Abstract
In the present study, we investigated genetic divergence between complete autologous HIV-1 env genes amplified directly from plasma of two antiretroviral-naive, slow progressing Indian patients with broad neutralizing antibody response. All the envelope (Env) clones obtained from one patient (LT1) belonged to subtype C; the second patient (LT5) harbored quasispecies comprised of pure B, C, and B/C recombinants with distinct breakpoints indicative of dual infection with genetically distinct strains. Further characterization of these Envs would provide insight into the biological properties under strong humoral immune response.
The persistence of HIV-1 in the presence of immense humoral immune pressure leads to a remarkable increase in diversity in its envelope protein. With progression in the disease course, individuals with human immunodeficiency virus type 1 (HIV-1) infection usually harbor numerous genetically related viral variants termed quasispecies.1,2 Such variants are believed to either determine disease progression3–6 by overcoming the host immune response and/or develop resistance to antiretroviral therapy agents.7–11 The HIV-1 envelope (Env), which plays a key role in the interaction with cellular receptors and coreceptors in the viral entry process, remains exposed on the virus surface under incessant host-selective pressure, particularly that of the autologous neutralizing antibodies. The continual evolution of viral quasispecies by mutation poses an impediment in successful recognition both by virus-specific cellular and humoral immune mechanisms.12,13 In addition, viral sequence diversity also takes complex shape through recombination, particularly in cases of dual infection11,14–19 by different subtypes. Studies of Env diversity would provide information on selective forces such as humoral immunity that might influence the rate of progression of disease and also would help in the identification of determinants on the Env protein that modulate viral response to immune pressure such as neutralizing antibodies. Here, we investigated the genetic properties of env genes representing viral quasispecies amplified from plasma of two antiretroviral treatment (ART)-naive slow progressing Indian patients with broadly neutralizing antibody response.
First, the neutralization potential of plasma specimens obtained from the two slow progressing patients (NARI-LT1 and NARI-LT5) was examined against 28 Env-pseudotyped viruses comprising tier-1, tier-2, and tier-3 viruses as described previously.20 Plasma samples were diluted in growth media (DMEM supplemented with 5% fetal bovine sera; Invitrogen Inc.) starting from a 1:20 dilution and incubated with Env-pseudotyped viruses for 1 h at 37°C. Subsequently 1×104 TZM-bl cells21 were added to this mixture in 96-well tray tissue culture plates supplemented with 25 μg/ml DEAE Dextran (Sigma Inc.) and further incubated for additional 2 days at 37°C in a CO2 incubator. The degree of neutralization of Env-pseudotyped viruses of TZM-bl cells in the presence of LT1 and LT5 plasma was determined by measuring the reduction in relative luminescence units (RLU) as described earlier.20 As shown in Table 1, the majority of viruses tested here were significantly neutralized by LT5 plasma; LT1 plasma also showed substantial neutralization potential albeit to a lesser extent than LT5 plasma. Of viruses, 16/28 showed 50% neutralization in 1:100 LT1 plasma dilutions while 11/28 viruses showed 50% neutralization at 1:500 dilutions and up to 1:5361 dilutions. On the other hand, 21/28 viruses showed 50% neutralization to LT5 plasma at 1:100 [including PVO.03 (tier 3 virus) and JRFL (tier 2 virus)], while 12/28 viruses showed 50% neutralization at 1:500 and up to 1:8210. Overall, LT5 plasma was found to be a better neutralizer than LT1, although both of them showed broad neutralizing property against the viruses tested here.
Table 1.
Neutralization Properties of LT1 and LT5 Plasma to Heterologous Env-Pseudotyped Viruses
| |
|
IC50 values (reciprocal dilutions) |
|
|---|---|---|---|
| Origin | Envelope | LT1 | LT5 |
| India Clade C | 2.J8 | 1500 | 772 |
| 2.J9 | 882 | 723 | |
| 3.J6 | 3064 | 4190 | |
| 4.J2 | 5361 | 8210 | |
| 4.J22 | 1530 | 2890 | |
| 4.J27 | 819 | 3880 | |
| 5.J41 | 893 | 3398 | |
| 7.J16 | <50 | 1650 | |
| 7.J20 | <50 | 1530 | |
| 11.J25 | 609 | 730 | |
| 11.J28 | 480 | 464 | |
| African Clade C | Du156 | 343 | 181 |
| Du172 | 274 | 124 | |
| Du422 | 115 | 90 | |
| Zm109 | 38 | 74 | |
| Zm197 | <20 | 123 | |
| CAP45 | 815 | 462 | |
| Clade B | QH0692 | 33 | <20 |
| SC422661 | 32 | 85 | |
| PVO.04 | <20 | 147 | |
| AC10.0.29 | 36 | <20 | |
| RHPA4259.7 | 40 | 138 | |
| 6535.3 | 651 | 3400 | |
| JRFL | <20 | 195 | |
| Clade A | Q461.E2 | 162 | 128 |
| Q482.d12 | <20 | <20 | |
| Recombinant | CRF02_AG | 62 | 84 |
We next examined the genetic properties of Env obtained from these two patients harboring broadly neutralizing antibodies (Table 2). Complete gp160 was amplified from reverse transcribed plasma viral RNA of both samples in the presence of high fidelity proofreading polymerase, Platinum Taq (Invitrogen Inc.). The gp160 amplicons were cloned in either pcDNA3.1TOPO (Invitrogen, Inc.) or in pSVIIIenv as described previously20 (Table 2). For patient LT1, Env clones were obtained from two different time points—2007 and 2009. Due to unavailability, we were able obtain Envs from patient LT5 at only one time point (2007). More than one Env clone was obtained from each plasma specimen. DNA sequences of patient Env clones were obtained through Genetic Analyzer 3130XL (Applied Biosystems, Life Technologies Inc.) using Big Dye terminator as described previously.22 The genetic properties are summarized in Table 1. We found that in regard to patient LT1, all the Envs were found to have comparable potential N-linked glycosylation sites (PNLGS) (between 26 and 27); however, for patient LT5, the PNLGs ranged from 25 to 34, indicating a variation in the glycosylation pattern in viral quasispecies in this patient. The V1V2 loop length also varied between the LT5 Envs, while it remained comparable in that of patient LT1. The V3 loop charge of LT1 Envs ranges from +3 to +5 and all possessed the GPGQ motif at the tip of the V3 loop; however, in case of LT5 Envs, the V3 loop charge ranged from +2 to +7 and six of them (LT5.J3, LT5.J10, LT5.J11, LT5.J13, LT5.J24, and LT5.J25) were found to contain the GPGR motif. Out of 10 LT1 Env clones, all but two (LT1_07.J10 and LT1_07.J15) showed infectivity in TZM-bl cells that express CD4, CCR5, and CXCR4. On the other hand, only 4/14 Env clones (LT5.J3b, LT5.J4b, LT5.J7b, and LT5.J12) obtained from LT5 plasma showed infectivity in TZM-bl cells. Other Envs showed infectivity below an acceptable range. All the functional Env clones from both LT1 and LT5 were found to be CCR5 tropic (Table 1). Nonetheless, LT5 Envs that contained the GPGR motif in the V3 loop and possessed a net positive charge of greater than 6 in the V3 loop revealed the presence of viral quasispecies that were presumably CXCR4 using. Importantly, two of the Env clones (LT5.J6 and J28) showed premature stop codons, indicating the presence of defective Envs in this patient.
Table 2.
Genetic Properties of Patient Env Clones
| |
|
|
|
|
|
|
Loop length |
|
|
|
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Year of infection | Year of collection | ART Status | CD4 count (cu/mm3) | Env clones | Vector | V1V2 | V3 | V4 | V5 | gp41 | PNLGS (gp160) | V3 loop charge | V3 tip | CoR usage |
| 97139 (LT1) | 1997 | 2007 | Naïve | 736 | LT1_07.J1 | pSVIIIenv | 62 | 35 | 28 | 11 | 352 | 27 | 3 | GPGQ | CCR5 |
| LT1_07.J4 | pSVIIIenv | 62 | 36 | 28 | 11 | 352 | 27 | 5 | GPGQ | CCR5 | |||||
| LT1_07.J26 | pSVIIIenv | 62 | 35 | 28 | 11 | 352 | 26 | 3 | GPGQ | CCR5 | |||||
| LT1_07.J10 | pSVIIIenv | 62 | 35 | 29 | 12 | 352 | 27 | 3 | GPGQ | NF | |||||
| LT1_07.J15 | pSVIIIenv | 62 | 35 | 29 | 11 | 352 | 27 | 3 | GPGQ | NF | |||||
| 2009 | Naïve | 508 | LT1_09.J3 | pSVIIIenv | 65 | 35 | 21 | 11 | 352 | 27 | 4 | GPGQ | CCR5 | ||
| LT1_09.J6 | pSVIIIenv | 62 | 35 | 28 | 11 | 352 | 27 | 3 | GPGQ | CCR5 | |||||
| LT1_09.J8 | pSVIIIenv | 65 | 35 | 21 | 11 | 352 | 27 | 4 | GPGQ | CCR5 | |||||
| LT1_09.J9 | pSVIIIenv | 62 | 35 | 28 | 11 | 352 | 27 | 3 | GPGQ | CCR5 | |||||
| 991566 (LT5) | 1999 | 2007 | Naïve | 720 | LT5.J3 | pcDNA3.1 | 65 | 35 | 35 | 13 | 345 | 26 | 7 | GPGR | NF |
| LT5.J3b | pSVIIIenv | 76 | 35 | 26 | 13 | 352 | 32 | 3 | GPGQ | CCR5 | |||||
| LT5.J4 | pcDNA3.1 | 71 | 35 | 26 | 14 | 345 | 32 | 2 | GPGQ | NF | |||||
| LT5.J4b | pcDNA3.1 | 65 | 35 | 32 | 13 | 345 | 25 | 4 | GPGQ | CCR5 | |||||
| LT5.J7b | pSVIIIenv | 68 | 35 | 26 | 14 | 352 | 32 | 4 | GPGQ | CCR5 | |||||
| LT5.J10 | pcDNA3.1 | 65 | 34 | 35 | 13 | 345 | 26 | 6 | GPGR | NF | |||||
| LT5.J11 | pcDNA3.1 | 65 | 35 | 35 | 13 | 345 | 26 | 6 | GPGR | NF | |||||
| LT5.J12 | pcDNA3.1 | 74 | 35 | 26 | 13 | 345 | 34 | 3 | GPGQ | CCR5 | |||||
| LT5.J13 | pcDNA3.1 | 65 | 35 | 35 | 13 | 345 | 26 | 6 | GPGR | NF | |||||
| LT5.J20 | pcDNA3.1 | 73 | 35 | 26 | 12 | 345 | 34 | 3 | GPGQ | NF | |||||
| LT5.J24 | pcDNA3.1 | 68 | 35 | 35 | 13 | 345 | 26 | 6 | GPGR | NF | |||||
| LT5.J25 | pcDNA3.1 | 65 | 35 | 35 | 13 | 345 | 27 | 6 | GPGR | NF | |||||
| LT5.J26 | pcDNA3.1 | 74 | 35 | 26 | 12 | 345 | 34 | 3 | GPGQ | NF | |||||
| LT5.J28 | pcDNA3.1 | 74 | 35 | 26 | 12 | 345 | 34 | 3 | GPGQ | NF | |||||
PNLGS, potential N-linked glycosylation sites; CoR, coreceptor usage; NF, nonfunctional Env clones; ART, antiretroviral therapy.
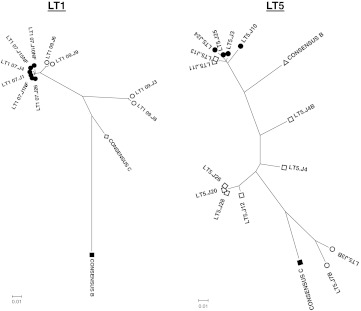
For analysis of intrapatient genetic divergence of Env clones, the deduced amino acid sequences were subjected to phylogenetic analysis using the neighbor-joining method with Kimura two-parameter using MEGA software (version 4). As shown in Fig. 1, for patient LT1, Envs obtained at the first time point (2007) clustered closely, while those obtained after 2 years (2009) were found to diverge away from Envs obtained in 2007. Interestingly, two of the Env clones (LT1-09.J3 and J8) obtained at the second time point (2009) showed significant deviation from the other Env clones obtained at the same time point. Indeed, all the Env clones belonged to subtype C. On the other hand, Env clones obtained from patient LT5 were found to be composed of pure clades B and C and B/C recombinants as identified by the Recombinant Identification Program (RIP; www.hiv.lanl.gov) leading to greater intrapatient genetic diversity in comparison to LT-1 Envs. While pure clade C (LT5.J3B and LT5.J7B) and pure clade B (LT5.J3, LT5.J24, and LT5.J25) Envs clustered closely with the respective subtype reference strains in the phylogenetic tree (Fig. 1), others varied considerably.
FIG. 1.
Genetic relatedness of autologous Envs. Deduced amino acid sequences of Env clones were used to prepare bootstrapped phylogenetic trees using MEGA4.1.
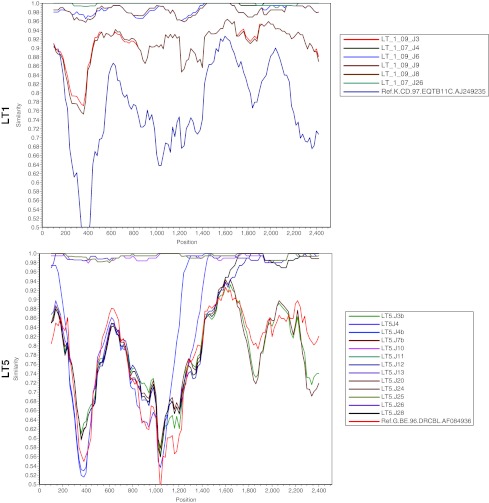
We further examined the percent similarity of nucleotide sequences of autologous Env clones using SimPlot analysis.23 As shown in Fig. 2, in case of LT1 Envs, LT1-09.J3 and LT1-09.J8 were found to differ significantly from the other LT1 Envs, suggesting considerable evolution of Env during this period. SimPlot analysis of LT-5 revealed that LT5.J4 and LT5.J4B were found to be clearly distinct between nucleotide positions 1000 and 1200. Although the LT1 complete Envs form a monophyletic cluster, when examined for domain-wise divergence, Envs obtained in 2009 (after 2 years) were found to diverge with respect to the variable loops and gp41 (data not shown). With regard to patient LT5, significant divergence in Env sequences in variable loops and gp41 was also found (data not shown). The divergence in LT5 Envs was presumably due to differences in subtypes and recombination in Env.
FIG. 2.
SimPlot analysis of complete gp160 nucleotides.
We next examined whether Env clones obtained from both LT1 and LT5 patients despite having genetic distances represented descendants closest to a common ancestor of the members of the group. For this, deduced amino acid of gp120 sequences of the Env clones was aligned with limited clade B and clade C global gp120 sequences and bootstrapped phylogenetic trees were constructed using the neighbor-joining method using MEGA (Version 4). As shown in Fig. 3, LT1 Env clones clustered closely to the Indian clade C, indicating that they evolved from a common ancestor. However, we found that LT5 Envs clustered with both subtype C and subtype B gp120 sequences; clade B sequences were close to Thai B sequences and clade C were closely clustered near Indian subtype C. The Env clone LT5.J4B gp160 sequence was found to diverge from the rest of the LT5 Envs. Our observation indicated that this particular patient LT5 was dually infected with different strains, possibly with Thai B and Indian C strains, and had likely undergone recombination during the course of infection.
FIG. 3.
Genetic relatedness of LT1 and LT5 Envs with global clade C and clade B gp120 protein sequences. Deduced amino acid sequences of LT1 and LT5 Envs were aligned with limited gp120 Env sequences obtained from the HIV database in Los Alamos (www.hiv.lanl.gov/content/nab-reference-strains/html/home.htm).
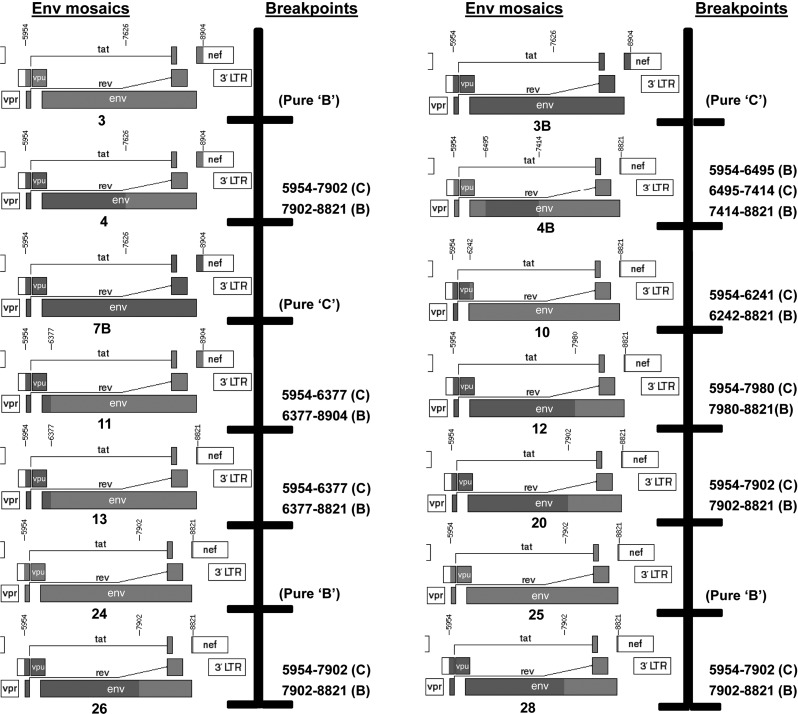
To further analyze the genetic relatedness of recombinant LT5 Envs, we studied the breakpoints. The amino acid sequences were thus subjected to breakpoint analysis through the jpHMM (jumping profile Hidden Markov Model) web server at GOBICS (http://jphmm.gobics.de) and using the breakpoints, the recombinant structures of each Env were drawn (Fig. 4) using the recombinant HIV-1 drawing tool (http://www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html) available at the HIV Los Alamos database (www.hiv.lanl.gov) to precisely dissect the position of recombination. As shown in Fig. 4, LT5.J3, J24 and J25 were found to possess Env of pure subtype B while only LT5.J3B and 7B contained pure subtype C Env. The rest of the autologous Envs were found to be mosaic in nature with distinct breakpoints between subtypes B and C. LT5.J4B was found to possess three breakpoints [5954–6495 (subtype B), 6495–7414 (subtype C), and 7414–8821 (subtype B)]; this recombination pattern was likely responsible for its significant deviation from other autologous Envs in the phylogenetic tree.
FIG. 4.
Analysis of breakpoints in LT5 Env clones. Deduced amino acids of LT5 Env clones were analyzed for breakpoints of clade B and clade C using the jpHMM web server at GOBICS (http://jphmm.gobics.de) and using the breakpoints; the recombinant structures of each Env were drawn using the recombinant HIV-1 drawing tool (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html) available from the HIV Los Alamos database (www.hiv.lanl.gov).
In conclusion, we report attributes of env genes amplified directly from plasma of two ART-naive HIV-positive slow progressing individuals. The two individuals were found to possess neutralizing plasma with considerable breadth. Both LT1 and LT5 plasma showed a fair degree of potency but subtype bias toward clade C Envs, especially LT1 plasma. Nonetheless, LT5 plasma was found to provide 50% neutralization with tier-2 and tier-3 Envs (JRFL and PVO.04), indicating that patient LT5 indeed harbored broad neutralizing antibody.
Phylogenetic tree of LT-5 sequences (Fig. 3) revealed that the Env variants were distinctly separated and clustered with epidemiologically unlinked reference viruses. Additionally, a multiregion hybridization assay (MHAbce v2)24 using LT-5 plasma also indicated recombination events in Env, suggesting that recombinant Env quasispecies constitute a major portion in the plasma (data not shown). It is unclear as to what impact dual infection has on disease progression. Results of previous studies25–30 suggested that superinfection by two different strains tends to result in faster disease progression and it was hypothesized that dual infection facilitates/accelerates viral adaptation and exploitation of cellular niches that would take many years to develop from a homogeneous infecting strain. Interestingly, in our study, we found the presence of distinct subtype B and C strains in the viral quasispecies in plasma in an antiretroviral-naive patient infected for more than 8 years. We also obtained evidence of Env clones from patient LT5 with premature stop codons. This indicated that this particular patient was infected with strains that gave rise to few defective virions. Similar observations were reported by Braibant et al.31 and Wang et al.32,33
Our findings are significant as these env genes represent viral quasispecies that were in circulation and were under immense humoral immune pressure, in contrast to the reports by Braibant et al. 31 in which they characterized Envs from proviral DNA that may not necessarily represent circulating viruses. Further analysis of these Env variants obtained from both the patients would help characterize factors that modulate replication and also identify important targets for virus neutralization.
GenBank Accession Numbers
The GenBank accession numbers of the LT5.J3, LT5.J4b, LT5.J12, LT5.J13, LT5.J20, and LT5.J26 envelopes are FJ515874, FJ515875, FJ515876, FJ515877, FJ515879, and FJ515878, respectively, while that of LT1_07.J1, LT1_07.J4, LT1_07.J26, LT1_07.J10, LT1_07.J15, LT1_09.J3, LT1_09.J6, LT1_09.J8, LT1_09.J9, LT5.J3B, LT5.J4, LT5.J7B, LT5.J10, LT5.J11, LT5.J24, LT5.J25 and LT5.J28 are JN400529, JN400530, JN400531, JN400532, JN400533, JN400534, JN400535, JN400536, JN400537, JN400538, JN400539, JN400540, JN400541, JN400542, JN400543, JN400544 and JN400545 respectively.
Acknowledgments
We thank NARI clinics for providing us with patient materials. This work was supported in part by a grant from Department of Biotechnology (DBT), Government of India (BT/PR7829/Med/14/1133/2006 and BT/PR12853/MED/29/141/2009) to J.B. The African clade C (Du and CAP) as well as clade B primary envelope plasmids were kindly provided by Dr. David Montefiori, Duke University, Durham, North Carolina, while JRFL envelope plasmid was kindly provided by Dr. Paul Clapham, University of Massachusetts Medical School, Worcester, Massachusetts. We thank Dr. Julie Overbaugh, FHCRC, Seattle, Washington and the NIH AIDS Research Reagents and Reference Program for providing us with subtype A envelope plasmids and the TZM-bl cell line. We thank Indian Council of Medical Research for providing S.M. with a Junior Research Fellowship. R.R. is a recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Government of India. We thank members of our laboratory for help and support.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Goodenow M. Huet T. Saurin W, et al. HIV-1 isolates are rapidly evolving quasispecies: Evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immune Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 2.Malim MH. Emerman M. HIV-1 sequence variation: Drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 3.Asjo B. Morfeldt-Manson L. Albert J, et al. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 4.Cheng-Mayer C. Seto D. Tateno M, et al. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 5.Tersmette M. Gruters RA. de Wolf F, et al. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: Studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anzala OA. Nagelkerke NJ. Bwayo JJ, et al. Rapid progression to disease in African sex workers with human immunodeficiency virus type 1 infection. J Infect Dis. 1995;171:686–689. doi: 10.1093/infdis/171.3.686. [DOI] [PubMed] [Google Scholar]
- 7.Albert J. Abrahamsson B. Nagy K. Aurelius E, et al. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Arendrup M. Sonnerborg A. Svennerholm B, et al. Neutralizing antibody response during human immunodeficiency virus type 1 infection: type and group specificity and viral escape. J Gen Virol. 1993;74(Pt 5):855–863. doi: 10.1099/0022-1317-74-5-855. [DOI] [PubMed] [Google Scholar]
- 9.Nowak MA. Anderson RM. McLean AR, et al. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 10.Wolfs TF. Zwart G. Bakker M, et al. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology. 1991;185:195–205. doi: 10.1016/0042-6822(91)90767-6. [DOI] [PubMed] [Google Scholar]
- 11.Diaz RS. Sabino EC. Mayer A, et al. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. The Transfusion Safety Study Group. J Virol. 1995;69:3273–3281. doi: 10.1128/jvi.69.6.3273-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brander C. Frahm N. Walker BD. The challenges of host and viral diversity in HIV vaccine design. Curr Opin Immunol. 2006;18:430–437. doi: 10.1016/j.coi.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Walker BD. Burton DR. Toward an AIDS vaccine. Science. 2008;320:760–764. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- 14.Thomson MM. Perez-Alvarez L. Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;2:461–471. doi: 10.1016/s1473-3099(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 15.Gao F. Robertson DL. Carruthers CD, et al. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabino EC. Shpaer EG. Morgado MG, et al. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68:6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson DL. Sharp PM. McCutchan FE, et al. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 18.Kampinga GA. Simonon A. Van de Perre P, et al. Primary infections with HIV-1 of women and their offspring in Rwanda: Findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology. 1997;227:63–76. doi: 10.1006/viro.1996.8318. [DOI] [PubMed] [Google Scholar]
- 19.Fang G. Weiser B. Kuiken C, et al. Recombination following superinfection by HIV-1. AIDS. 2004;18:153–159. doi: 10.1097/00002030-200401230-00003. [DOI] [PubMed] [Google Scholar]
- 20.Ringe R. Thakar M. Bhattacharya J. Variations in autologous neutralization and CD4 dependence of b12 resistant HIV-1 clade C env clones obtained at different time points from antiretroviral naive Indian patients with recent infection. Retrovirology. 2010;7:76. doi: 10.1186/1742-4690-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M. Gao F. Mascola JR, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakhashe S. Tripathy S. Paranjape R, et al. Characterization of B/C recombinants of near full-length HIV type 1 from northeastern India with mosaics identical to ARE195FL but with a different ancestral origin. AIDS Res Hum Retroviruses. 2008;24:92–99. doi: 10.1089/aid.2007.0214. [DOI] [PubMed] [Google Scholar]
- 23.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kijak GH. Tovanabutra S. Sanders-Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v.2. Virology. 2007;358:178–191. doi: 10.1016/j.virol.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb GS. Nickle DC. Jensen MA, et al. Dual HIV-1 infection associated with rapid disease progression. Lancet. 2004;363:619–622. doi: 10.1016/S0140-6736(04)15596-7. [DOI] [PubMed] [Google Scholar]
- 26.Chohan B. Lavreys L. Rainwater SM, et al. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J Virol. 2005;79:10701–10708. doi: 10.1128/JVI.79.16.10701-10708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piantadosi A. Chohan B. Chohan V, et al. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 2007;3:e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piantadosi A. Humes D. Chohan B, et al. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Templeton AR. Kramer MG. Jarvis J, et al. Multiple-infection and recombination in HIV-1 within a longitudinal cohort of women. Retrovirology. 2009;6:54. doi: 10.1186/1742-4690-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCutchan FE. Hoelscher M. Tovanabutra S, et al. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: Evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol. 2005;79:11693–11704. doi: 10.1128/JVI.79.18.11693-11704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braibant M. Agut H. Rouzioux C, et al. Characteristics of the env genes of HIV type 1 quasispecies in long-term nonprogressors with broadly neutralizing antibodies. J Acquir Immune Defic Syndr. 2008;47:274–284. doi: 10.1097/QAI.0b013e318162cac2. [DOI] [PubMed] [Google Scholar]
- 32.Wang B. Dyer WB. Zaunders JJ, et al. Comprehensive analyses of a unique HIV-1-infected nonprogressor reveal a complex association of immunobiological mechanisms in the context of replication-incompetent infection. Virology. 2002;304:246–264. doi: 10.1006/viro.2002.1706. [DOI] [PubMed] [Google Scholar]
- 33.Wang B. Mikhail M. Dyer WB, et al. First demonstration of a lack of viral sequence evolution in a nonprogressor, defining replication-incompetent HIV-1 infection. Virology. 2003;312:135–150. doi: 10.1016/s0042-6822(03)00159-4. [DOI] [PubMed] [Google Scholar]