogenetic signal below the phylum level, did not yield a satisfactory fit (Fig. 5) (Puigbò et al. 2009). Thus, the CC model appears to be a more appropriate representation of the early phases of evolution of archaea and bacteria than the BBB model. In other words, the signal of apparent vertical inheritance (a central trend in the FOL) is detectable even for the earliest stages of evolution of each prokaryotic domain, although given the high level of inconsistency, the determination of the correct tree topology of the deepest branches in the tree is problematic at best. This analysis does not rule out a BBB as the generative mechanism underlying the divergence of archaea and bacteria, but this model cannot be tested using the approach described above because of the absence of an outgroup.
Figure 5.
Simulations of a BBB at different phylogenetic depths and with different numbers of HGT events. Each panel is a plot of the mean tree inconsistency versus phylogenetic depth (as in Fig. 4). The empirical dependence is shown by the thick blue line, and the results of simulations with 1 to 200 HGT events are shown by thin lines along a color gradient. (A) BBB simulated at depth 0.6, (B) BBB simulated at depth 0.7, (C) BBB simulated at depth 0.8. (Modified from Puigbò et al. 2009.)
THE TRENDS IN THE FOREST OF LIFE
Recent developments in prokaryotic genomics reveal the ubiquity of HGT and overthrow the “strong” TOL concept under which all (or the substantial majority) of the genes would tell a consistent story of genome evolution (the species tree, or the TOL) if analyzed with appropriate methods (“uprooting the tree of life”) (Doolittle 1999a, 2000; Pennisi 1999; Gogarten et al. 2002; Wolf et al. 2002; Gogarten and Townsend 2005; Doolittle and Bapteste 2007; Koonin 2009a). Is there any hope to salvage the TOL as a statistical central trend (Wolf et al. 2002)? The results of a comprehensive comparative analysis of phylogenetic trees for prokaryotic genes described here suggest that such a trend does exist.
The results of the FOL analysis are twofold. On the one hand, we observed high levels of inconsistency among the trees in the FOL, owing mostly to extensive HGT, as demonstrated more directly by the observations of numerous likely transfers of genes between archaea and bacteria. On the other hand, we also detected a distinct signal of a consensus topology that was particularly strong among the NUTs. Although the NUTs showed a substantial amount of apparent HGT, the transfer events seemed to be distributed randomly and did not obscure the apparent vertical signal. Moreover, the topology of the NUTs was quite similar to those of numerous other trees in the FOL, so although the NUTs certainly cannot represent the FOL completely, this set of largely congruent, nearly universal trees is a reasonable candidate for representing a central trend. However, the opposite side of the coin is that the consistency between the trees in the FOL is high at the external branches of the trees and abruptly drops, almost to the level of random trees, at greater phylogenetic depths that correspond to the radiation of archaeal and bacterial phyla. This observation casts doubt on the reality of a central trend in the FOL and suggests the possibility that the early phases of evolution might have been non-tree-like (a BBB; Koonin 2007). We addressed this problem directly by simulating evolution under the compressed cladogenesis model (Rokas et al. 2005; Rokas and Carroll 2006) and under the BBB model and found that the CC scenario better fits the observed dependence between tree inconsistency and phylogenetic depth. Thus, a consistent phylogenetic signal seems to be discernible throughout the evolution of archaea and bacteria, although, under the CC model, the prospect of unequivocally resolving the relationships between the major archaeal and bacterial clades is bleak.
The detected central trend in the FOL is most likely to represent vertical inheritance permeating the entire history of archaea and bacteria. A contribution from “highways” of HGT (i.e., preferential HGT between certain groups of archaea and bacteria, in particular, those closely related) that could mimic vertical evolution cannot be ruled out (Gogarten et al. 2002). However, the lack of significant clustering within the group of NUTs and the comparable high levels of similarity between the NUTs and different clusters of trees in the FOL suggest that the trend, even if relatively weak, is primarily vertical.
In the following sections, we take a more general, conceptual standpoint to discuss the status of the TOL in light of these findings and additional considerations.
A TREE IS AN ISOMORPHOUS REPRESENTATION OF REPLICATION HISTORY
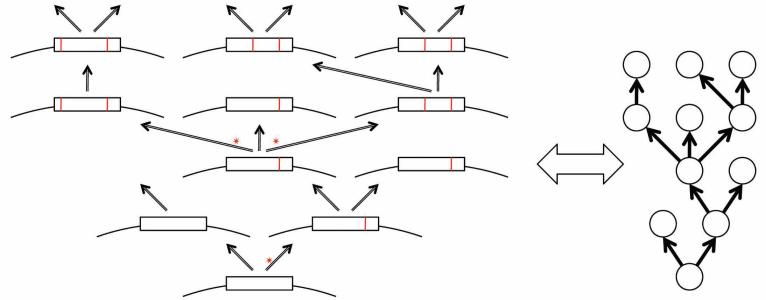
Replication of nucleic acids with an error rate below the mutational meltdown threshold is both a necessary condition and the direct cause of evolution by random drift and natural selection (Eigen 1971; Koonin and Wolf 2009). Crucially, replication and the ensuing evolution are inherently tree-like processes: A replicating molecule gives rise to two (semiconservative replication of double-stranded DNA that occurs in all cellular organisms and many viruses) or multiple (conservative replication of viruses with single-stranded DNA or single-stranded RNA genomes) copies with errors, resulting in a tree-like process of divergence (Fig. 6). In graph-theoretical terms, such a process can be isomorphously represented with a directed acyclic graph known as arborescence, which is a generalized tree where multifurcations are allowed and all edges are directed away from the root (Fig. 6) (Evans and Minieka 1992). As a result of occasional extinction of one or both progeny molecules, some of the vertices in the resulting graph emit no edges, but this does not violate the definition of an arborescence (Fig. 6) (hereafter, for the sake of simplicity, we speak of trees rather than of arborescences).
Figure 6.
A tree (arborescence) is an isomorphous representation of the error-prone replication process. An idealized scheme of the replication history of a general replicating unit (GRU) includes both bifurcations and a multifurcation (shown by asterisks). Fixed mutations are shown by red lines. (Reprinted from Koonin and Wolf 2009.)
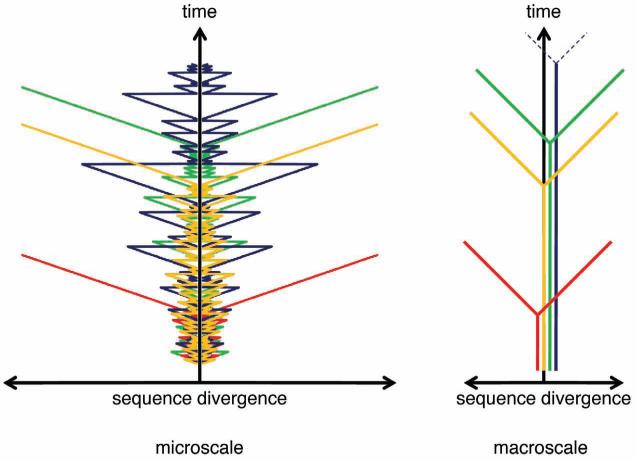
A major complication to the tree-like character of evolution is recombination that, if common, would turn the tree-like representation of the history of a replicating lineage into a network. Is there a fundamental “atomic” level of genetic organization at which recombination is negligible? In the case of homologous recombination that is extensive during coreplication of closely related sequences, in particular, in eukaryotes that engage in regular sex, and in “quasi-sexual” prokaryotes (Feil et al. 2001; Spratt et al. 2001; Turner and Feil 2007; Doolittle and Zhaxybayeva 2009), the atomic unit, effectively, is a single base pair which of course is not a level at which any analysis can be conducted. In contrast, homologous recombination between distantly related sequences is impossible, so HGT between diverse prokaryotes involves only nonhomologous (illegitimate) recombination complemented by more specific routes such as dissemination via bacteriophages and plasmids. Unlike the case of homologous recombination, there is a strong preference for evolutionary fixation of nonhomologous recombination events outside genes because preservation of the integrity of a gene after nonhomologous recombination is unlikely. An important exception is fusion and shuffling of domains in multidomain proteins (Basu et al. 2009). Consequently, the evolutionary history of a gene or domain is reticulate on the microscale owing to homologous recombination but is essentially tree-like on the macroscale (Fig. 7).
Figure 7.
Evolution of a GRU is reticulate on the microscale but tree-like on the macroscale. The scheme schematically shows the evolution of four GRUs designated by different colors. The divergence history of each GRU was simulated under the model of random homologous recombination, with the probability of recombination exponentially decreasing with sequence divergence. At each simulation step, the two daughter GRUs diverge by a constant amount (clock-like divergence) and either undergo homologous recombination (which brings the difference between the two back to zero) or not, preserving the existing state of divergence. After a number of short periods of divergence and recombination, the GRUs stochastically diverge far enough for recombination to become extremely unlikely, after which point they continue diverging without recombination. At a macroscale, this process looks like a simple bifurcation in the tree graph. (Reprinted from Koonin and Wolf 2009.)
It was argued that a tree can well describe relationships that have nothing to do with common descent, so “tree thinking” was deemed not to be a priori relevant in biology (Doolittle and Bapteste 2007). Although technically valid, this argument seems to miss the crucial point that a tree is a necessary formal consequence of the descent history of replicating nucleic acids and the ensuing evolution. Therefore, trees cannot be banished from evolutionary biology for the simple reason that they are intrinsic to the evolutionary process. Then, the main pertinent question becomes What are the fundamental units whose evolution should be represented by trees? In the practice of evolutionary biology, trees are most often built for individual genes or for sets of genes that are believed to evolve coherently. However, it is typically stated or implied that the ultimate goal is a species (organismal) tree. In our opinion, the lack of clarity about the basic unit to which tree analysis applies is the source of the entire controversy around the TOL.
GENERAL REPLICATING UNITS: FUNDAMENTAL AGENCY OF (TREE-LIKE) EVOLUTION
Conceptually, the fundamental unit of evolution can be most appropriately defined as the smallest portion genetic material with a distinct evolutionary identity, i.e., one that evolves independently of other such units, at least, during some periods of evolution. We denote such fundamental units of evolution general replicating units (GRUs) because their key characteristic is the potential of differential reproduction that makes them subject to selection independently of other GRUs. Two distinct classes of GRUs can be defined:
Bona fide selfish elements such as viruses, viroids, transposons, and plasmids. All of these elements encode some of the information required for their replication and are united through their ability to promote their evolutionary success by exploiting resources of other organisms (Koonin et al. 2006).
Quasi-independent elements that do not encode devices for their own replication but possess distinct selective value and, in that capacity, can be transferred between ensembles of GRUs (genomes) and promote their own replication along with the rest of the genome. Essentially, any functional gene or even a portion of a gene encoding a distinct protein domain with an independent functional role fits this definition.
The concept of GRUs is, in part, derived from the “selfish gene” idea of Dawkins (1976) and the selfish operon hypothesis of Lawrence and Roth (1996; Lawrence 1999). These concepts seem to generate some confusion by assigning “selfishness” to genetic elements that do not actually contribute to their own replication at the mechanistic level. It seems that the partitioning of GRUs into two distinct classes eliminates this tension, with the understanding that some of the GRUs of the first class (such as large viruses and megaplasmids) could contain multiple GRUs of the second class.
Given the extensive HGT in the prokaryotic world, any gene or a portion of a gene encoding a distinct domain possesses a degree of independence and can be fixed in the recipient population even if the conferred advantage is relatively small, or even neutral (Novozhilov et al. 2005). Therefore, the prokaryotic genetic universe appears to be a consortium of GRUs with varying degrees of independence, some of which form ensembles that evolve as a physical and functional unity during extended time intervals and are more commonly known as genomes of viruses, plasmids, and cellular life forms (Koonin and Wolf 2008).
Additional motivation for the GRU concept comes from theoretical research and simple logical considerations on precellular evolution. It appears inconceivable that the first replicating elements were comparable in size and complexity to those of modern prokaryotic genomes. Evolution of life must have started with ensembles of relatively small GRUs, some of which would provide the means for the replication of others that in turn would provide other benefits, for instance, precursor synthesis, resulting in symbiotic relationships; fully selfish elements would necessarily parasitize on such ensembles. Physical joining of GRUs would be beneficial in many cases, provided sufficient replication fidelity. Qualitative and quantitative models of this early, collective phase in the evolution of life were developed (Szathmary and Demeter 1987; Zintzaras et al. 2002; Koonin and Martin 2005; Wolf and Koonin 2007; Takeuchi et al. 2008).
As we argued previously, this precellular stage of evolution could be considered virus-like in many respects, and the principal classes of extant viruses and other selfish elements, most likely, emerged already at that stage (Koonin et al. 2006; Koonin 2009b). There is an ongoing debate regarding the place of this collective stage of evolution in the history of life, and in particular, whether the last universal cellular ancestor (LUCA) was a typical cell, a cell with a fragmented genome, or a precellular ensemble of genetic elements (Koonin and Martin 2005; Forterre 2006; Glansdorff et al. 2008). Regardless of the ultimate outcome of this debate, in principle, there seems to be no reasonable doubt as to the reality of the collective stage. Furthermore, extensive mixing and matching of GRUs (which may or may not be called HGT, depending on whether this stage is envisaged as cellular) might be not only an inherent feature of this evolutionary stage, but also a prerequisite of a rapid increase in genetic and organizational complexity of life forms (Woese 1998, 2002; Koonin and Martin 2005; Koonin et al. 2006; Koonin 2009b).
Considering the virtual inevitability of an early collective stage of evolution and the extensive HGT that permeates modern prokaryotic world, the entire evolution of prokaryotes can be viewed as a dynamic process that plays out on the network of GRUs, although relatively stable genomes consisting of hundreds and thousands of GRUs, of course, are major components of that network (Koonin and Wolf 2008). Accordingly, GRUs should be construed as fundamental units of evolution, whereas all other levels of genetic organization are more properly viewed as derived.
CONCLUSIONS
Considering that GRUs appear to be fundamental units of evolution and that a tree is a necessary form of description of the evolution of any GRU, the adequate representation of evolution of life as a whole is the full compendium of GRU-specific trees, i.e., the FOL. This being the case, the notion of a species tree becomes if not obsolete at least secondary, being applicable to some phases of evolution of some groups of organisms but not in general. This conclusion does not imply that there is no order in the FOL and that signals of coherence among the trees are not to be sought. Such patterns are indeed discernible (Galtier and Daubin 2008), and as described above, the central trend, even if relatively weak, seems to correspond to the signal of vertical inheritance detectable in the nearly universal trees (Puigbò et al. 2009). Further study of different trends detectable in the FOL is expected to clarify the relationship between vertical and horizontal transmission of genetic material in the evolution of prokaryotes.
ACKNOWLEDGMENTS
The authors thank Liran Carmel (Hebrew University, Israel) for helpful discussions of multidimensional analysis and clustering. E.V.K. thanks Michael Gelfand, Andrei Mironov, and members of the Moscow Seminar on Bioinformatics for an inspiring discussion. The authors’ research is supported by the Department of Health and Human Services intramural program (National Institutes of Health, National Library of Medicine).
REFERENCES
- Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu MK, Poliakov E, Rogozin IB. Domain mobility in proteins: Functional and evolutionary implications. Brief Bioinform. 2009;10:205–216. doi: 10.1093/bib/bbn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Bapteste E, Moreira D, Philippe H. Eubacterial phylogeny based on translational apparatus proteins. Trends Genet. 2002;18:1–5. doi: 10.1016/s0168-9525(01)02522-7. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection. 1st ed. Murray; London: 1859. [Google Scholar]
- Dawkins R. The selfish gene. Oxford University Press; Oxford: 1976. [Google Scholar]
- Doolittle WF. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999a;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Lateral genomics. Trends Cell Biol. 1999b;9:M5–M8. [PubMed] [Google Scholar]
- Doolittle WF. Uprooting the tree of life. Sci Am. 2000;282:90–95. doi: 10.1038/scientificamerican0200-90. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Boucher Y, Nesbo CL, Douady CJ, Andersson JO, Roger AJ. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Evans JR, Minieka E. Optimization algorithms for networks and graphs. Taylor and Francis; New York: 1992. [Google Scholar]
- Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, Goldstein R, Hood DW, Kalia A, Moore CE, et al. Recombination within natural populations of pathogenic bacteria: Short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc Natl Acad Sci. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Daubin V. Dealing with incongruence in phylogenomic analyses. Philos Trans R Soc Lond B Biol Sci. 2008;363:4023–4029. doi: 10.1098/rstb.2008.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansdorff N, Xu Y, Labedan B. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP. The early evolution of cellular life. Trends Ecol Evol. 1995;10:147–151. doi: 10.1016/s0169-5347(00)89024-2. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Hilario E, Gogarten JP. Horizontal transfer of ATPase genes: The tree of life becomes a net of life. Biosystems. 1993;31:111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. The biological big bang model for the major transitions in evolution. Biol Direct. 2007;2:21. doi: 10.1186/1745-6150-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009a;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. On the origin of cells and viruses: Primordial virus world scenario. Ann NY Acad Sci. 2009b doi: 10.1111/j.1749-6632.2009.04992.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: The bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. The fundamental units, processes and patterns of evolution, and the Tree of Life conundrum. Biol Direct. 2009 doi: 10.1186/1745-6150-4-33. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Dolja VV. The ancient virus world and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. The net of life: Reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG. Something for everyone. Horizontal gene transfer in evolution. EMBO Rep. 2000;1:92–95. doi: 10.1093/embo-reports/kvd042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. Horizontal gene transfer: A critical view. Proc Natl Acad Sci. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Lawrence J. Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Hendrickson H. Lateral gene transfer: When will adolescence end? Mol Microbiol. 2003;50:739–749. doi: 10.1046/j.1365-2958.2003.03778.x. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. Mosaic bacterial chromosomes: A challenge en route to a tree of genomes. Bioessays. 1999;21:99–104. doi: 10.1002/(SICI)1521-1878(199902)21:2<99::AID-BIES3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novozhilov AS, Karev GP, Koonin EV. Mathematical modeling of evolution of horizontally transferred genes. Mol Biol Evol. 2005;22:1721–1732. doi: 10.1093/molbev/msi167. [DOI] [PubMed] [Google Scholar]
- O'Malley MA, Boucher Y. Paradigm change in evolutionary microbiology. Stud Hist Philos Biol Biomed Sci. 2005;36:183–208. doi: 10.1016/j.shpsc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45:325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Is it time to uproot the tree of life? Science. 1999;284:1305–1307. doi: 10.1126/science.284.5418.1305. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Garcia-Vallvé S, McInerney JO. TOPD/FMTS: A new software to compare phylogenetic trees. Bioinformatics. 2007;23:1556–1558. doi: 10.1093/bioinformatics/btm135. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Wolf YI, Koonin EV. Search for a “Tree of Life” in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Kruger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- Spratt BG, Hanage WP, Feil EJ. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr Opin Microbiol. 2001;4:602–606. doi: 10.1016/s1369-5274(00)00257-5. [DOI] [PubMed] [Google Scholar]
- Szathmary E, Demeter L. Group selection of early replicators and the origin of life. J Theor Biol. 1987;128:463–486. doi: 10.1016/s0022-5193(87)80191-1. [DOI] [PubMed] [Google Scholar]
- Takeuchi N, Salazar L, Poole AM, Hogeweg P. The evolution of strand preference in simulated RNA replicators with strand displacement: Implications for the origin of transcription. Biol Direct. 2008;3:33. doi: 10.1186/1745-6150-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Feil EJ. The secret life of the multilocus sequence type. Int J Antimicrob Agents. 2007;29:129–135. doi: 10.1016/j.ijantimicag.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Virchow RLK. Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre. Hirschwald; Berlin: 1858. [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. The universal ancestor. Proc Natl Acad Sci. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. On the evolution of cells. Proc Natl Acad Sci. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct. 2007;2:14. doi: 10.1186/1745-6150-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Lapierre P, Gogarten JP. Genome mosaicism and organismal lineages. Trends Genet. 2004;20:254–260. doi: 10.1016/j.tig.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Santos M, Szathmary E. “Living” under the challenge of information decay: The stochastic corrector model vs. hypercycles. J Theor Biol. 2002;217:167–181. doi: 10.1006/jtbi.2002.3026. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Molecular evolution. In: Kasha M, Pullman B, editors. Horizons in biochemistry. Academic; New York: 1962. pp. 189–225. [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence of proteins. In: Bryson V, Vogel HJ, editors. Evolving gene and proteins. Academic; New York: 1965. pp. 97–165. [Google Scholar]