Abstract
Synthesis of accurately initiated transcripts has been reconstituted with RNA polymerase II and a set of five transcription factors purified from rat liver. In addition to three previously identified factors α, βγ, and δ (Conaway, R. C, and Conaway, J. W. (1989) Proc. Natl. Acad. Sci. U. S. A. 86, 7356–7360), transcription in the reconstituted liver system requires two novel factors designated τ and ε. These five transcription factors comprise two functional classes: (i) promoter recognition factors (τ and ε), which interact with template DNA to facilitate formation of a stable initial complex that is subsequently recognized and bound by RNA polymerase II, and (ii) RNA chain initiation factors (α, βγ, and δ), which do not participate in formation of the initial complex, but which are essential for transcription initiation.
Initiation of messenger RNA synthesis in mammalian cells is an elaborate process governed by interactions between RNA polymerase II, multiple accessory transcription factors, and promoter DNA (1 – 13). Efforts to elucidate the mechanism of initiation have been hampered by lack of a highly purified, reconstituted transcription system suitable for detailed enzymological studies. In no case has initiation been reconstituted in vitro with homogeneous proteins.
Despite this limitation, a working model of initiation has emerged from analyses of partially purified HeLa cell transcription systems. According to this model, an accessory transcription factor, contained in a fraction designated TFIID (14–17), [DB] (3, 18), or BTF1 (4, 10), first binds directly to the TATA region of promoters to form a stable initial complex. RNA polymerase II, assisted by additional accessory factors, then recognizes and assembles with this nucleoprotein complex to form a functional preinitiation complex that is capable of initiating RNA synthesis upon addition of ribonucleoside triphosphates (4, 10, 14–20).
The mammalian promoter binding transcription factor(s) has been the focus of intense research, not only because it plays a crucial role in initiation but also because it is believed to be a target for regulation by proteins that control gene expression through interactions with upstream promoter elements and enhancers (14, 21–23). Although biochemical studies have begun to shed light on the role this factor plays in initiation, numerous questions have yet to be resolved. It is not known, for example, which promoter sequences are recognized and bound by the promoter binding factor(s) or how variations in promoter structure affect binding. Furthermore, it is not known how the promoter binding factor(s) interacts with RNA polymerase II and the other accessory factors during transcription initiation. Because this mammalian factor(s) has not been completely purified, it has been impossible to establish its structure unequivocally. In fact, quite different sizes have been reported for the partially purified factor from HeLa cells; TFIID is reported to be a protein of 120 to 140 kDa (15), whereas [DB] is reported to be much larger at 17 S (3). In as much as the mammalian promoter binding factor(s) have only been partially purified, it is not clear whether, in mammals, stable binding to promoters is mediated by a single factor or whether it is accomplished by the concerted action of multiple factors.
In some cases, an additional transcription factor, designated either TFIIA (1, 15), [AB] (3, 24, 25), or STF (4, 10, 26), has been reported to stimulate promoter binding by the factor(s) in TFIID, [DB], or BTF1, respectively; however, the nature of this auxiliary factor is presently not clear. Egly et al. (26) purified STF from both calf thymus and HeLa cells. They showed that a 43-kDa polypeptide contained transcription activity and argued that this transcription factor was most likely actin. Samuels and Sharp (24) purified [AB] from calf thymus and found it to be a multisubunit protein composed of 19-kDa and 13-kDa polypeptides, with a native molecular mass of 25 to 35 kDa. Reinberg et al. (15) reported that partially purified preparations of TFIIA from HeLa cells contain a transcription factor with a native molecular mass of approximately 82 kDa, significantly larger than either STF or [AB]. These results are not inconsistent with the possibility that these investigators are describing different transcription factors. In addition, the requirement for TFIIA in transcription initiation is at present controversial. Some studies report that TFIIA is indeed required for transcription (15). Other studies, however, have found that TFIIA/[AB] is not essential, but stimulates initiation from the AdML promoter (25). Recently, it has been reported that the requirement for TFIIA has been variable (17), and that transcription from at least some promoters is not dependent on TFIIA at all (21). Thus, many questions concerning the natures of the transcription factors that mediate promoter recognition by mammalian RNA polymerase II remain to be answered.
In an effort to address these questions, we sought to purify the promoter binding factor(s) from rat liver, which has proven to be an ideal source for the purification of other RNA polymerase II transcription factors (8, 11, 13). We previously described fractionation of rat liver and identification of an enzyme fraction, designated D (8), which is required for initiation and which contains a factor(s) that interact stably with the TATA region of promoters (11). The functional and chromatographic properties of the transcription factors in fractions D from liver and TFIID, [DB], and BTF1 from HeLa cells are similar; in fact, fraction D was found to substitute for fraction TFIID/[DB] in runoff transcription assays.1 We have purified fraction D further and report here that it contains two distinct transcription factors that are both required for formation of the first stable intermediate in assembly of a functional preinitiation complex. The apparent sizes of the two liver factors, which have been designated ε and τ, are remarkably similar to the sizes reported for the HeLa cell transcription factors in fractions TFIID and [DB], respectively.
EXPERIMENTAL PROCEDURES
Materials
Male Sprague-Dawley rats (200–300 g) were purchased from Simonson. Unlabeled ultrapure ribonucleoside 5′-triphosphates were from Pharmacia LKB Biotechnology Inc. [α-32P]CTP at 800 Ci/ mmol was obtained from Du Pont-New England Nuclear. Bovine serum albumin (reagent grade) was from ICN Immunobiologicals. Phenylmethylsulfonyl fluoride (PMSF)2 was obtained from Sigma and was dissolved in dimethyl sulfoxide to 1 M. Antipain and leupeptin were also from Sigma and were dissolved in water to 25 mg/ml.
Chromatography and Buffers
Phosphocellulose (P11) and DEAE-cellulose (DE52) were purchased from Whatman. Carboxymethyl-Sephadex (C-25) was from Pharmacia LKB Biotechnology Inc., and Ultrogel AcA 22 was from IBF Biotechnics. TSK DEAE-NPR was obtained from Hewlett-Packard, and 4000SW Spherogel TSK was from Beckman. TMSD was 10 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 0.25 M sucrose, 0.5 mM DTT, 0.5 mM PMSF. Buffer A was 20 mM Hepes-NaOH, pH 7.9, 1 mM EDTA, 1 mM DTT, 20% (v/v) glycerol, 0.5 mM PMSF. Buffer B was 50 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 1 mM DTT, 20% (v/v) glycerol, 0.5 mM PMSF. Buffer C was 40 mM Tris-HCl, pH 7.9, 0.5 mM EDTA, 1 mM DTT, 10% (v/v) glycerol. Buffer F was 40 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 mM DTT, 10% (v/v) glycerol.
Preparation of the Nuclear Extract
Fifty rats were lightly anesthetized with ether and killed by decapitation. All further steps were carried out at 4 °C. The livers were removed, rinsed in TMSD, minced, suspended in TMSD to a final volume of 1,500 ml, homogenized by two passes through a continuous flow homogenizer (27), and centrifuged at 800 × g for 10 min. The crude nuclear pellet was washed twice more by resuspension in TMSD and centrifugation at 800 × g for 10 min, suspended in TMSD to 2,000 ml, and extracted with 0.33 M (NH4)2SO4. (NH4)2SO4 (0.186 g of (NH4)2SO4/ml) was then added slowly to the supernatant (Fraction I) to 40% of saturation. After addition of 1 μl of 1 N NaOH per g of (NH4)2SO4, the suspension was centrifuged at 12,000 × g for 45 min. The precipitate was resuspended in buffer A containing leupeptin and antipain at 10 μg/ml each and dialyzed against buffer A to a conductivity equivalent to that of 0.2 M KCl in buffer A (Fraction II).
Preparation of Transcription Factor τ
Fraction II was centrifuged at 7,500 × g for 10 min, and the supernatant was mixed with 100 ml of phosphocellulose in a 5-cm diameter column and allowed to sit for 30 min. The slurry was then filtered and washed at 100 ml per h with buffer A containing 0.5 M KCl until the column eluated contained less than 0.1 mg/ml protein. The column was eluted with buffer A containing 1.0 M KCl, and the active fractions were pooled and dialyzed against buffer A to a conductivity equivalent to that of 35 mM (NH4)2SO4 in buffer A (Fraction III). Fraction III was applied to a 3-ml carboxymethyl-Sephadex (C-25) column equilibrated with buffer A containing 35 mM (NH4)2SO4. The column was eluted stepwise with buffer A containing 140 mM (NH4)2SO4, and the active fractions were pooled (Fraction IV). Solid (NH4)2SO4 (0.35 g/ml of fraction) was added slowly to Fraction IV with stirring. After the addition of 1 μl of 1 N NaOH per g of (NH4)2SO4, the suspension was stirred for 30 min more and then centrifuged at 40,000 × g for 45 min. The pellet was dissolved in 200 μl of buffer F and applied to a 4000SW Spherogel TSK HPLC column (7.5 × 600 mm) equilibrated with buffer F containing 0.5 M KCl. The column was eluted at 0.5 ml/min, and 0.5-ml fractions were collected. Active fractions were pooled, dialyzed against buffer A to a conductivity equivalent to that of 0.1 M KCl in buffer A, and stored at −80 °C (Fraction V).
Preparation of RNA Polymerase II and Transcription Factors α, βγ, δ, and ε
Transcription factors α (8), βγ (11), and δ (13) were purified from rat liver as previously described. RNA polymerase II was purified from the nuclear extract (Fraction I) by ammonium sulfate fractionation, followed by chromatography on successive phosphocellulose and DEAE-cellulose columns as previously described (8), except that, in some preparations, protein was adsorbed to phosphocellulose at 0.1 M KCl rather than at 0.2 M KCl. Transcription factor ε co-fractionates with the polymerase through these purification steps. Active fractions from DEAE-cellulose were concentrated by precipitation with 0.35 g of (NH4)2SO4/ml; RNA polymerase II and ε were then resolved by Ultrogel AcA 22 gel filtration on a column (1.5 × 60 cm) equilibrated in buffer A containing 1.0 M KCl. The polymerase was further purified by TSK DEAE-NPR HPLC on a column (4.6 × 35 mm) eluted at 0.6 ml/min with a 9-ml gradient from 0.1 to 0.5 M KCl in buffer C. As judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, RNA polymerase II, which was assayed as described (28), was more than 95% form IIA (29).
Assay of Runoff Transcription
Except where indicated in the figure legends, assays were performed as described (8) with 0.1 μg of NdeI-digested pDN-AdML (30) or pN4 (31), 2 ng of α (Fraction V), 10 ng of βγ (Fraction V), 40 ng of δ (Fraction VI), 80 ng of ε, 60 ng of τ (Fraction V), and 0.01 unit of RNA polymerase II. To limit transcription to one round of initiation per promoter, RNA synthesis was initiated by the addition of 50 μM ATP, 50 μM UTP, 10 μM CTP, 10 μCi of [α-32P]CTP, and 7 mM MgCl2; after 2–3 min, heparin and GTP were added to each reaction mixture to final concentrations of 10 μg/ml and 50 μM, respectively. Runoff transcripts were analyzed by 6% polyacrylamide, 7 M urea gel electrophoresis as described (8). Transcription was quantitated by densitometry of autoradiograms.
Protein Determination
Protein was measured with the protein dye assay (Bio-Rad) according to the manufacturer’s instructions. Bovine serum albumin was the standard.
RESULTS
The Reconstituted Liver Transcription System
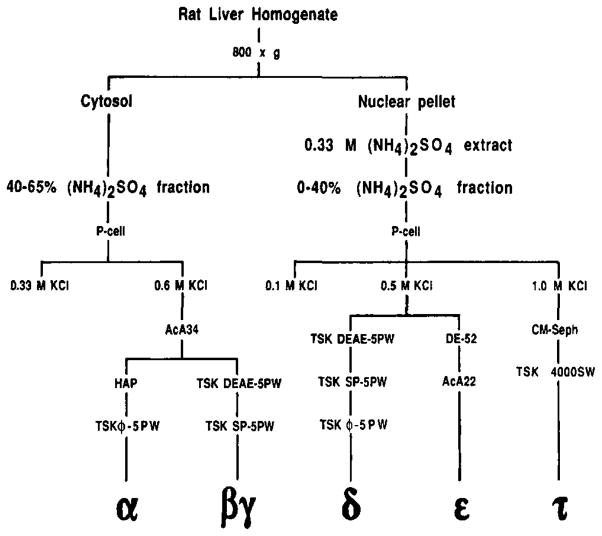
We have fractionated rat liver and identified a set of five transcription factors that are essential for synthesis of accurately initiated transcripts by RNA polymerase II. These transcription factors were initially resolved into nuclear and cytoplasmic protein fractions (Fig. 1). The cytoplasmic fraction was previously shown to contain two transcription factors designated α and βγ (8,11). α is a 35-kDa protein, and βγ is a 250-kDa protein composed of 67-kDa (β) and 31-kDa (γ) polypeptides. The nuclear protein fraction contains three distinct transcription factors designated δ, ε, and τ. δ is a factor of approximately 230 kDa with an associated DNA-dependent ATPase (dA-TPase) activity that is strongly stimulated by the TATA region of promoters (13). In this report, we describe transcription factors τ and ε and show that they play an integral role in promoter recognition by RNA polymerase II.
Fig. 1. Resolution and purification of transcription factors from rat liver.
P-cell, phosphocellulose; CM-Seph, carboxymethyl-Sephadex; HAP, hydroxylapatite; φ, phenyl.
Purification and Properties of Transcription Factors τ and ε
Transcription factors τ and ε were purified by their ability to reconstitute synthesis of a 260-nucleotide runoff transcript from the adenovirus 2 major late (AdML) promoter in the presence of saturating amounts of RNA polymerase II and the remaining transcription factors. Our standard template is pDN-AdML (30), which includes AdML promoter sequences from positions −50 to +10, but which lacks sequences that mediate stimulation of transcription by USF/MLTF (14, 32) from positions −51 to −63. A similar template has been used previously for purification of RNA polymerase II transcription factors (for example, see Refs. 11, 13–15, 19, and 33).
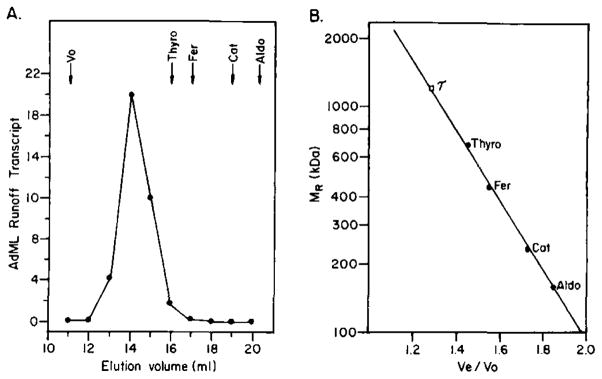
Transcription factor τ was purified more than 3000-fold from a 0.33 M ammonium sulfate extract of crude rat liver nuclei by ammonium sulfate fractionation, followed by chromatography on successive phosphocellulose, carboxymethyl-Sephadex, and TSK 4000SW columns (Table I), τ has a native molecular mass of nearly 1300 kDa when measured by size exclusion HPLC through TSK 4000SW Spherogel in 0.5 M KCl (Fig. 2) or by gel filtration through Ultrogel AcA 22 in 1.0 M KCl (data not shown). It will be important to discover whether transcription factor τ is actually a 1300-kDa protein or whether it has an unusual shape or exists in a complex with other cellular components. Furthermore, it is worth noting that the total yield of activity of τ is reproducibly 5 to 10-fold lower than the total yield of activity of any of the other accessory factors. It is not yet clear whether, in the cell, τ is less abundant than the other factors or whether a significant portion of τ activity is lost during preparation of the nuclear extract.
Table I.
Purification of transcription factor τ from 0.5 kg of rat liver
| Fraction | Protein | Activitya | Specific activity | Yield |
|---|---|---|---|---|
|
| ||||
| mg | units | units/mg | % | |
| I. Nuclear extract | 7,110 | |||
| II. 40% (NH4)2SO4 fraction | 1,876 | 32,000 | 17 | 100b |
| III. Phosphocellulose | 11 | 13,400 | 1,220 | 43 |
| IV. Carboxymethyl-Sephadex | 1 | 7,400 | 7,400 | 23 |
| V. TSK 4000SW | 0.08 | 4,500 | 56,250 | 14 |
One unit is the amount of τ required for half-maximal runoff transcription in the presence of saturating amounts of RNA polymerase II and the remaining transcription factors.
Yield is based on fraction II since activity could not be reliably measured in Fraction I.
Fig. 2. TSK 4000SW size exclusion HPLC of τ.
Runoff transcription reactions with pDN-AdML as template and chromatography were performed as described under “Experimental Procedures.” In this and subsequent figures, AdML Runoff Transcript refers to the relative synthesis, per transcription reaction, of the 260-nucleotide runoff transcript synthesized from the AdML promoter in pDN-AdML (expressed in arbitrary units determined by densitometry of autoradiograms). Standards for size exclusion chromatography were bacteriophage λ DNA (V0); thyroglobulin (Thyro), 670 kDa; ferritin (Fer), 440 kDa; catalase (Cat), 232 kDa; aldolase (Aldo), 158 kDa.
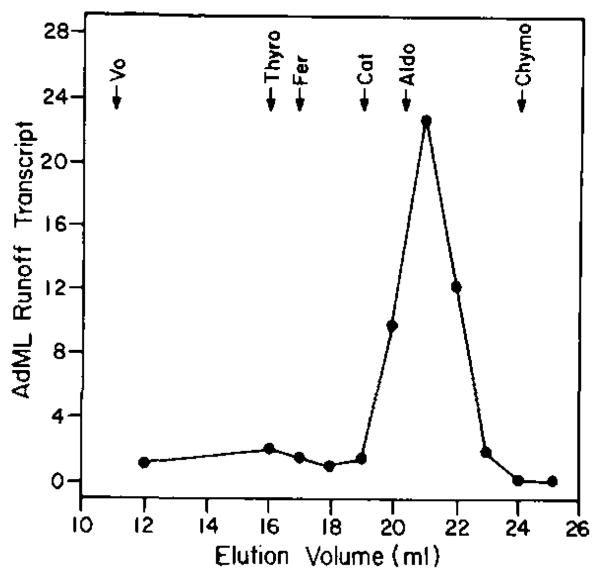
The bulk of transcription factor ε is found in partially purified preparations of RNA polymerase II. ε co-fractionates with RNA polymerase II through chromatography on successive phosphocellulose, DEAE-cellulose, and DEAE-Sephadex columns; ε can be quantitatively separated from the polymerase, however, by Ultrogel AcA 22 gel filtration, ε is also present with transcription factor τ in fraction D (8); the two factors are separated from one another by gel filtration during the purification of τ from fraction D. ε exhibits a native molecular mass of approximately 140 kDa when measured by size exclusion HPLC through TSK 4000SW Spherogel in 0.5 M KCl (Fig. 3).
Fig. 3. TSK 4000SW size exclusion HPLC of ε.
Runoff transcription reactions with pDN-AdML as template and chromatography were performed as described for τ under “Experimental Procedures.” Standards for size exclusion chromatography were bacteriophage λ DNA (V0), thyroglobulin (Thyro), 670 kDa; ferritin (Fer), 440 kDa; catalase (Cat), 232 kDa; aldolase (Aldo), 158 kDa; chymotrypsinogen A (Chymo), 25 kDa.
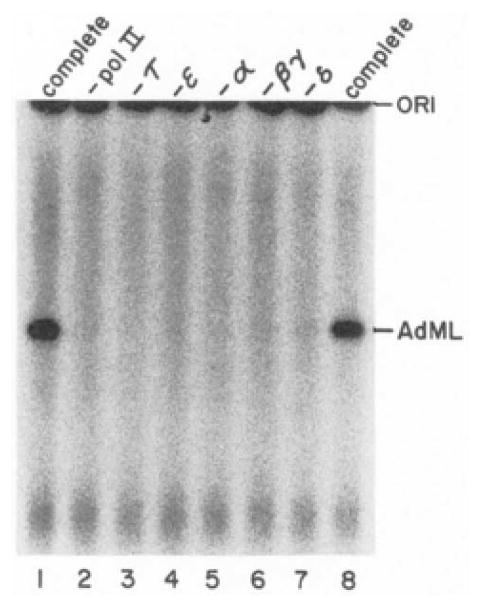
τ and ε, as well as RNA polymerase II and transcription factors α, βγ, and δ, are all required for synthesis of full length runoff transcripts from the AdML promoter (Fig. 4). Thus, both τ and ε perform essential functions in the reconstituted liver transcription system.
Fig. 4. Transcription depends on RNA polymerase II and transcription factors α, βγ, δ, ε; and τ.
Runoff transcription reactions with pDN-AdML as template were performed as described under “Experimental Procedures,” except that RNA polymerase II and transcription factors were omitted from reaction mixtures as indicated, pol II, polymerase II.
τ and ε Act Together at a Distinct Stage in Formation of a Functional Preinitiation Complex
Two observations suggested that τ and ε function in a stage preceding RNA synthesis. First, preincubation of RNA polymerase II and transcription factors with template DNA, prior to addition of ribonucleoside triphosphates, relieved a lag in the synthesis of accurately initiated transcripts from the AdML promoter (data not shown). Transcription was significantly reduced if either τ or ε was omitted from the preincubation. Second, preincubation of RNA polymerase II and transcription factors with template DNA, prior to addition of ribonucleoside triphosphates, rendered synthesis of runoff transcripts from the AdML promoter resistant to inhibition by heparin, which was added to reaction mixtures 2 min after addition of the rihonucleoside triphosphates. When τ and ε were omitted from this preincubation, no heparin-resistant transcription was detected (data not shown). Because heparin inhibits initiation but not elongation of runoff transcripts (10, 15, 19, 26, 34), these results suggested that τ and ε participate in the time-dependent assembly of a functional preinitiation complex prior to RNA synthesis.
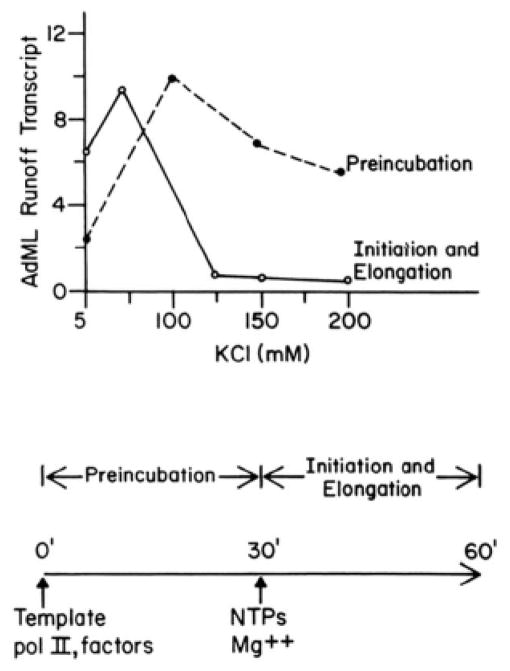
Two distinct stages in formation of the functional preinitiation complex could be resolved by manipulating the ionic strength of reaction mixtures. As shown in Fig. 5, runoff transcription from the AdML promoter is maximal when preincubation of RNA polymerase II, transcription factors, and template DNA is carried out between 100 and 200 mM KCl and when the subsequent stages of RNA chain initiation and elongation are carried out between 60 and 70 mM KCl. Kinetic measurements reveal that the rate-limiting step in formation of the functional preinitiation complex takes place during the high salt preincubation; this step is complete is approximately 20 min at saturating levels of RNA polymerase II, transcription factors, and template DNA (Fig. 6). An additional preincubation at low salt is necessary for maximal transcription; RNA polymerase II and transcription factors must be preincubated for 3 to 4 min in order to render subsequent transcription resistant to inhibition by heparin added 1–2 min after the ribonucleoside triphosphates (data not shown). Adjustment of the ionic strength therefore provides a convenient means of separating the biochemical events that precede transcription initiation into “high salt” and “low salt” stages. These stages resemble the “template committment” and “rapid start complex formation” stages previously defined by Hawley and Roeder (35, 36) on the basis of differential sensitivity to the detergent Sarkosyl.
Fig. 5. Resolution of two stages in runoff transcription.
Runoff transcription reactions with pDN-AdML as template were performed as described under “Experimental Procedures,” except that RNA polymerase II, transcription factors, and template were all added to reaction mixtures together, transcription was initiated by addition of all four ribonucleoside triphosphates, and heparin was not added to the reactions. The concentration of KCl was varied as indicated in the figure. When the concentration of KCl was varied during the Preincubation stage, the concentration of KCl in the Initiation and Elongation stage was held constant at 70 mM (– – –). When the concentration of KCl was varied during the Initiation and Elongation stage, the concentration of KCl in the Preincubation stage was held constant at 150 mM (——). pol II, RNA polymerase II,
Fig. 6. Time course of the high salt preincubation.
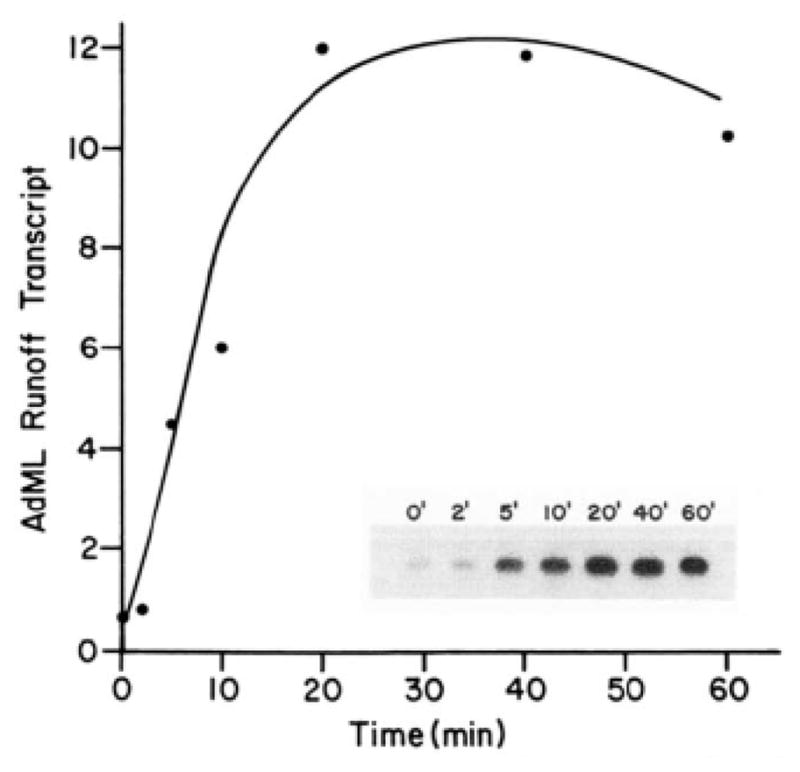
Runoff transcription reactions with pDN-AdML as template were performed as described under “Experimental Procedures,” except that RNA polymerase II and transcription factors were all added at the beginning of the high salt preincubation. The length of the high salt preincubation was varied as indicated.
To determine when τ and ε act during formation of the functional preinitiation complex, order of addition experiments were performed. The results, which are shown in Fig. 7 and Table II, reveal that τ, ε;, and template DNA must all be present in the high salt preincubation for maximal runoff transcription from the AdML promoter. Transcription is stimulated slightly if RNA polymerase II is also included in this preincubation; neither α, βγ, nor δ is required in this stage of transcription. Thus, both τ and ε function in the high salt preincubation stage.
Fig. 7. τ and ε function in the high salt preincubation stage.
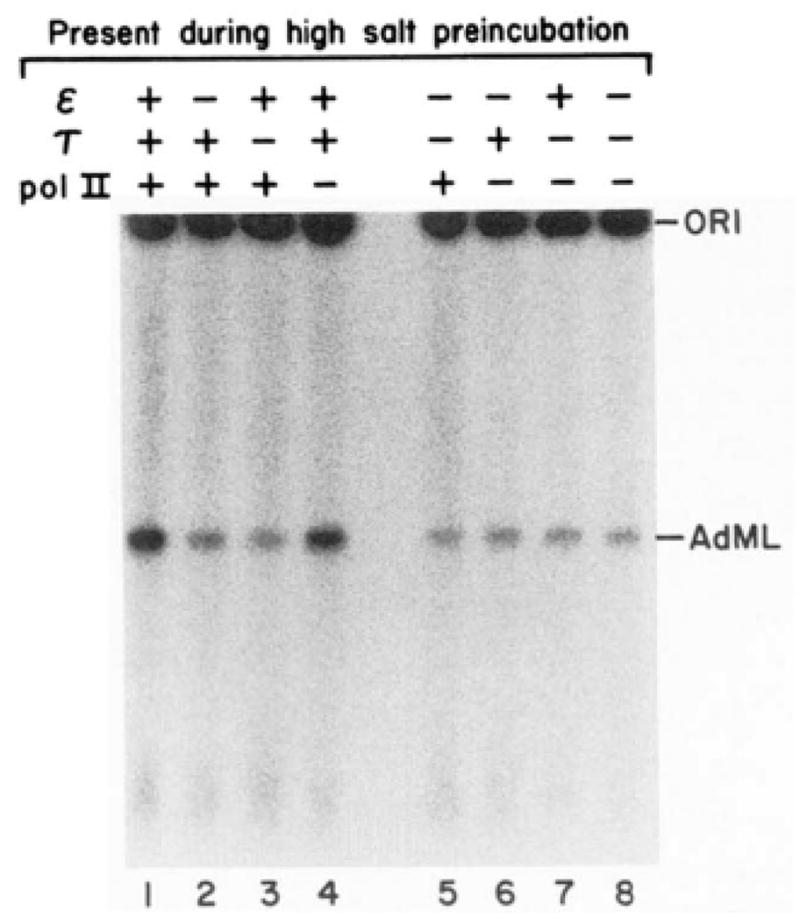
Runoff transcription reactions were performed as described under “Experimental Procedures” and in Table II. pol II, RNA polymerase II.
Table II. Requirements for the high salt preincubation stage.
Runoff transcription reactions were performed as described under “Experimental Procedures” with pDN-AdML as template. The high salt preincubation was carried out for 20 min at 150 mM KCl; the low salt preincubation was carried out for 20 min at 60 mM KCl. Reaction components were added in either the high salt or low salt preincubations as indicated. Transcription was quantitated by densitometry of autoradiograms, pol II, RNA polymerase II; NTPs, ribonucleoside triphosphates.
| Component added
|
Transcription | |
|---|---|---|
| High salt preincubation | Low salt preincubation | |
|
| ||
| % maximum | ||
| τ, ε, δ, α, βγ, pol II, template | None | 100 |
| Template | τ, ε, δ, α, βγ, pol II | 16 |
| τ, ε, δ, α, βγ, pol II | Template | 17 |
| τ, ε, pol II, template | δ, α, βγ | 100 |
| τ, ε, template | δ,α, βγ, pol II | 76 |
| ε, pol II, template | τ, δ, α, βγ | 18 |
| τ, pol II, template | ε, δ, α, βγ | 24 |
| pol II, template | τ, ε, δ, α, βγ | 16 |
| τ, template | ε, δ, α, βγ, pol II | 21 |
| ε, template | τ, δ, α, βγ, pol II | 18 |
| τ, ε, pol II, template, MgCl2, NTPsa | δ, α, βγ | 91 |
MgCl2, was maintained at 7 mM throughout this entire reaction; ATP, CTP, UTP, and GTP were maintained at 10, 10, 50, and 50 μM, respectively.
τ and ε Act Cooperatively to Facilitate Formation of the First Stable Intermediate in Assembly of the Functional Preinitiation Complex
A template challenge assay (4, 18, 35–37) was used to identify the transcription factors needed for formation of the first stable intermediate in assembly of the functional preinitiation complex. Two different templates containing the AdML promoter were used. One template was pN4 (31), linearized with NdeI at a site approximately 340 nucleotides downstream of the cap site. The other template was pDN-AdML (30), linearized with NdeI at a site approximately 260 nucleotides downstream of the cap site. Various combinations of RNA polymerase II and transcription factors were first preincubated for 20 min with either pN4, or pDN-AdML (preincubation 1); to begin preincubation 2, the other template was added to reaction mixtures. After 20 min, transcription was initiated by addition of ribonucleoside triphosphates and magnesium. If a stable complex assembles on the first template during preincubation 1, preferential synthesis of runoff transcripts from the promoter on that template should be observed. On the other hand, if a stable complex fails to assemble on the first template, the transcription factors should distribute equally on the two templates during preincubation 2, and equivalent runoff transcription from both promoters should be observed. As shown in Fig. 8, preferential transcription of pN4 was observed only when both τ and ε were included in preincubation 1. When RNA polymerase II was included with τ and ε in preincubation 1, only a slight stimulation of runoff transcription from pN4, was observed. When either α, βγ, or δ was included with τ and ε in preincubation 1, runoff transcription was unaffected (data not shown). Thus, τ and ε act in concert to facilitate formation of the first stable intermediate, or the “initial complex,” in assembly of the functional preinitiation complex.
Fig. 8. τ and ε are both required for template committment.
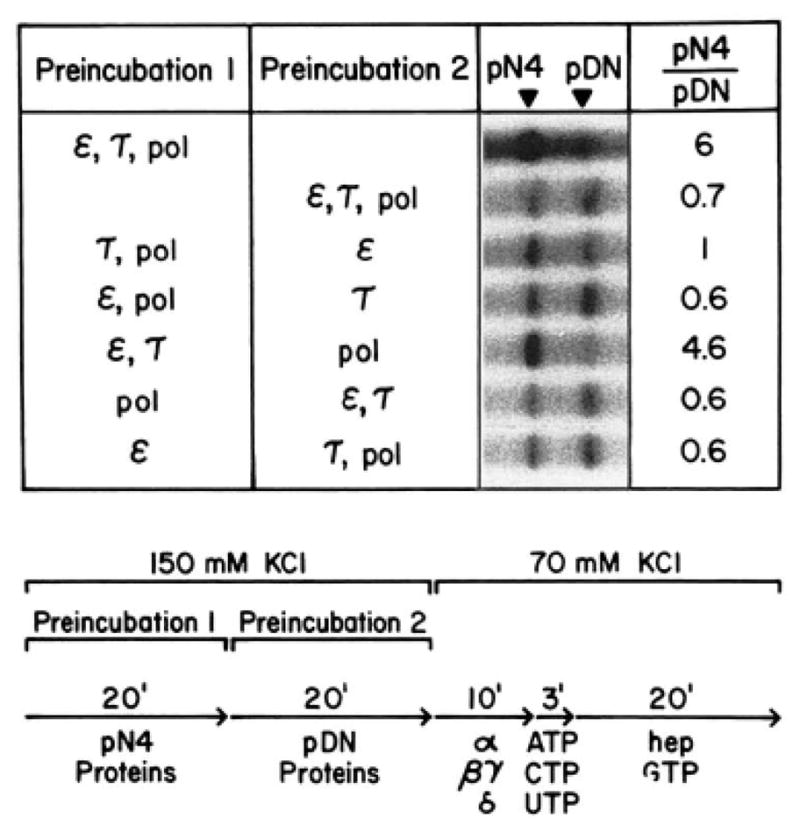
Runoff transcription reactions were performed as described under “Experimental Procedures.” Incubations were as described in the figure, pol, RNA polymerase II; pDN, pDN-AdML; hep, heparin.
RNA Polymerase II Associates Stably with Templates Containing a Preassembled Initial Complex, in the Absence of Transcription Factors α, βγ, and δ
A modification of the template challenge assay was used to investigate the mechanism by which RNA polymerase II interacts with its promoter (see Fig. 9). Initial complexes were preassembled on pN4 in reaction mixture I and on pDN-AdML in reaction mixture II, by preincubating them for 20 min with transcription factors τ and ε (preincubation 1). Various combinations of RNA polymerase II, α, βγ, and δ were then added to reaction mixtures I and II, which were subsequently incubated for 20 min (preincubation 2). Following this incubation, reaction mixtures I and II were combined to begin preincubation 3. After 20 min, transcription was initiated by addition of ribonucleoside triphosphates and magnesium. If RNA polymerase II can stably associate with pN4 during preincubation 2, preferential synthesis of runoff transcripts from the promoter on that template should be observed. On the other hand, if RNA polymerase II fails to associate stably with pN4 it should distribute equally on the two templates during preincubation 3, and equivalent runoff transcription from both promoters should be observed. The results shown in Fig. 9 suggest that RNA polymerase II is capable of associating stably with the initial complex, even in the absence of α, βγ, and δ. Neither α nor δ had a significant effect on the interaction of RNA polymerase II with templates containing an initial complex. Surprisingly, in the absence of α and δ, βγ appeared to antagonize the association of RNA polymerase II with template DNA.
Fig. 9. RNA polymerase II associates stably with DNA templates containing a preassembled initial complex.
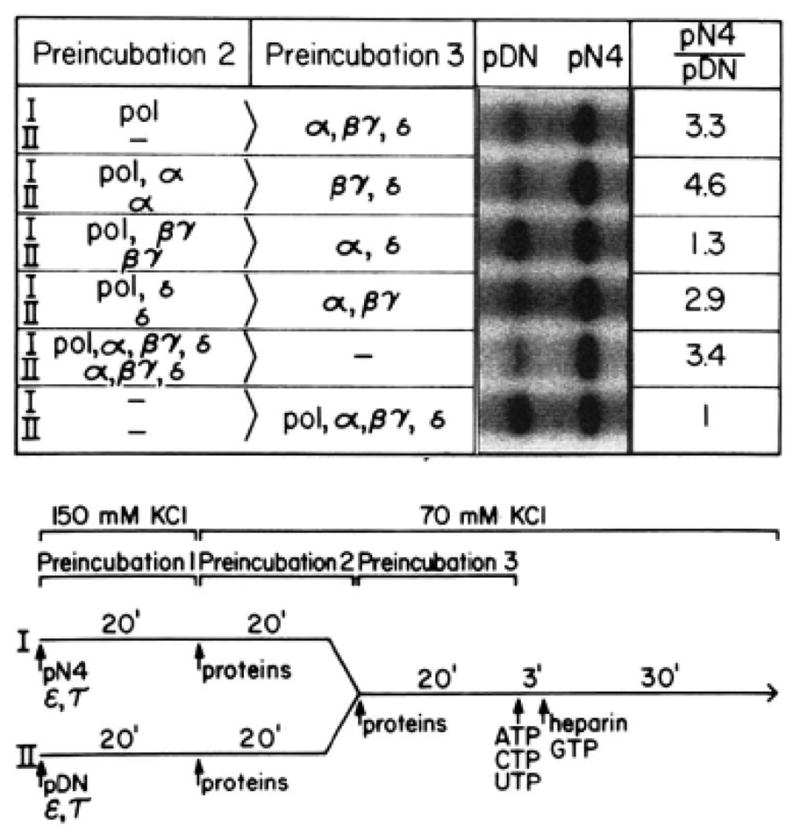
Runoff transcription reactions were performed as described under “Experimental Procedures.” Incubations were as described in the figure, pol, RNA polymerase II; pDN, pDN-AdML.
DISCUSSION
We have used a highly purified transcription system derived from rat liver to investigate the mechanism by which RNA polymerase II recognizes and assembles with its promoter during formation of an active transcription complex. Synthesis of accurately initiated transcripts in the liver system depends on RNA polymerase II and a set of five transcription factors designated α, βγ, δ, ε, and τ. Our findings support the model that transcription factors τ and ε interact with template DNA to facilitate assembly of a stable initial complex that ultimately serves as a recognition site for RNA polymerase II.
Results of template challenge experiments indicate that both τ and ε are essential for formation of the initial complex and that, alone, neither factor interacts stably with template under conditions optimal for initial complex formation. It is not clear, however, whether both τ and ε become stably associated with template or whether one factor merely facilitates binding of the other to the template. Furthermore, the template challenge assays used in this study do not make clear whether τ and ε recognize a specific DNA sequence, such as the promoter, or whether they interact with template in a sequence-independent manner.
Recently, a 25-kDa yeast transcription factor designated either TFIID (38–43) or BTF1 (44, 45), which is reported to substitute for fractions containing TFIID, [DB], and BTF1 in HeLa cell transcription systems, has been purified and cloned. We observe that this factor, which binds to the TATA-box element within the adenovirus 2 major late promoter, will replace τ but not ε in reconstituted transcription reactions in the rat liver system.1 This observation suggests that τ might be capable of interacting with the TATA-box element. In the accompanying paper (46), we present evidence that initial complex formation, which requires both τ and ε, is dependent upon a complex set of promoter sequences including not only the TATA-box element, but also the cap site region and sequences in between.
τ and ε are reminiscent of HeLa cell transcription factors TFIID (14–17), [DB] (3, 18), or BTF1 (4, 10) and TFIIA (1, 15), [AB] (3, 24, 25), or STF (4, 10, 26), respectively; however, whereas τ shares both functional and some chromatographic properties with TFIID and [DB],ε has rather different properties than either TFIIA, [AB], or STF. Like τ, TFIID, [DB], and BTF1 are eluted from phosphocellulose between 0.5 and 1.0 M KCl. More significant, we observe that rat liver fraction D (8), which contains both τ and ε, will substitute for TFIID[DB] in runoff transcription reactions reconstituted with crude fractions from HeLa cells.1 Unlike ε, TFIIA, [AB], and STF all flow through phosphocellulose at 0.1 M KCl. Moreover, we observe that neither ε nor τ can be replaced in runoff transcription reactions by a crude preparation of HeLa cell TFIIA/[AB] (0.1 M KCl phosphocellulose flow through).1 These results argue that ε may be different from either TFIIA, [AB], or STF.
In further studies, we have used a modification of the template challenge assay (4, 18, 37) to investigate the interaction of RNA polymerase II with templates containing a preassembled initial complex. Our results indicate that RNA polymerase II associates stably with these templates, even in the absence of transcription factors α, βγ, and δ. Moreover, we observe that transcription factors α and δ do not significantly affect the binding of RNA polymerase II to template in this stage. In contrast, in the absence of α and δ, transcription factor βγ appears to antagonize the association of polymerase with template.
It is not yet clear whether RNA polymerase II recognizes and interacts specifically with the initial complex in the absence of α, βγ, and δ. Our results do not rule out the possibility that polymerase first binds elsewhere on the template and is subsequently guided to the promoter on that template by one or all of the remaining factors. It is likely, in fact, that RNA polymerase II is not in its final preinitiation configuration at the promoter until all of the transcription factors are present, since α, βγ, and δ are also required for RNA synthesis. In the accompanying paper (34), we discuss these results further in light of two previously proposed models for the binding of RNA polymerase II to the initial complex.
Acknowledgments
We thank Eleanor Travis for expert technical assistance, Monica Coverson and Marilyn Cannefax for preparing the manuscript, and Raquelle Keegan for artwork.
Footnotes
This work was supported in part by Grant GM41628 from the National Institutes of Health.
J. W. Conaway and R. C. Conaway, unpublished observations.
The abbreviations used are: PMSF, phenylmethylsulfonyl fluoride; DTT, dithiothreitol; USF, upstream stimulatory factor; MLTF, major late transcription factor; HPLC, high pressure liquid chromatography; Hepes, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid.
References
- 1.Matsui T, Segall J, Weil PA, Roeder RG. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 2.Tsai SY, Tsai MJ, Kops LE, Minghetti PP, O’Malley BW. J Biol Chem. 1981;256:13055–13059. [PubMed] [Google Scholar]
- 3.Samuels M, Fire A, Sharp PA. J Biol Chem. 1982;257:14419–14427. [PubMed] [Google Scholar]
- 4.Davison BL, Egly JM, Mulvihill ER, Chambon P. Nature. 1983;301:680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- 5.Dynan WS, Tjian R. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- 6.Parker CS, Topol J. Cell. 1984;36:357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- 7.Tolunay HE, Yang L, Safer B, French-Anderson W. Mol Cell Biol. 1984;4:17–22. doi: 10.1128/mcb.4.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conaway JW, Bond MW, Conaway RC. J Biol Chem. 1987;262:8293–8297. [PubMed] [Google Scholar]
- 9.Price DH, Sluder AE, Greenleaf AL. J Biol Chem. 1987;262:3244–3255. [PubMed] [Google Scholar]
- 10.Zheng XM, Moncollin V, Egly JM, Chambon P. Cell. 1987;50:361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]
- 11.Conaway JW, Conaway RC. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 12.Lue NF, Kornberg RD. Proc Natl Acad Sci U S A. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conaway RC, Conaway JW. Proc Natl Acad Sci U S A. 1989;86:7356–7360. doi: 10.1073/pnas.86.19.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawadogo M, Roeder RG. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 15.Reinberg D, Horikoshi M, Roeder RG. J Biol Chem. 1987;262:3322–3330. [PubMed] [Google Scholar]
- 16.Nakajima N, Horikoshi M, Roeder RG. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Dyke MW, Roeder RG, Sawadogo M. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 18.Fire A, Samuels M, Sharp PA. J Biol Chem. 1984;259:2509–2516. [PubMed] [Google Scholar]
- 19.Reinberg D, Roeder RG. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 20.Carthew RW, Samuels M, Sharp PA. J Biol Chem. 1988;263:17128–17135. [PubMed] [Google Scholar]
- 21.Horikoshi M, Hai T, Lin YS, Green MR, Roeder RG. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 22.Hai T, Horikoshi M, Roeder RG, Green MR. Cell. 1988;54:1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- 23.Abmayr SM, Workman JL, Roeder RG. Genes Dev. 1988;2:542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- 24.Samuels M, Sharp PA. J Biol Chem. 1986;261:2003–2013. [PubMed] [Google Scholar]
- 25.Buratowski S, Hahn S, Guarente L, Sharp PA. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 26.Egly JM, Miyamoto NG, Moncollin V, Chambon P. EMBO J. 1984;3:2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler DM, Pettit FH. Biochemistry. 1966;5:2932–2938. doi: 10.1021/bi00873a024. [DOI] [PubMed] [Google Scholar]
- 28.Hodo HG, Blatti SP. Biochemistry. 1977;16:2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 29.Dahmus ME. J Biol Chem. 1983;258:3956–3960. [PubMed] [Google Scholar]
- 30.Conaway RC, Conaway JW. J Biol Chem. 1988;263:2962–2968. [PubMed] [Google Scholar]
- 31.Lorch Y, LaPointe JW, Kornberg RD. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 32.Carthew RW, Chodosh LA, Sharp PA. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- 33.Sawadogo M, Roeder RG. Proc Natl Acad Sci U S A. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conaway RC, Conaway JW. J Biol Chem. 1990;265:7559–7563. [PubMed] [Google Scholar]
- 35.Hawley DK, Roeder RG. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 36.Hawley DK„, Roeder RG. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 37.Kadesch TR, Rosenberg S, Chamberlin MJ. J Mol Biol. 1982;155:1–29. doi: 10.1016/0022-2836(82)90489-2. [DOI] [PubMed] [Google Scholar]
- 38.Buratowski S, Hahn S, Sharp PA, Guarente L. Nature. 1988;334:37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- 39.Hahn S, Buratowski S, Sharp PA, Guarente L. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 40.Horikoshi M, Wang CK, Fujii H, Cromlish JA, Weil PA, Roeder RG. Proc Natl Acad Sci U S A. 1989;86:4843–4847. doi: 10.1073/pnas.86.13.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horikoshi M, Wang CK, Fujii H, Cromlish JA, Weil PA, Roeder RG. Nature. 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 42.Eisenmann DM, Dollard C, Winston F. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt MC, Kao CC, Pei R, Berk AJ. Proc Natl Acad Sci U S A. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavallini B, Huet J, Plassat JL, Sentenac A, Egly JM, Chambon P. Nature. 1988;334:77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- 45.Cavallini B, Faus I, Matthes H, Chipoulet JM, Windsor B, Egly JM, Chambon P. Proc Natl Acad Sci U S A. 1989;86:9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conaway JW, Travis E, Conaway RC. J Biol Chem. 1990;265:7564–7569. [PubMed] [Google Scholar]