Abstract
Low–molecular-weight metabolites produced by intestinal microbiota play a direct role in health and disease. In this study, we analyzed the colonic luminal metabolome using capillary electrophoresis mass spectrometry with time-of-flight (CE-TOFMS) —a novel technique for analyzing and differentially displaying metabolic profiles— in order to clarify the metabolite profiles in the intestinal lumen. CE-TOFMS identified 179 metabolites from the colonic luminal metabolome and 48 metabolites were present in significantly higher concentrations and/or incidence in the germ-free (GF) mice than in the Ex-GF mice (p < 0.05), 77 metabolites were present in significantly lower concentrations and/or incidence in the GF mice than in the Ex-GF mice (p < 0.05), and 56 metabolites showed no differences in the concentration or incidence between GF and Ex-GF mice. These indicate that intestinal microbiota highly influenced the colonic luminal metabolome and a comprehensive understanding of intestinal luminal metabolome is critical for clarifying host-intestinal bacterial interactions.
At least 1,000 bacterial species have been found to inhabit the human intestine, and 1014 individual bacterial cells of at least 160 different species inhabit each individual's intestine1, which is 10 times greater than the total number of somatic and germ cells in the human body. Intestinal microbiota play a fundamentally important role in health and disease such as inflammatory bowel disease2, allergy3,4, and colon cancer5. Recently, the relationship between intestinal microbiota and systemic phenomena, such as obesity6 and autism7 have been reported. The lifespan of germ-free (GF) mice is longer than that of conventional mice8, showing that intestinal microbiota contribute to lifespan. In the latter half of the 1990s, many molecular-biological approaches using 16S rRNA gene sequencing have been performed for the analysis of intestinal microbiota9. More recently, metagenomic techniques have been used to characterize both the composition and the potential physiological effects of the microbial community (microbiome)10,11. Some reports have demonstrated that the gut microbiome influences the metabolic profiling of organs, blood, and urine of the host12,13.
Low–molecular-weight metabolites produced by intestinal microbiota are absorbed constantly from the intestinal lumen and carried to systemic circulation; they play a direct role in health and disease. There are few reports concerning the function of metabolites produced by intestinal microbiota. At present, some research studies are targeting specific metabolites, such as short chain fatty acids (SCFAs)14 and polyamines (PAs)15,16, but not global metabolites (metabolome). Metabolomics is the rapidly evolving field of the comprehensive measurement of, ideally, all metabolites in a biological fluid17,18. To the best of our knowledge, although metabolomic approaches have been employed for the biochemical characterization of metabolic changes in blood19 or urine20 triggered by gut microbiota, there is little knowledge about the intestinal luminal metabolome, which may be source of metabolites in blood and urine. Nicholson and co-workers21,22 suggested analyzing intestinal microbial-host metabolic interactions by metabolomic analysis using 1H nuclear magnetic resonance (NMR) spectroscopy and ultra-performance liquid chromatography-mass spectrometry (UPLC-MS). The dynamics of fecal volatile metabolites were changed by the administration of prebiotics and probiotics based on the results of gas chromatography mass spectrometry solid phase micro-extraction23. However, these metabolome analyses focused on specific metabolites, such as bile acids metabolism, and did not focus on a wide spectrum of intestinal luminal metabolites. In addition, in these studies21,22, they analyzed metabolome-containing bacterial intercellular metabolites because the sample was prepared by sonication of feces. For clarifying the relationship between health/disease and intestinal bacterial metabolites, only free bacterial metabolites in the intestinal luminal content should be analyzed. On the other hand, none of the previous researches have taken food ingredients into consideration. Therefore, there is little information available regarding the intestinal luminal metabolome produced by intestinal microbiota.
Large-scale metabolome analysis methods are based on gas chromatography mass spectrometry (GC-MS), liquid chromatography mass spectrometry (LC-MS), NMR, Fourier-transform ion cyclotron resonance mass spectrometry, and capillary electrophoresis mass spectrometry (CE-MS). Each of these methods has its own merits and demerits. For example, although NMR can identify metabolites exactly, its sensitivity is lower than that of other methods. GC-MS is suitable only for volatile metabolites. Although LC-MS detects relatively wide-spectrum metabolites, chromatographic separation is not satisfactory. The utility of CE-MS in metabolomics has recently been recognized; this is more sensitive and detects a wider spectrum than LC-MS24. In particular, CE with time-of-flight mass spectrometry (CE-TOFMS) is a novel strategy for analyzing and differentially displaying metabolic profiles25. The major advantages of CE-TOFMS include its extremely high resolution, high throughput, and ability to simultaneously quantify all charged low-molecular-weight compounds in a sample. Because this can determine the majority of metabolic intermediates, many metabolites in the cellular/bacterial metabolic pathways can be thus observed18. A comprehensive understanding of the metabolite profile in the intestinal lumen by using CE-TOFMS would be critical for elucidating the host-intestinal bacterial interaction.
Here, using CE-TOFMS, we analyzed the colonic luminal metabolome obtained from GF mice and ex-germ free (Ex-GF) mice, divided from brothers bred from same parents, harboring Specific pathogen-free (SPF) mice intestinal microbiota and demonstrated the large effect of intestinal microbiota on the colonic luminal metabolome.
Results
The difference in colonic luminal metabolome between GF and Ex-GF mice
From the colonic luminal metabolome in both of GF and Ex-GF mice, CE-TOFMS identified 179 metabolites. Principal component analysis (PCA) and hierarchical clustering, showing patterns of metabolites, are shown in Fig. 1a and b, respectively. A remarkable difference was observed in the colonic luminal metabolome between GF mice and Ex-GF mice, showing that intestinal microbiota highly influences the colonic luminal metabolome, as expected. From the pellet, 250 metabolites (ingredients) were detected.
Figure 1. The difference in the colonic metabolomes between GF mice and Ex-GF mice.
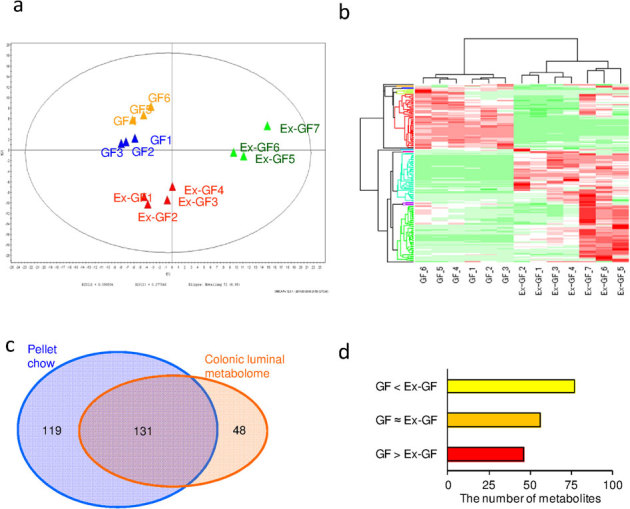
(a) PCA of the profiling data from the colonic metabolome. (b) Hierarchical clustering showing patterns of metabolites. Red and green indicate high and low concentrations of metabolites, respectively. (c) Venn diagram of the metabolites noted in the pellet and the colonic metabolome. (d) The number of colonic luminal metabolites in the group GF > Ex−GF, GF≈Ex−GF, and GF > Ex−GF.
Of the 179 metabolites in the colonic luminal metabolome, 131 metabolites were common to those detected from the pellet, and 48 metabolites were detected only from the colonic luminal metabolome (Fig. 1c). Further, 119 metabolites were detected only from the pellet (Supplemental Table S1), indicating that these were perfectly absorbed in the upper part of the intestine. Of the 179 metabolites in the colonic luminal metabolome, 48 metabolites were present in significantly (p < 0.05) higher concentrations and/or incidence in the GF mice than in the Ex-GF mice (group GF > Ex-GF), 77 metabolites were present in significantly (p < 0.05) lower concentrations and/or incidence in the GF mice than in the Ex-GF mice (group GF < Ex-GF), and 56 metabolites showed no differences in the concentration or incidence between GF and Ex-GF mice (group GF ≈ Ex-GF) (Fig. 1d). These metabolites were classified into 10 categories as follows: amino acid (AA) derivatives, AA metabolism relatives, basic AAs, carbohydrate metabolism intermediates, central carbon metabolism intermediates, co-enzyme/co-enzyme derivatives, lipid metabolism relatives, nucleic acid relatives, peptides, and others. The details of the metabolites belonging to group GF > Ex-GF, group GF ≈ Ex-GF, and group GF < Ex-GF are shown in Supplemental Table S2 and Supplemental Fig. S1, Supplemental Table S3 and Supplemental Fig. S2, and Supplemental Table S4 and Supplemental Fig. S3, respectively. In the group GF > Ex-GF, carbohydrate metabolism intermediates, such as glucosamine, were characteristically detected. In the group GF ≈ Ex-GF, basic AAs and peptides were characteristically detected. In the group GF < Ex-GF, AA metabolism relatives, central carbon metabolism intermediated, co-enzyme/co-enzyme derivatives, and others (unclassified) were characteristically detected.
Comparison of colonic luminal metabolites in GF and Ex-GF mice
For further understanding, we classified metabolites into 7 groups as follows: (A) metabolites that belong to group GF > Ex-GF but not detected from the pellet, (B) metabolites that belong to group GF > Ex-GF and detected from the pellet, (C) metabolites that belong to group GF ≈ Ex-GF but not detected from the pellet, (D) metabolites that belong to group GF ≈ Ex-GF and detected from the pellet, (E) metabolites that belong to group GF < Ex-GF but not detected from the pellet, (F) metabolites that belong to group GF < Ex-GF and detected from the pellet, and (G) metabolites (ingredients) detected from only the pellet (Fig. 2). Of the 179 metabolites in the colonic luminal metabolome, the concentration of 60 metabolites involved in primary metabolisms could be measured; further, the absolute concentration of these metabolites in the pellet, colonic lumen of GF mice, and that of Ex-GF mice were compared (Fig. 3). In Fig. 3, we have shown the estimated value for the metabolite's (ingredient) concentration of the pellet in the colonic lumen. The average solid content of the pellet and colonic content were 93.45 % and 35.06 %, respectively. We consider that the pellet reached the colon with dilution by drinking water and digestive juices without absorption/hydrolyzation; we estimated that the pellet was diluted 2.665-fold by drinking water and digestive juices (2.665 = 93.45/35.06). Thus, the metabolite concentration of the pellet was calculated using the following formula:
Figure 2. Classification of metabolites detected from mice and pellets.
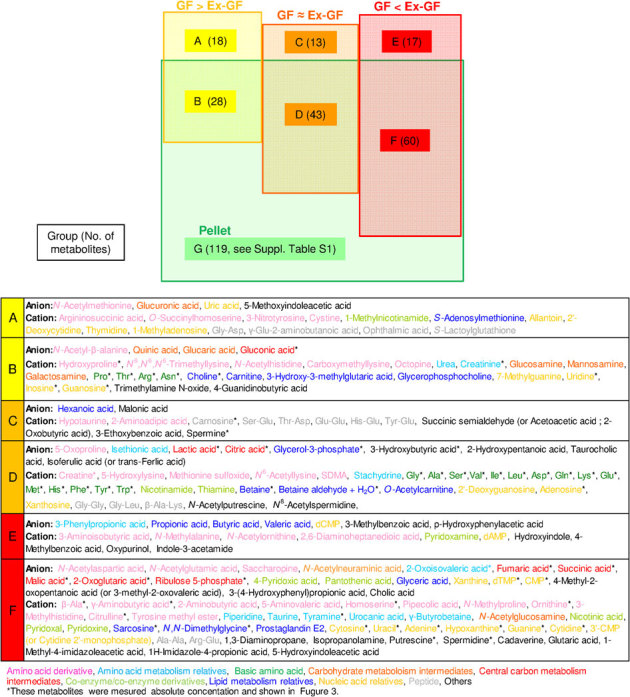
The metabolites in each group are classified as follows: (A) Metabolites produced by host and absorbed/hydrolyzed by colonic microbiota. (B) Metabolites produced by host or derived from pellet and absorbed/hydrolyzed by colonic microbiota. (C) Metabolites produced by host and not influenced by colonic microbiota. (D) Metabolites produced by host or derived from pellet and not influenced by colonic microbiota. (E) Metabolites produced by colonic microbiota. (F) Metabolites produced by colonic microbiota or derived from pellet and the absorption of which was possibly inhibited in the colon. (G) Ingredients in the pellet absorbed by the host.
Figure 3. Absolute quantitative comparison of metabolites in the pellet and the colonic lumen of GF mice and Ex-GF mice (nmol/g of feces).
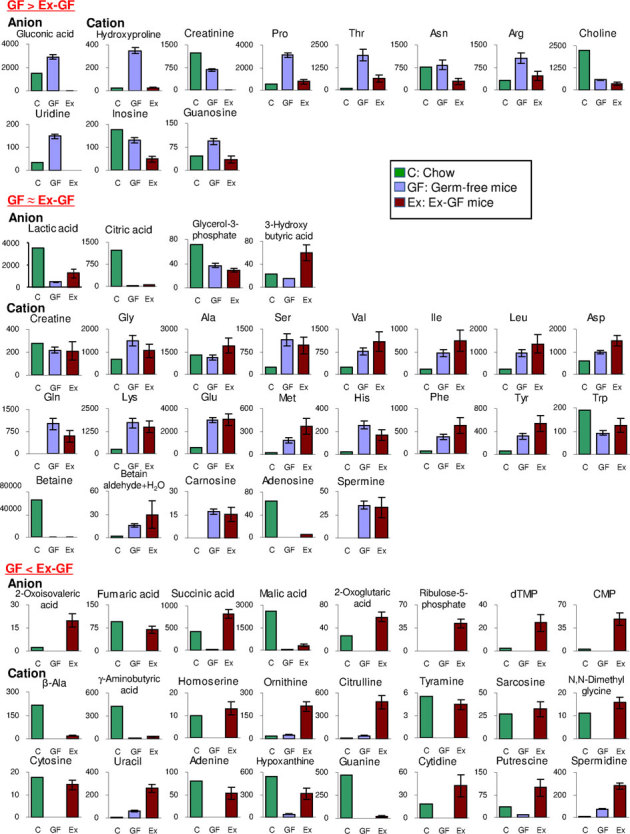
Metabolites belonging to group GF > Ex−GF, GF ≈ Ex−GF, and GF < Ex−GF are shown in the upper, middle, and bottom panels, respectively. Data are represented as mean ± SEM. Estimated values of pellet ingredients in the colonic lumen are calculated as follows: Estimated values of ingredients (nmol/g) = measured concentration in pellet by CE-TOFMS/2.665 [ = pellet solid content (93.451%)/solid content of colonic content (33.06%)].
Equivalent value (nmol/g) = value measured by CE-TOFMS/2.665
Eighteen metabolites belonging to group A were those derived from host/digestion and absorbed/hydrolyzed by intestinal microbiota. Twenty-eight metabolites belonging to group B were derived from the pellet or host/digestion and absorbed/hydrolyzed by intestinal microbiota. The concentrations of hydroxyprolin, Pro, Thr, Arg, and uridine in the intestinal lumen of GF mice were obviously higher than those in the pellet (Fig. 3 upper panel). In contrast, the concentrations of creatinine, Asn, choline, and inosine in the intestinal lumen of GF mice were equal or lower than those in the pellet (Fig. 3 upper panel). Thirteen metabolites belonging to group C were derived from host/digestion and not absorbed/hydrolyzed by intestinal microbiota. In this group, peptides were characteristically detected. Forty-three metabolites belonging to group D were derived from pellet or host/digestion and not absorbed/hydrolyzed by intestinal microbiota. In this group, basic AAs were characteristically detected. The concentration of these AAs, except Trp, in GF mice was obviously higher than that in the pellet (Fig. 3 middle panel). SCFAs, i.e., butyric acid and propionic acid, were characteristically detected. Seventeen metabolites belonging to group E were those produced by intestinal microbiota. Sixty metabolites belonging to group F were those produced by intestinal microbiota or derived from the pellet and their absorption/hydrolyzation by intestinal microbiota was inhibited. However, to the best of our knowledge, no report has described that intestinal microbiota suppress the absorption/hydrolyzation of low-molecular metabolites (ingredients) derived from foods. Therefore, we believe that the metabolites in groups E and F were simply produced by intestinal microbiota. Primary amines, β-Ala, tyamine, γ-aminobutyric acid (GABA), and PAs (putrescine, spermidine, and cadaverine), along with the bases adenine, guanine, cytosine, and uracil, were characteristically detected in group F (Fig. 3 bottom panel).
Colonic microbiota of Ex-GF mice
The PCA diagram and dendrogram of colonic microbiota of Ex-GF mice based on the T-RFLP pattern are shown in Fig. 4a and 4b, respectively. The colonic microbiota of Ex-GF mice were divided into two clusters—Ex-GF 1, 2, 3, and 4 (first sons) and Ex-GF 5, 6, and 7 (second sons)—along with the result of PCA of the colonic luminal metabolome (Fig. 1a). The position of Ex-GF 7 was separated from the other brothers from the same mother in both PCA and the dendrogram of colonic microbiota as well as the PCA of the metabolome, indicating that intestinal microbiota influenced the intestinal luminal metabolome. The number of predominant bacterial genera and groups was determined by real-time PCR. Figure 4c shows the cell numbers for predominant bacterial genera and groups in the Ex-GF 1–4 (first sons) and Ex-GF 5–7 (second sons) generations. The populations of Enterococcus spp. and Lactobacillus spp. in Ex-GF 1–4 were significantly higher than those in Ex-GF 5–7 (p < 0.05). The population of Enterobacteriaceae in Ex-GF 1–4 tended to be lower than that in Ex-GF 5–7 (p = 0.082).
Figure 4. Colonic microbiota in GF mice and Ex−GF mice.
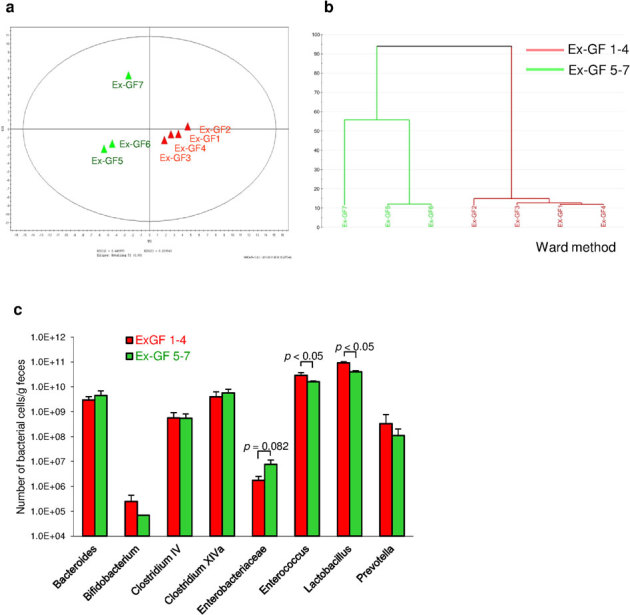
(a) PCA of the profiling data from colonic microbiota. (b) The dendrogram of the T-RFLP profiles of colonic microbiota from GF mice and Ex-GF mice. (c) The number of predominant bacterial genera and groups in the Ex-GF 1, 2, 3, and 4 (first sons) and Ex-GF 5, 6, and 7 (second sons) clusters.
Relationships between predominant bacteria and metabolites
The relationship between predominant bacteria populations and the concentration of metabolites that there are difference between Ex-GF 1–4 (first sons) and Ex-GF 5–7 (second sons) is shown in Table 1. The number of Enterobacteriaceae, Enterococcus spp., and Lactobacillus spp. was positively correlated with the concentration of 9, 1, and 1 of the 25 metabolites, respectively. In contrast, the number of Enterobacteriaceae, Enterococcus spp., and Lactobacillus spp. was inversely correlated with the concentration of 1, 4, and 20 out of the 25 metabolites, respectively. The number of Bacteroides spp., Bifidobacterium spp., Clostridium cluster IV, Clostridium subcluster XIVa, and Prevotella spp. was not correlated with the concentration of any of the metabolites.
Table 1. Interrelation between the number of predominant intestinal bacteria and metabolite concentrations.
| Metabolites1) | Predominant bacterial groups | |||||||
|---|---|---|---|---|---|---|---|---|
| GF < Ex-GF | Bacteroides | Bifidobacterium | Clostridium IV | Clostridium XVIa | Enterobacteriaceae | Enterococcus | Lactobacillus | Prevotella |
| Ornithine | 0.308 | −0.345 | −0.136 | 0.238 | 0.722 | −0.619 | −0.934 ** | −0.278 |
| 3-Methylhistidine | 0.415 | −0.549 | 0.250 | 0.529 | 0.767 * | −0.790 * | −0.982 *** | 0.022 |
| Homoserine | 0.597 | −0.256 | 0.550 | 0.723 | 0.633 | −0.896 | −0.964 | 0.125 |
| γ-Aminobutyric acid | 0.484 | −0.330 | 0.075 | 0.419 | 0.655 | −0.782 * | −0.932 ** | −0.212 |
| N-Methylalanine | 0.400 | −0.271 | 0.024 | 0.416 | 0.800 * | −0.650 | −0.893 ** | −0.327 |
| Pipecolic acid | 0.251 | −0.645 | 0.324 | 0.589 | 0.827 * | −0.753 | −0.955 *** | 0.012 |
| Urocanic acid | 0.689 | −0.760 | 0.306 | 0.534 | 0.444 | −0.728 | −0.824 * | 0.000 |
| 2-Oxoisovaleric acid | 0.501 | −0.672 | −0.311 | 0.070 | 0.520 | −0.648 | −0.916 ** | −0.177 |
| Piperidine | 0.217 | −0.673 | 0.128 | 0.483 | 0.865 * | −0.508 | −0.853 * | −0.285 |
| Glyceric acid | 0.217 | −0.575 | 0.243 | 0.560 | 0.901 ** | −0.689 | −0.934 ** | −0.123 |
| N,N-Dimethylglycine | 0.648 | −0.061 | −0.308 | −0.011 | 0.378 | −0.488 | −0.759 * | −0.126 |
| Propionic acid | −0.630 | 0.925 | −0.002 | −0.313 | −0.342 | 0.482 | 0.744 | 0.326 |
| Hypoxanthine | 0.608 | −0.515 | 0.310 | 0.530 | 0.463 | −0.826 * | −0.847 * | 0.029 |
| Cytosine | 0.496 | −0.465 | −0.024 | 0.309 | 0.657 | −0.669 | −0.953 *** | −0.135 |
| Adenine | −0.306 | 0.428 | −0.159 | −0.479 | −0.771 * | 0.828 * | 0.998 *** | 0.117 |
| 4-Methyl-2- oxopentanoic acid2) | 0.639 | −0.665 | 0.123 | 0.413 | 0.532 | −0.706 | −0.908 ** | −0.081 |
| Pantothenic acid | 0.476 | −0.476 | 0.271 | 0.601 | 0.827 * | −0.713 | −0.884 ** | −0.150 |
| Isopropanolamine | 0.425 | −0.484 | 0.843 | 0.908 | 0.824 | −0.887 | −0.888 | 0.168 |
| Glutaric acid | 0.317 | −0.471 | 0.262 | 0.609 | 0.903 ** | −0.741 | −0.928 ** | −0.194 |
| Pyridoxine | 0.269 | −0.328 | −0.063 | 0.311 | 0.880 * | −0.630 * | −0.862 * | −0.270 |
| GF > Ex−GF | ||||||||
| Hydroxyproline | 0.554 | −0.470 | 0.154 | 0.448 | 0.724 | −0.712 | −0.941 ** | 0.003 |
| N-Acetylhistidine | 0.136 | −0.030 | 0.374 | −0.052 | −0.676 | 0.326 | 0.616 | 0.716 |
| Asn | 0.534 | −0.507 | 0.105 | 0.366 | 0.663 | −0.701 | −0.962 ** | 0.302 |
| Arg | 0.573 | −0.623 | 0.105 | 0.445 | 0.510 | −0.671 | −0.810 * | −0.334 |
| Glucosamine | 0.268 | 0.049 | −0.170 | 0.224 | 0.769 * | −0.589 | −0.841 * | −0.357 |
The metabolites were selected based on the difference in their concentrations between the cluster Ex-GF 1-4 (first sons) and Ex-GF 5−7 (second sons).
1) a. Amino acid derivative, b. Amino acid metabolism relatives, c. Basic amino acid, d. Lipid metabolism relatives, e. Nucleic acid relatives, f. Carb ohydrate metaboloism intermediates, g. Others.
2)This was idintified as 4-Methyl-2-oxopentanoic acid or 3-Methyl-2-oxovaleric acid.
Interrelation between intestinal bacteria and metabolites (*P <0.05, **P < 0.01, ***P < 0.001) h. positive correlation, i. inversely correlation.
Discussion
In this study, by comparing the colonic luminal metabolome of GF mice and Ex-GF mice using CE-TOFMS, we obtained a novel profile of numerous metabolites in the intestinal lumen without comparison in the past21,22,23. In other studies, 10–60 metabolites have been detected from fecal samples by NMR and LC-MS, but in this study, 179 metabolites were detected by CE-TOFMS, which is 5 times higher than that of past studies. This is attributable to the extremely high resolution provided by CE-TOFMS24,25. The present results indicate that CE-TOFMS is the most suitable method for comprehensive and large-scale metabolome analysis in the intestinal luminal environment. Understanding the input of metabolites from the pellet enabled us to consider the impact of intestinal microbiota on the colonic luminal metabolome. We compared the results of the present study with the general knowledge in our field as follows.
Almost all basic AAs—except Pro, Thr, Asn, and Arg—belonged to group GF ≈ Ex-GF (Supplemental Table S3) and were obviously present in higher concentrations in the colonic lumen of both GF and Ex-GF mice than in the pellet, indicating that free basic AAs in colonic lumen are not derived from the pellet directly, but are produced by the digestion of proteins contained in the pellet and/or intestinal mucus in the upper part of the intestine. This also suggests that AAs arising from digestion in the upper part of the intestine are not entirely absorbed there, resulting in the entry of unabsorbed AAs into the colon. We have believed that AAs are utilized as sources of nitrogen by intestinal microbiota; for example, AAs arising from digestion are deaminated/degraded by intestinal microbiota, and NH3 degraded from AAs may be utilized for microbial cell synthesis, while microbial metabolism of AAs results in the production of SCFAs and gases in the colon26,27. However, 15 basic AAs—except Pro, Thr, Asn, and Arg—were not significantly absorbed/degraded by the intestinal microbiota. Based on these results, we consider that the theory concerning the relationship between AAs and intestinal microbiota in the colon needs to be reassessed. Further studies are required to clarify why intestinal bacteria utilize only Pro, Thr, Asn, and Arg.
Although most peptides were not absorbed/degraded by intestinal microbiota similar to most of the AAs, 4 peptides—Gly-Asp, γ-Glu-2-aminobutanoic acid, ophthalmic acid, and S-lactoylglutathione—were produced by host/digestion and/or degraded perfectly by intestinal microbiota, while 2 peptides, Arg-Glu and Ala-Ala, were produced by intestinal microbiota (Supplemental Fig. S4). Interestingly, γ-Glu-2-aminobutanoic acid and ophthalmic acid are biosynthesized by hepatic glutathione consumption induced by oxidative stress, with serum ophthalmic acid being a sensitive indicator of hepatic glutathione depletion28. After birth, oxygen in the colon appears to be consumed first by colonized aerobes, such as Enterobacteriaceae and Streptococcus29; therefore, we suppose that a little oxygen exists in the colonic lumen in GF mice and colonocytes are influenced by oxidative stress. Arg-Gln inhibits retinal neovascularization in the mouse model of oxygen-induced retinopathy30, indicating that intestinal microbiota produce functional peptides.
Most primary amines biosynthesized by decarboxylation from AAs have physiological functions. Although no differences in the concentrations of most of the precursors were noted between the GF mice and the Ex-GF mice or even if the concentrations of the precursors in the GF mice were higher than those in the Ex-GF mice, the concentrations of all primary amines in the Ex-GF mice were higher than those in the GF mice (Supplemental Fig. S5). In the colon, β-Ala, cadaverine, putrescine, tyramine, and GABA are synthesized by colonic bacterial decarboxylase from free AAs, Asp, Lys, ornithine, Tyr, and Glu, respectively. These are also biosynthesized in the host cells. Further research is warranted in order to confirm whether a certain proportion of these primary amines produced by intestinal microbiota are absorbed by colonocytes, with the remaining portion circulating in the blood.
PAs, such as putrescine, spermidine, and spermine, are one of the most important metabolites produced by intestinal microbiota affecting the health and disease of the host31. PAs are organic cations required for cell growth and differentiation and for the synthesis of DNA, RNA, and proteins32; they are constantly absorbed as energy sources from the intestinal lumen33. PAs have many functions, such as the maturation and maintenance of intestinal mucosal barrier34,35, anti-inflammatory actions36, antimutagenicity37, and autophagy38. Although we have demonstrated that colonic microbiota are involved in regulating colonic luminal PA concentrations39,40, which entity influenced colonic luminal PA concentrations to a greater extent—intestinal microbiota or colonocytes—remained unclear. This study demonstrates that the intestinal luminal concentrations of putrescine and spermidine are mainly dependent on colonic microbiota. However, spermine concentrations were not influenced by intestinal microbiota. Spermine has the strongest physiological function and toxicity among PAs, and its concentration appears to be strictly controlled by both colonic microbiota and colonocytes. We mapped the results for each PA onto the PA metabolic pathways of Escherichia coli (Supplemental Fig. S6). Here, agmatine was not detected although Arg decarboxylation is the dominant pathway for PA biosynthesis in human gut microbiota41. On the other hand, the pathway via which putrescine is decarboxylated from ornithine produced from Arg appeared to be available. Further studies are required to clarify the PA metabolic pathways of intestinal microbiota.
Prostaglandin E2 (PGE2) was detected only in Ex-GF mice. PGE2 is an interleukin-10 (IL-10)-independent innate immune suppressor. A recent study reported PGE2 and suppressor of cytokine signalling 1 (SOCS1) as an essential mediator of immune tolerance in the intestine, and that these may act as an alternative intestinal tolerance mechanism distinct from IL-10 and regulatory T cells by potentially interacting together42. Therefore, we suppose that the intestinal microbiota contain activation factors for innate immunity, similar to inflammation.
These findings were reconfirmed by the present CE-TOFMS analysis. SCFAs, such as propionic acid and butyric acid, that belonged to group E are produced by intestinal microbiota. Urea was detected only from GF mice, showing that urea is hydrolyzed by the urease of intestinal microbiota43. Taurocholic acid, a major conjugated bile acid in mice44, was detected from the intestinal lumen of both GF mice and Ex-GF mice, although the concentrations of taurine and cholic acid in the colonic lumen of Ex-GF mice were significantly higher than those in GF mice, proving that conjugated bile acids not absorbed in the ileum are reconjugated by intestinal microbiota (Supplemental Fig. S7). CE-TOFMS could not detect secondary bile acids such as deoxycholic acid, a strongly toxic acid. However, since the reconjugation of bile acids is the first step in the production of secondary bile acids, this is one of the harmful influences of intestinal microbiota.
We tried to test the association between the number of clusters of colonic microbiota of Ex-GF mice (i.e., Ex-GF1-4 and Ex-GF5-7) and the concentration of metabolites, using genera- or group-specific primer sets in order to comprehensively analyze uncultured bacteria. Enterobacteriaceae, Enterococus spp., and Lactobacillus spp. were found to influence metabolite concentrations. To the best of our knowledge, this finding is novel because none of the published studies on microbiota and metabolite correlation have taken into account uncultured bacteria. Especially, Enterobacteriaceae thrive in an inflamed environment45,46 and are known to be present in increased amounts in the elderly47. The relationship between Enterobacteriaceae, metabolites, and inflanmmation/ageing warrants further study.
These discussions center on a comparison between general knowledge and the data obtained in the present study. However, the effects of almost all metabolites in the colon are unclear. Even for GABA, which is known as a neurotransmitter, it remains unknown whether colonic GABA significantly affects health or disease. Although several metabolites might not contribute to health and disease, some metabolites might be important findings for researchers in another field. We consider that these data have implications for researchers in a number of fields, such as medicine, immunology, physiology, pharmacology, bacteriology, nutrition, and life sciences.
Methods
Mice
Germfree BALB/c mice were purchased originally from Japan Clea Inc. (Tokyo, Japan), and were bred in the Department of Infectious Diseases, Tokai University School of Medicine, Kanagawa, Japan. We divided male mice bred from sister-brother mating into two groups, GF mice (first sons: GF 1–3, second sons: GF-4–6) and Ex-GF mice (first sons: Ex-GF 1–4, Ex-GF 5–7), twice (Supplemental Fig. S8). They were housed in Trexler-Type flexible film plastic isolators with sterilized Clean tip (CLEA Japan, Inc., Tokyo) as bedding and given sterilized water and sterilized commercial CL-2 pellets (CLEA Japan, Inc.) ad libitum. The diet was sterilized with an autoclave (121°C, 30 min). Surveillance for bacterial contamination was performed by periodic bacteriological examination of feces throughout the experiments. Ex-GF mice were inoculated at 4 weeks of age into the stomach by a metal catheter with 0.5 mL of a 10−1 suspension of feces obtained from SPF BALB/c mice. The protocols approved by the Kyodo Milk Animal Use Committee (Permit Number: 2009-02) and all experimental procedures were performed according to the guidelines of the Animal Care Committee of Tokai University.
Preparation of colonic luminal metabolome
Seven-week-old mice were sacrificed by cervical dislocation and the colon was resected via an abdominal incision. The colon was opened using a longitudinal incision, and the colonic contents were obtained and stored at –80°C until use. Frozen colonic contents (approximately 100 mg) were diluted 5-fold with Dulbecco's Phosphate Buffered Saline (D-PBS) (GIBCO) and extracted three times by intense mixing for 1 min and letting them stand for 5 min on the icebox. At 1 min after extraction, the upper aqueous portion without precipitation at the bottom was collected and centrifuged (12,000 × g for 10 min at 4°C), and 100 μL of supernatant was centrifugally filtered through a 5-kDa cutoff filter Ultrafree-MC (Millipore). The filtrate was stored at –80°C until use. We confirmed the absence of intracellular metabolites derived from bacteria destroyed by this extraction, in the supernatant (Supplemental Figure S9). Because this extraction was conducted to obtain only water-soluble metabolites, non-water-soluble metabolites were not extracted efficiently. Even if the extraction efficiencies for each metabolite vary, this extraction method is suitable for relative quantification, since all the samples were extracted using the same procedure at the same time. The precipitate was washed by 1 mL D-PBS twice, stored at –80°C, and used for the analysis of intestinal microbiota.
Preparation of pellet for metabolome analysis
Sterilized commercial CL-2 pellets (CLEA Japan, Inc.) was crushed by mortar and prepared as described in the colonic luminal metabolome section above.
CE-TOFMS
The metabolomics measurement and data processing was performed as described previously with an Agilent Capillary Electrophoresis System48. Then, 20 μL of fecal solution was mixed with 80 μL of 250 μM each of methionine sulphone (MetSul) and CSA. The measurement of extracted metabolites in both positive and negative modes was performed by using commercial electrophoresis buffer (Solution ID H3301-1001 for the cation mode and H3302-1021 for the anion mode; Human Metabolome Technologies Inc., Tsuruoka, Japan). The alignment of detected peaks was performed according to the m/z value and normalized migration time. Then, peak areas were normalized against those of the internal standards MetSul and D-camphor-10-sulfonic acid (CSA) for cationic and anionic metabolites, respectively. Annotation tables were produced from measurement of standard compounds and were aligned with the datasets according to similar m/z value and normalized migration time. We confirmed that this method has sufficient reproducibility for analysis of the intestinal luminal metabolome by using human feces (Supplemental Figure S10). Principal component analysis (PCA) and clustering analysis was processed by SIMCA-P+ (Umetrics), respectively.
Colonic bacterial DNA extraction
The precipitate obtained by the first centrifugation in the procedure of the preparation of the colonic luminal metabolome was washed by 1 mL D-PBS twice, stored at –80°C, and used for the analysis of intestinal microbiota. This bacterial DNA was isolated using the methods described by Matsuki et al.49 with some modifications. Briefly, bacterial suspension with lysis buffer was treated at 70°C for 10 min in a water bath and vortexed vigorously for 60 s with a Micro Smash MS-100 (Tomy Digital Biology Co., Ltd., Tokyo, Japan) at 4,000 rpm.
T-RFLP analysis
T-RFLP analysis was performed as described in our previous report40 with some modifications. Briefly, 27F primer (5′-AGAGTTTGATCCTGGCTCAG-3′) labelled at the 5′-end with 6-carboxyfluorescein was used instead of the 529F primer. Then 50 ng of the purified PCR product was digested using 0.25 units of HaeIII or AluI (Takara) at 37°C for 3 h. The PCR products and the restriction digest products were purified by GenElute PCR Clean-Up Kit (Sigma). The terminal-restriction fragment (T-RF) length was determined using a 1200LIZ standard size marker (Applied Biosystems) in a 3130 Genetic Analyzer (Applied Biosystems) and with the aid of Gene Mapper software v.4.0 (Applied Biosystems). PCA was processed by SIMCA-P+ (Umetrics).
Real-time PCR for quantitative determination of bacterial cell numbers
Real-time PCR for the quantification of predominant bacterial genera and groups was performed using the StepOne Real-time PCR system (Applied Biosystems), as described in our previous report50. The primer sets used are shown in Supplemental Table S5 as well as in our previous report31.
Data analysis and statistics
PCA in metabolome and colonic microbiota was processed by SIMCA-P+ (Umetrics). Clustering analysis in metabolome was processed by MATLAB 2008a (MathWorks, MA, USA). Differences in relative quantity between GF mice and Ex-GF mice were evaluated for individual metabolites by Welch's t-test. The relationship between metabolite concentrations and the number of predominant bacterial cells was examined by linear regression analysis. The incidences of the individual metabolites in GF mice and Ex-GF mice were compared by Fisher's exact test. StatMate IV (ATMS Co. Ltd., Tokyo, Japan) was used to conduct these analyses.
Author Contributions
Author contribution M.M., S.K., Y.A., Y.K., and Y.B. designed this study. Y.A. performed animal experiments. T.O. analyzed the metabolome. M.M, R.K, T.O., and E.S. analyzed the data. M.M wrote the paper.
Supplementary Material
Supplementary information
Acknowledgments
This study was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), JAPAN. We thank Mr Koji Muramatsu for the preparation of colonic luminal metabolome and bacterial DNA from colonic content.
Footnotes
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), JAPAN. This work was funded by Kyodo Milk Industry Co. Ltd and Human Metabolome Technologies, Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M. Matsumoto and E. Sawaki are employees of Kyodo Milk Industry Co. Ltd. and had a role in study design, data analysis, preparation of the manuscript, and decision to publish the manuscript. T. Ooga is employee of Human Metabolome Technologies, Inc. and had a role in data analysis and decision to publish the manuscript. All of the other authors declare that they have no conflict of interest.
References
- Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G. W. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol 1 Suppl 1, S15–18 (2008). [DOI] [PubMed] [Google Scholar]
- Kirjavainen P. V., Arvola T., Salminen S. J. & Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut 51, 51–55 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Stobberingh E. E., van den Brandt P. A. & Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62, 1223–1236 (2007). [DOI] [PubMed] [Google Scholar]
- Sobhani I. et al. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS One 6, e16393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- Song Y., Liu C. & Finegold S. M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol 70, 6459–6465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon H. A., Bruckner-Kardoss E. & Wostmann B. S. Aging in germ-free mice: life tables and lesions observed at natural death. J Gerontol 21, 380–387 (1966). [DOI] [PubMed] [Google Scholar]
- Suau A. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65, 4799–4807 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14, 169–181 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol 4, 219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S. P. et al. Colonization-induced host-gut microbial metabolic interaction. MBio 2, e00271–00210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen P. B. & Clausen M. R. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl 216, 132–148 (1996). [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Kakizoe K. & Benno Y. Comparison of fecal microbiota and polyamine concentration in adult patients with intractable atopic dermatitis and healthy adults. Microbiol Immunol 51, 37–46 (2007). [DOI] [PubMed] [Google Scholar]
- Matsumoto M. & Benno Y. Consumption of Bifidobacterium lactis LKM512 yogurt reduces gut mutagenicity by increasing gut polyamine contents in healthy adult subjects. Mutat. Res. 568, 147–153 (2004). [DOI] [PubMed] [Google Scholar]
- Raamsdonk L. M. et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol 19, 45–50 (2001). [DOI] [PubMed] [Google Scholar]
- Ohashi Y. et al. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst 4, 135–147 (2008). [DOI] [PubMed] [Google Scholar]
- Wikoff W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106, 3698–3703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A 105, 2117–2122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. P. et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol 3, 112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. P. et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol 4, 157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali B. et al. Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 10, 4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T. et al. Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J Chromatogr A 1159, 125–133 (2007). [DOI] [PubMed] [Google Scholar]
- Monton M. R. & Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J Chromatogr A 1168, 237–246; discussion 236, (2007). [DOI] [PubMed] [Google Scholar]
- Bergen W. G. & Wu G. Intestinal nitrogen recycling and utilization in health and disease. J Nutr 139, 821–825 (2009). [DOI] [PubMed] [Google Scholar]
- Metges C. C. Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr 130, 1857S–1864S (2000). [DOI] [PubMed] [Google Scholar]
- Soga T. et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem 281, 16768–16776 (2006). [DOI] [PubMed] [Google Scholar]
- Mitsuoka T. & Hayakawa K. Die Faekalflora bei Menschen. I. Mitteilung: Die Zusammensetzung der Faekalflora der verschiedenen Altersgruppen. Zbl. Bakt. Hyg. I Abt. Orig. A 223, 333–342 (1972). [PubMed] [Google Scholar]
- Neu J. et al. The dipeptide Arg-Gln inhibits retinal neovascularization in the mouse model of oxygen-induced retinopathy. Invest Ophthalmol Vis Sci 47, 3151–3155 (2006). [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Kurihara S., Kibe R., Ashida H. & Benno Y. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS One 6, e23652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E. & McCann P. P. Polyamine metabolism and function. Am. J. Physiol. 243, C212–C221 (1982). [DOI] [PubMed] [Google Scholar]
- Murphy G. M. Polyamines in the human gut. Eur J Gastroenterol Hepatol 13, 1011–1014 (2001). [DOI] [PubMed] [Google Scholar]
- Loser C., Eisel A., Harms D. & Folsch U. R. Dietary polyamines are essential luminal growth factors for small intestinal and colonic mucosal growth and development. Gut 44, 12–16 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux G. D., Marton L. J. & Baylin S. B. Ornithine decarboxylase is important in intestinal mucosal maturation and recovery from injury in rats. Science 210, 195–198 (1980). [DOI] [PubMed] [Google Scholar]
- Zhang M. et al. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: A counterregulatory mechanism that restrains the immune response. J. Exp. Med. 185, 1759–1768 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. H. & Shankel D. M. Antimutagens against spontaneous and induced reversion of a lacZ frameshift mutation in E. coli K-12 strain ND-160. Mutat. Res. 202, 19–23 (1988). [DOI] [PubMed] [Google Scholar]
- Eisenberg T. et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11, 1305–1314 (2009). [DOI] [PubMed] [Google Scholar]
- Matsumoto M. & Benno Y. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol Immunol 51, 25–35 (2007). [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Sakamoto M. & Benno Y. Dynamics of fecal microbiota in hospitalized elderly fed probiotic LKM512 yogurt. Microbiol Immunol 53, 421–432 (2009). [DOI] [PubMed] [Google Scholar]
- Burrell M., Hanfrey C. C., Murray E. J., Stanley-Wall N. R. & Michael A. J. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem 285, 39224–39238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T. et al. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat Commun 2, 190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozny M. A., Bryant M. P., Holdeman L. V. & Moore W. E. Urease assay and urease-producing species of anaerobes in the bovine rumen and human feces. Appl Environ Microbiol 33, 1097–1104 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch M. H. Bile salts, intestinal microflora and enterohepatic circulation. Dig Liver Dis 34 Suppl 2, S54–57 (2002). [DOI] [PubMed] [Google Scholar]
- Pedron T. & Sansonetti P. Commensals, bacterial pathogens and intestinal inflammation: an intriguing menage a trois. Cell host & microbe 3, 344–347 (2008). [DOI] [PubMed] [Google Scholar]
- Sansonetti P. J. & Medzhitov R. Learning tolerance while fighting ignorance. Cell 138, 416–420 (2009). [DOI] [PubMed] [Google Scholar]
- Guigoz Y., Dore J. & Schiffrin E. J. The inflammatory status of old age can be nurtured from the intestinal environment. Current opinion in clinical nutrition and metabolic care 11, 13–20 (2008). [DOI] [PubMed] [Google Scholar]
- Ooga T. et al. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol Biosyst 7, 1217–1223 (2011). [DOI] [PubMed] [Google Scholar]
- Matsuki T. et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70, 167–173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Sakamoto M. & Benno Y. Dynamics of fecal microbiota in hospitalized elderly fed probiotic LKM512 yogurt. Microbiol Immunol 53, 421–432 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


