Abstract
The CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) modules are adaptive immunity systems that are present in many archaea and bacteria. These defence systems are encoded by operons that have an extraordinarily diverse architecture and a high rate of evolution for both the cas genes and the unique spacer content. Here, we provide an updated analysis of the evolutionary relationships between CRISPR–Cas systems and Cas proteins. Three major types of CRISPR–Cas system are delineated, with a further division into several subtypes and a few chimeric variants. Given the complexity of the genomic architectures and the extremely dynamic evolution of the CRISPR–Cas systems, a unified classification of these systems should be based on multiple criteria. Accordingly, we propose a `polythetic' classification that integrates the phylogenies of the most common cas genes, the sequence and organization of the CRISPR repeats and the architecture of the CRISPR–cas loci.
The CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) modules are adaptive immunity systems that are encoded by most archaea and many bacteria and that act against invading genetic elements1–6, such as viruses and plasmids (Supplementary information S1 (table)). Distinct arrays of short repeats interspersed with unique spacers have been recognized in bacterial and archaeal genomes for years, and although it was proposed that these repeat arrays could have an important common function7, the nature of that function has been elucidated only recently. Independently, Cas proteins that are encoded by putative operons adjacent to CRISPR sequences were analysed in detail with computational methods and found to contain domains that are characteristic of several nucleases, a helicase, a polymerase and various RNA-binding proteins8. It was initially speculated that these proteins constitute a novel DNA repair system9, but the observation that some of the unique CRISPR spacers are almost identical to fragments of virus and plasmid genes led to the hypothesis that CRISPR–Cas systems might be involved in defence against selfish elements10–12. On the basis of these findings and a comprehensive computational re-analysis of the Cas proteins13,14, a model was proposed14 that drew an analogy between the CRISPR–Cas system of archaea and bacteria and the RNA interference (RNAi) mechanisms of eukaryotes15. However, unlike the eukaryotic RNAi systems, the CRISPR–Cas system integrates a small piece of DNA derived from foreign nucleic acid into the CRISPR locus of the host genome as the first step in the series of events that leads to immunity against the invader14. The hypothesis that the CRISPR–Cas system plays a part in defence against invading DNA has been validated by the demonstration that integration of a short phage-specific sequence into the CRISPR locus of the lactic acid bacterium Streptococcus thermophilus conferred resistance to the cognate phage16. In these experiments, resistance to the phage was abrogated by as little as a single mismatch between the CRISPR insert (referred to as the spacer) and the target phage sequence16, although recent studies with archaeal CRISPR–Cas systems revealed a lower stringency of spacer–target complementarity17,18.
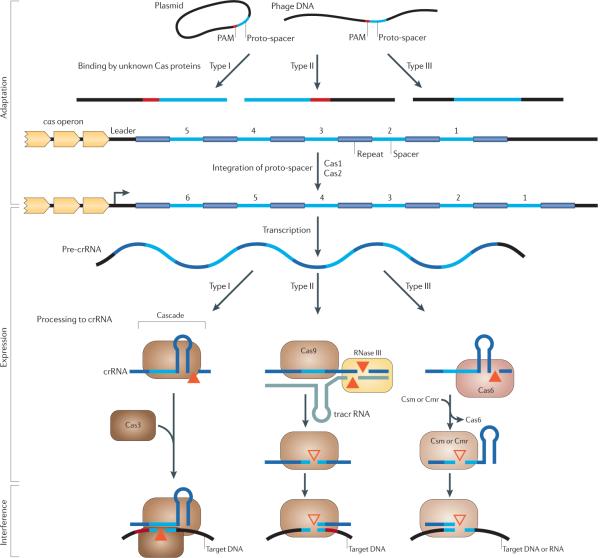
The CRISPR–Cas systems mediate immunity to invading genetic elements via a three-stage process — adaptation, expression and interference (FIG. 1) — that can be divided into two distinct, quasi-independent subsystems: the highly conserved `information processing' subsystem, which includes the adaptation stage, and the `executive' subsystem, which includes the expression and interference stages. Whereas the proteins involved in the information processing subsystem (Cas1 and Cas2) are likely to be highly conserved, the proteins of the executive subsystem vary greatly between different organisms1–3,6,19.
Figure 1. The three stages of CRISPR–Cas action.
CRISPR-Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems act in three stages: adaptation, expression and interference. In type I and type II CRISPR-Cas systems, but not in type III systems, the selection of proto-spacers in invading nucleic acid probably depends on a proto-spacer-adjacent motif (PAM)22,30,31, but how the PAM or the nucleic acid is recognized is still unclear. After the initial recognition step, Cas1 and Cas2 most probably incorporate the proto-spacers into the CRISPR locus to form spacers. During the expression stage, the CRISPR locus containing the spacers is expressed, producing a long primary CRISPR transcript (the precrRNA). The CRISPR-associated complex for antiviral defence (Cascade) complex binds the pre-crRNA, which is then cleaved by the Cas6e or Cas6f subunits (in subtype I-E or I-F, respectively), resulting in crRNAs with a typical 8-nucleotide repeat fragment on the 5′ end and the remainder of the repeat fragment, which generally forms a hairpin structure, on the 3′ flank. Type II systems use a trans-encoded small RNA (tracrRNA) that pairs with the repeat fragment of the pre-crRNA, followed by cleavage within the repeats by the housekeeping RNase III in the presence of Cas9 (formerly known as Csn1 or Csx12). Subsequent maturation might occur by cleavage at a fixed distance within the spacers25, probably catalysed by Cas9. In type III systems, Cas6 is responsible for the processing step, but the crRNAs seem to be transferred to a distinct Cas complex (called Csm in subtype III-A systems and Cmr in subtype III-B systems). In subtype III-B systems, the 3′ end of the crRNA is trimmed further28. During the interference step, the invading nucleic acid is cleaved. In type I systems, the crRNA guides the Cascade complex to targets that contain the complementary DNA, and the Cas3 subunit is probably responsible for cleaving the invading DNA21. The PAM probably also plays an important part in target recognition in type I systems. In type II and type III systems, no Cas3 orthologue is involved (TABLE 2). In type II systems, Cas9 loaded with crRNA probably directly targets invading DNA, in a process that requires the PAM26. The two subtypes of CRISPR–Cas type III systems target either DNA (subtype III-A systems31) or RNA (subtype III-B systems28). In type III systems, a chromosomal CRISPR locus and an invading DNA fragment are distinguished by either base pairing to the 5′ repeat fragment of the mature crRNA (resulting in no interference) or no base pairing (resulting in interference)30. Filled triangles represent experimentally characterized nucleases, and unfilled triangles represent nucleases that have not yet been identified.
During the adaptation stage, short pieces of DNA homologous to virus or plasmid sequences are integrated into the CRISPR loci16,20,21. Viral challenge typically triggers insertion of a single virus-derived resistance-conferring spacer, with a characteristic length of approximately 30 bp, at the leader side of a CRISPR locus; acquisition of multiple spacers from the same phage is less frequent, as are internal insertions. Each integration event is accompanied by the duplication of a repeat and thus creates a new spacer-repeat unit. The selection of spacer precursors (proto-spacers) from the invading DNA appears to be determined by the recognition of proto-spacer-adjacent motifs (PAMs) (FIG. 1); PAMs are usually only several nucleotides long and differ between variants of the CRISPR–Cas system22,23. There is currently no direct evidence for a mechanism of spacer acquisition, although the most highly conserved Cas proteins, Cas1 and Cas2, are the prime candidates for proteins with key roles in this process16,24.
The second stage in CRISPR–Cas-mediated immunity is expression (FIG. 1), during which the long primary transcript of a CRISPR locus (pre-crRNA) is generated and processed into short crRNAs. The processing step is catalysed by endoribonucleases that either operate as a subunit of a larger complex (such as the CRISPR-associated complex for antiviral defence (Cascade) in Escherichia coli) (FIG. 1) or as a single enzyme (such as Cas6 in the archaeon Pyrococcus furiosus). Recently, an intriguing variant was discovered in Streptococcus pyo-genes in which a trans-encoded small RNA (tracrRNA) acts as a guide for the processing of pre-crRNA, which in this organism is catalysed by RNase III in the presence of Csn1 (also known as Cas9; see below)25. In the case of the Cascade complex of type I CRISPR-Cas systems24,26, the mature crRNA remains associated with the complex after the initial endonuclease cleavage (FIG. 1), whereas in P. furiosus the crRNA, processed by Cas6, is passed on to a distinct Cas protein complex (the Cascade complex of type III systems, Cmr-type; see below), where it is processed further at the 3′ end by unknown nucleases27–29.
The third step is interference (FIG. 1), during which the foreign DNA or RNA is targeted and cleaved within the proto-spacer sequence6,20,21. The crRNAs guide the respective complexes of Cas proteins, such as the E. coli Cascade complex, to the complementary virus or plasmid target sequences that match the spacers. In E. coli, the cleavage is probably catalysed by the HD endonuclease domain of the Cas3 protein24. Furthermore, the PAMs seem to play an important part in the interference process23,30. In S. thermo philus and E. coli, targeting either strand of the phage DNA confers immunity to the cognate phage, an observation that is best compatible with DNA being the target16,24,26. Furthermore, insertion of a self-splicing intron into the proto-spacer sequence of the target gene renders the corresponding plasmid resistant to CRISPR-mediated immunity in Staphylococcus epidermidis, indicating that it is the invading DNA rather than the corresponding mRNA that is targeted in this species31. In addition, the hyperthermophilic archaeon Sulfolobus solfataricus targets the DNA of Sulfolobus spindle-shaped virus 1 (SSV1), as CRISPR-mediated immunity does not depend on transcription of the target gene18. However, in vitro experiments with the CRISPR–Cas system from P. furiosus showed that in this species the crRNA targets the foreign mRNA instead28. These findings emphasize the remarkable mechanistic and functional diversity of CRISPR–Cas systems, although the full range of their activities remains to be determined. Various Cas proteins might participate in either one stage or multiple stages of CRISPR–Cas system action, most probably as protein complexes6.
Several Cas proteins have been shown to possess RNase and/or DNase activity, often in agreement with the bioinformatic predictions. This includes the two universal core Cas proteins: Cas1, a metal-dependent DNase that has no sequence specificity and has been proposed to be involved in the integration of the spacer DNA into the CRISPR cassette32, and Cas2, a metal-dependent endoribonuclease for which the role in the CRISPR–Cas mechanism remains unclear33. Repeat-associated mysterious proteins (RAMPs) (see below), which form a large superfamily of Cas proteins, contain at least one RNA recognition motif (RRM; also known as a ferredoxin-fold domain) and a characteristic glycine-rich loop14. Some of the RAMPs have been shown to possess sequence- or structure-specific RNase activity that is involved in the processing of pre-crRNA transcripts24,26,27.
Extensive bioinformatic analyses have shown that the genomes of various CRISPR-containing organisms encode approximately 65 distinct sets of orthologous Cas proteins, which can be classified into 23–45 families, depending on the classification criteria13,14. Furthermore, eight distinct subtypes of the CRISPR–Cas system (CASS1–CASS8) have been delineated on the basis of the composition and architecture of the cas operons and on Cas1 phylogeny13,14.
The diversity of CRISPR–Cas systems identified in newly sequenced genomes is rapidly increasing1,4 — in a representative set of 703 archaeal and bacterial genomes, 310 (44%) encode one or more CRISPR–Cas modules (TABLE 1; Supplementary Information S1 (table)) — hence, an urgent need exists for a unified classification and nomenclature of the cas genes. In this Opinion article, we summarize the shortcomings of the existing classifications and nomenclature of the CRISPR–Cas systems and propose a new, `polythetic' classification that combines information from phylogenetic and comparative genomic analyses.
Table 1.
Taxonomic distribution of three CRISPR-Cas system types
| Taxonomic group | Genomes analyzed | Genomes containing cas1 | Proportion of genomes containing cas1 | Genomes containing a type I system (cas7 and cas3) | Genomes containing a type II system (cas9) | Genomes containing a type III system (cas10) |
|---|---|---|---|---|---|---|
| Archaea | ||||||
| Crenarchaeota | 17 | 15 | 0.88 | 15 | 0 | 16 |
| Euryarchaeota | 47 | 37 | 0.79 | 33 | 0 | 23 |
| All Archaea | 67 | 54 | 0.81 | 50 | 0 | 40 |
| Bacteria | ||||||
| Actinobacteria | 72 | 26 | 0.36 | 28 | 15 | 8 |
| Aquificae | 7 | 5 | 0.71 | 7 | 1 | 4 |
| Bacteroidetes-Chlorobi group | 32 | 16 | 0.50 | 14 | 2 | 6 |
| Chlamydiae-Verrucomicrobia group | 10 | 2 | 0.20 | 0 | 1 | 1 |
| Chloroflexi | 10 | 9 | 0.90 | 9 | 2 | 7 |
| Cyanobacteria | 14 | 7 | 0.50 | 7 | 1 | 7 |
| Firmicutes | 126 | 56 | 0.44 | 40 | 17 | 23 |
| Proteobacteria | 318 | 107 | 0.34 | 117 | 20 | 22 |
| Spirochaetes | 13 | 3 | 0.23 | 2 | 1 | 0 |
| Thermotogae | 11 | 10 | 0.91 | 10 | 0 | 9 |
| All Bacteria | 639 | 256 | 0.40 | 245 | 65 | 99 |
cas, CRISPR-associated protein gene; CRISPR, clustered regularly interspaced short palindromic repeats.
Existing CRISPR–1Cas classification
The original, widely used classification proposed in 2005 by Haft et al. was based on an analysis of 40 bacterial and archaeal genomes, the topology of the Cas1 phylo-genetic tree, and generalized cas operon organizations typified by the CRISPR–Cas systems that are present in eight genomes13. The names of four core cas genes were adopted as originally proposed by Jansen et al. in 2002 (REF. 8). Two other core genes, cas5 and cas6, were then added using the same principle, and names for genes encoding proteins specific to each of the eight CRISPR systems were proposed13. For example, the unique genes found in the E. coli system were denoted cse1 (CRISPR system of E. coli gene number 1), cse2, cse3, cse4 and cas5e (elsewhere, these E. coli genes were also labelled casA, casB, casE, casC and casD, respectively, which added to the confusion)24.
Although the original approach13 offered attractive simplicity, it did not take into account the distant relationships that have been shown to exist between many Cas proteins. For example, the proteins of COG1857 (see the clusters of orthologous groups of proteins (COGs) database34), which are present in the majority of CRISPR–Cas systems and are clearly orthologous14, have been given at least five different names: Cse4, Csd2, Csh2, Cst2 and Csa2 (TABLE 2). Furthermore, the currently used classification does not account for the complexity of the evolutionary relationships between the CRISPR–Cas systems in diverse bacteria and archaea. For example, the Ecoli and Ypest systems (named after E. coli str. K12 sub-str. MG1655 and various strains of Yersina pestis, in which they are the only CRISPR–Cas systems found) are clearly related, as indicated by the similarity of their operon organizations, the absence of cas4 and the phylogenetic clustering of Cas1, whereas the Apern, Tneap–Hmari and Dvulg systems (the only systems found in Aeropyrum pernix, Thermotoga neapolitana DSM 4359 and Haloarcula marismortui str. ATCC 43049, and Desulfovibrio vulgaris str. Hildenborough, respectively) are also related, as they share a common gene of the BH0338 family14. Conversely, extensive recombination within CRISPR–Cas operons has resulted in hybrid systems that cannot be assigned to any of the proposed groups despite the fact that they contain typical cas genes. The linkage between CRISPR–Cas groups and particular organisms can be misleading owing to the presence of multiple CRISPR–Cas systems in the same genome, the presence of different systems in different strains of a single species and the occurrence of hybrid systems.
Table 2.
Classification and nomenclature of CRISPR-associated genes*
| Proposed gene name‡ | System type or subtype | Name from Haft et al.§ | Name from Brouns et al.∥ | Structure of encoded protein (PDB accessions)¶ | Families (and superfamily) of encoded protein#** | Representatives |
|---|---|---|---|---|---|---|
| cas1 | • Type I | cas1 | cas1 | 3GOD,3LFX and 2YZS | COG1518 | SERP2463,SPy1047 and ygbT |
| • Type II | ||||||
| • Type III | ||||||
|
| ||||||
| cas2 | • Type I | cas2 | cas2 | 2IVY,2I8E and 3EXC | COG1343 and COG3512 | SERP2462, SPy1048, SPy1723 (N-terminal domain) and ygbF |
| • Type II | ||||||
| • Type III | ||||||
|
| ||||||
| cas3 ′ | • Type I‡‡ | cas3 | cas3 | NA | COG1203 | APE1232 and ygcB |
|
| ||||||
| cas3 ″ | • Subtype I-A | NA | NA | NA | COG2254 | APE1231 and BH0336 |
| • Subtype I-B | ||||||
|
| ||||||
| cas4 | • Subtype I-A | cas4 and csa1 | NA | NA | COG1468 | APE1239 and BH0340 |
| • Subtype I-B | ||||||
| • Subtype I-C | ||||||
| • Subtype I-D | ||||||
| • Subtype II-B | ||||||
|
| ||||||
| cas5 | • Subtype I-A | cas5a, cas5d, cas5e, cas5h, cas5p, cas5t and cmx5 | casD | 3KG4 | COG1688 (RAMP) | APE1234, BH0337, devS and ygcI |
| • Subtype I-B | ||||||
| • Subtype I-C | ||||||
| • Subtype I-E | ||||||
|
| ||||||
| cas6 | • Subtype I-A | cas6 and cmx6 | NA | 3I4H | COG1583 and COG5551 (RAMP) | PF1131 and slr7014 |
| • Subtype I-B | ||||||
| • Subtype I-D | ||||||
| • Subtype III-A | ||||||
| •Subtype III-B | ||||||
|
| ||||||
| cas6e | • Subtype I-E | cse3 | casE | 1WJ9 | (RAMP) | ygcH |
|
| ||||||
| cas6f | • Subtype I-F | csy4 | NA | 2XLJ | (RAMP) | y1727 |
|
| ||||||
| cas7 | • Subtype I-A | csa2,csd2,cse4, csh2, csp1 and cst2 | casC | NA | COG1857 and COG3649 (RAMP) | devR and ygcJ |
| • Subtype I-B | ||||||
| • Subtype I-C | ||||||
| • Subtype I-E | ||||||
|
| ||||||
| cas8a1 | • Subtype I-A‡‡ | cmx1, cst1, csx8, csx13 and CXXC-CXXC | NA | NA | BH0338-like | LA3191§§and PG2018§§ |
|
| ||||||
| cas8a2 | • Subtype I-A‡‡ | csa4 and csx9 | NA | NA | PH0918 | AF0070,AF1873, MJ0385,PF0637,PH0918 and SSO1401 |
|
| ||||||
| cas8b | • Subtype I-B‡‡ | csh1 and TM1802 | NA | NA | BH0338-like | MTH1090 and TM1802 |
|
| ||||||
| cas8c | • Subtype I-C‡‡ | csd1 and csp2 | NA | NA | BH0338-like | BH0338 |
|
| ||||||
| cas9 | • Type II‡‡ | csn1 and csx12 | NA | NA | COG3513 | FTN_0757 and SPy1046 |
|
| ||||||
| cas10 | • Type III‡‡ | cmr2, csm1 and csx1 1 | NA | NA | COG1353 | MTH326,Rv2823c§§ and TM 1794§§ |
|
| ||||||
| cas10d | • Subtype I-D‡‡ | csc3 | NA | NA | COG1353 | slr7011 |
|
| ||||||
| csy1 | • Subtype I-F‡‡ | csy1 | NA | NA | y1724-like | y1724 |
|
| ||||||
| csy2 | • Subtype I-F | csy2 | NA | NA | (RAMP) | y1725 |
|
| ||||||
| csy3 | • Subtype I-F | csy3 | NA | NA | (RAMP) | y1726 |
|
| ||||||
| cse1 | • Subtype I-E‡‡ | cse1 | casA | NA | YgcL-like | ygcL |
|
| ||||||
| cse2 | • Subtype I-E | cse2 | casB | 2ZCA | YgcK-like | ygcK |
|
| ||||||
| csc1 | • Subtype I-D | csc1 | NA | NA | alr1563-like(RAMP) | alr1563 |
|
| ||||||
| csc2 | • Subtype I-D | csc1 and csc2 | NA | NA | COG1337(RAMP) | slr7012 |
|
| ||||||
| csa5 | • Subtype I-A | csa5 | NA | NA | AF1870 | AF1870,MJ0380,PF0643 and SSO1398 |
|
| ||||||
| csn2 | • Subtype II-A | csn2 | NA | NA | SPy1049-like | SPy1049 |
|
| ||||||
| csm2 | • Subtype III-A‡‡ | csm2 | NA | NA | COG1421 | MTH1081 and SERP2460 |
|
| ||||||
| csm3 | • Subtype III-A | csc2 and csm3 | NA | NA | COG1337(RAMP) | MTH1080 and SERP2459 |
|
| ||||||
| csm4 | • Subtype III-A | csm4 | NA | NA | COG1567(RAMP) | MTH1079 and SERP2458 |
| csm5 | • Subtype III-A | csm5 | NA | NA | COG1332(RAMP) | MTH1078 and SERP2457 |
|
| ||||||
| csm6 | • Subtype III-A | APE2256 and csm6 | NA | 2WTE | COG1517 | APE2256 and SSO1445 |
|
| ||||||
| cmr1 | • Subtype III-B | cmr1 | NA | NA | COG1367(RAMP) | PF1130 |
|
| ||||||
| cmr3 | • Subtype III-B | cmr3 | NA | NA | COG1769(RAMP) | PF1128 |
|
| ||||||
| cmr4 | • Subtype III-B | cmr4 | NA | NA | COG1336(RAMP) | PF1126 |
|
| ||||||
| cmr5 | • Subtype III-B‡‡ | cmr5 | NA | 2ZOP and 2OEB | COG3337 | MTH324 and PF1125 |
|
| ||||||
| cmr6 | • Subtype III-B | cmr6 | NA | NA | COG1604(RAMP) | PF1124 |
|
| ||||||
| csb1 | • Subtype I-U | GSU0053 | NA | NA | (RAMP) | Balac_1306 and GSU0053 |
|
| ||||||
| csb2 | • Subtype I-U§§ | NA | NA | NA | (RAMP) | Balac 1305 and GSU0054 |
|
| ||||||
| csb3 | • Subtype I-U | NA | NA | NA | (RAMP) | Balac_1303# |
|
| ||||||
| csx17 | • Subtype I-U | NA | NA | NA | NA | Btus_2683 |
|
| ||||||
| csx14 | • Subtype I-U | NA | NA | NA | NA | GSU0052 |
|
| ||||||
| csx10 | • Subtype I-U | csx10 | NA | NA | (RAMP) | Caur_2274 |
|
| ||||||
| csx16 | • Subtype III-U | VVA1 548 | NA | NA | NA | VVA1 548 |
|
| ||||||
| csaX | • Subtype III-U | csaX | NA | NA | NA | SSO1438 |
|
| ||||||
| csx3 | • Subtype III-U | csx3 | NA | NA | NA | AF1864 |
|
| ||||||
| csx1 | • Subtype III-U | csa3,csx1,csx2,DXTHG, NE0113 and TIGR02710 | NA | 1XMX and 2I71 | COG1517 and COG4006 | MJ1666, NE0113, PF1127 and TM1812 |
|
| ||||||
| csx15 | • Unknown | NA | NA | NA | TTE2665 | TTE2665 |
|
| ||||||
| csf1 | • Type U | csf1 | NA | NA | NA | AFE_1038 |
|
| ||||||
| csf2 | • Type U | csf2 | NA | NA | (RAMP) | AFE_1039 |
|
| ||||||
| csf3 | • Type U | csf3 | NA | NA | (RAMP) | AFE_1040 |
|
| ||||||
| csf4 | • Type U | csf4 | NA | NA | NA | AFE_1037 |
N, amino; NA, not applicable; RAMP, repeat-associated mysterious protein.
Includes the names of all genes that have been shown to function within the CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) systems and/or are associated with CRISPR–cas loci in diverse genomes. Genes that are associated with CRISPR–cas loci in only one or a few closely related genomes are not included. Subsequent to their original publication13, Haft et al. introduced a number of new types of CRISPR-Cas systems as well as gene names that are included in the TIGRFAMs database50 but mostly fit into previously described gene and protein families.
The updated TIGRFAMs identifiers are given in Supplementary information S4 (table). The csx names are temporarily given to cas genes that cannot be confidently included in any of the large cas families but are currently not characterized in sufficient detail to rule out the possibility of such assignments in the future. Beginning with release 10.1 (ftp://ftp.jcvi.org/pub/data/TIGRFAMs/), the hidden Markov model (HMM)-based classifiers in TIGRFAMs assign polythetic names reflecting the nomenclature changes described here while retaining the narrower protein family granularities of the original nomenclature13.
See REF. 13. Most of the families correspond to those proposed by Makarova et al.14, with a few changes and additions.
See REF. 24.
All available structures are listed; see the Protein Data Bank (PDB).
Tentative predictions based on weak sequence similarity, sequence length and gene order in an operon.
See the clusters of orthologous groups of proteins (COGs) database.
These are signature genes for these CRISPR-Cas system types and subtypes.
Unclassified.
The inconsistencies between the nomenclature of the CRISPR–Cas systems and the names of Cas proteins are rapidly growing. In particular, many of these proteins are currently classified into families that do not have systematic names pointing to their involvement with a CRISPR–Cas system (such as the BH0338 family, the CXXC-CXXC family and the GSU0053 family, among many others).
Taken together, these problems substantially complicate the use of the current classification and nomenclature of CRISPR–Cas systems and motivate the effort behind the creation of a new, unifying, internally consistent and flexible classification scheme.
A new CRISPR–Cas classification
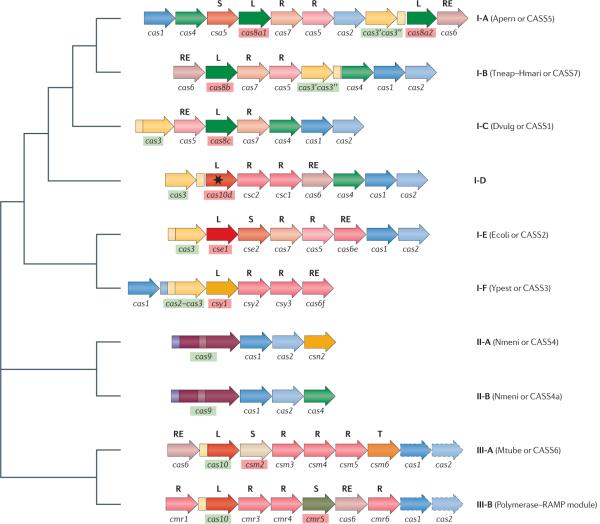
Here, we propose a new, polythetic classification of CRISPR–Cas systems in which the cas1 and cas2 genes constitute the core of three distinct types of system (FIG. 2; TABLE 2). Cas1 and Cas2 are present in all CRISPR–Cas systems that are predicted to be active, and are thought to be the information-processing subsystem that is involved in spacer integration during the adaptation stage.
Figure 2. The relationship of the three major types and ten sub-types of CRISPR systems.
The typical, simplest operon architectures are shown for each type and subtype of CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) system; numerous variations exist. Orthologous genes are colour coded and identified by a family name, as given in TABLE 2. The signature genes for CRISPR–Cas types are shown within green boxes, and those for sub-types are shown within red boxes. The letters above the genes show major categories of Cas proteins: large CRISPR-associated complex for antiviral defence (Cascade) subunits (L), small Cascade subunits (S), repeat-associated mysterious protein (RAMP) Cascade subunits (R), RAMP family RNases involved in crRNA processing (RE) (note that only those in subtypes I-E, I-F and III-B systems have been characterized), and transcriptional regulators (T). The star indicates a predicted inactivated polymerase with an HD domain. For subtype I-A systems, the cas8a1 and cas8a2 genes are typically mutually exclusive but both can be considered signature genes for the subtype. For type III systems, the cas1 and cas2 genes in dashed boxes are not associated with all type III polymerase–RAMP modules. In addition to previously published data, this schematic shows Cas7 (COG1857) as a member of the RAMP superfamily. For each CRISPR–Cas subtype (except for the newly identified subtype I-D), the old names from REFS 13,14 are indicated in parentheses. Figure is modified, with permission, from REF. 14 © (2006) BioMed Central.
Type I CRISPR–Cas systems
Typical type I loci contain the cas3 gene, which encodes a large protein with separate helicase and DNase activities35, in addition to genes encoding proteins that probably form Cascade-like complexes with different compositions24,26. These complexes contain numerous proteins that have been included in the RAMP superfamily, which encompasses the large Cas5 and Cas6 families, on the basis of extensive sequence and structure comparisons14 (see TABLE 2 for the available structures). Furthermore, the Cas7 (COG1857) proteins represent another distinct, large family within the RAMP superfamily, as detected by the HHPred method, which can detect distant sequence and structure similarities between proteins36 (Supplementary Information S2 (figure)). In addition, the complexes involved in the CRISPR–Cas function may contain large proteins such as Cse1 and BH0338-like families, as well as small α-helical proteins such as Cse2, or other, less conserved subunits.
In the Cascade complex, a RAMP protein with RNA endonuclease activity has been identified as the main enzyme that catalyses the processing of the long spacer–repeat-containing transcript into a mature crRNA24,26. In most cases, the catalytic RAMP proteins (Cas6, Cas6e and Cas6f; see TABLE 2) do not belong to the most prevalent Cas5 or Cas7 families of RAMPs and are often encoded in the periphery of the respective operon. However, the subtype I-C system (also known as Dvulg or CASS1) (FIG. 2; TABLE 2) might be an exception in which either Cas5 or Cas7 possesses RNase activity. The type I CRISPR–Cas systems seem to target DNA; target cleavage is catalysed by the HD nuclease domains of Cas3 (REF. 35). As the RecB nuclease domain of Cas4 is fused to Cas1 in several type I CRISPR–Cas systems, Cas4 could potentially play a part in spacer acquisition instead.
Type II CRISPR–Cas systems
The type II systems include the `HNH'-type system (Streptococcus-like; also known as the Nmeni subtype, for Neisseria meningitidis serogroup A str. Z2491, or CASS4), in which Cas9, a single, very large protein, seems to be sufficient for generating crRNA and cleaving the target DNA, in addition to the ubiquitous Cas1 and Cas2. Cas9 contains at least two nuclease domains, a RuvC-like nuclease domain near the amino terminus and the HNH (or McrA-like) nuclease domain in the middle of the protein, but the function of these domains remains to be elucidated. However, as the HNH nuclease domain is abundant in restriction enzymes and possesses endonuclease activity37,38, it is likely to be responsible for target cleavage. Furthermore, for the S. thermophilus type II CRISPR–Cas system, targeting of plasmid and phage DNA has been demonstrated in vivo20 and inactivation of Cas9 has been shown to abolish interference16.
Type II systems cleave the pre-crRNA through an unusual mechanism that involves duplex formation between a tracrRNA and part of the repeat in the pre-crRNA; the first cleavage in the pre-crRNA processing pathway subsequently occurs in this repeat region. This cleavage is catalysed by the housekeeping, double-stranded RNA-specific RNase III in the presence of Cas925.
Type III CRISPR–Cas systems
The type III CRISPR–Cas systems contain polymerase and RAMP modules in which at least some of the RAMPs seem to be involved in the processing of the spacer–repeat transcripts, analogous to the Cascade complex. Type III systems can be further divided into sub-types III-A (also known as Mtube or CASS6) and III-B (also known as the polymerase–RAMP module). Subtype III-A systems can target plasmids, as has been demonstrated in vivo for S. epidermidis31, and it seems plausible that the HD domain of the polymerase-like protein encoded in this subtype (COG1353) might be involved in the cleavage of target DNA. There is strong evidence that, at least in vitro, the type III-B CRISPR–Cas systems can target RNA, as shown with a subtype III-B system from P. furiosus28. It is intriguing that these two type III systems seem to target different nucleic acids, and this finding will require further study.
The only identified ribonucleases in the type III CRISPR–Cas systems, apart from the universal Cas2 protein, are RAMP proteins. Type III systems include at least two RAMPs in addition to Cas6, which is involved in CRISPR transcript processing. In many organisms, type III CRISPR–cas operons lack the cas1–cas2 gene pair; in all these cases, an additional CRISPR locus (of either type I or type II) is also present in the respective genome, indicating that Cas1 and Cas2 are probably provided in trans. In other organisms, the polymerase–RAMP modules are present in a single operon with cas1 and cas2, forming a module with the typical architecture in S. epidermidis and Mycobacterium tuberculosis (a type III-A module) and forming a distinct version in Halorhodospira halophila (a type III-B module). In these organisms, the type III operon is the only CRISPR–cas locus, suggesting that the polymerase–RAMP module forms a fully functional, autonomous type III system when combined with Cas1 and Cas2, which are likely to be involved in the incorporation of new spacers.
Unclassified CRISPR–Cas systems
Most of the CRISPR–cas loci can be readily classified into the proposed three types and their subtypes according to the presence of type-specific and subtype-specific signature genes (TABLE 2). However, for the loci that cannot be classified even at the type level, such as the CRISPR–Cas system in Acidithiobacillus ferrooxidans str. ATCC 23270 (discussed further below), we propose the name type U.
Distribution of the three types of CRISPR–Cas systems in the Archaea and the Bacteria
The three types of CRISPR systems show a distinctly non-uniform distribution among the major lineages of the Archaea and the Bacteria (TABLE 1). In particular, the type II systems have been found exclusively in the Bacteria so far, whereas type III systems are more common in the Archaea. The previously observed trend of over-representation of CRISPR in the Archaea compared to the Bacteria still holds14,39 (TABLE 1). Moreover, the majority of archaeal genomes carry more than one CRISPR–Cas system; typically, different modules within the same genome are unrelated.
CRISPR–Cas subtypes and their evolution
On the basis of the gene composition and architecture of the respective cas operons, the three basic types of CRISPR–Cas system can be further classified into subtypes that largely agree with the previously delineated variants13,14. Each of the subtypes contains a signature gene or genes that are represented almost exclusively in the given subtype and can be used to identify the subtype (FIG. 2; TABLE 2). To facilitate classification, a single signature gene was chosen for each subtype: in cases with several candidates, the longest gene was selected, as longer genes are typically more easily detectable in sequence searches than shorter genes. In addition, we introduce subtypes I-U, II-U and III-U for systems that lack currently defined subtype-specific signature genes but either might fit one of the established subtypes on the basis of further structure and sequence analysis, or potentially could become founders of new subtypes.
The ubiquitous, highly conserved Cas1 protein can be used as a scaffold to investigate the evolution of the CRISPR–Cas system (the other universal protein, Cas2, is too small to yield a well resolved tree). The phylogenetic tree of Cas1 includes several well-resolved branches that generally agree with the classification of CRISPR–Cas systems into subtypes I-A (Apern or CASS5), I-B (Tneap–Hmari or CASS7), I-C (Dvulg or CASS1), I-E (Ecoli or CASS2), I-F (Ypest or CASS3) and III-A (Mtube or CASS6), and type II (Nmeni or CASS4)14, with a few notable exceptions (FIG. 3; see Supplementary information S3 (box) for data to construct the complete tree). In particular, Cas1 proteins associated with the polymerase–RAMP module (the type III systems) appear in several unrelated positions in the tree (FIG. 3), suggesting that this module can operate with a variety of cas1 and cas2 genes both in cis and in trans.
Figure 3. Phylogenetic tree for Cas1 (COG1518) proteins.
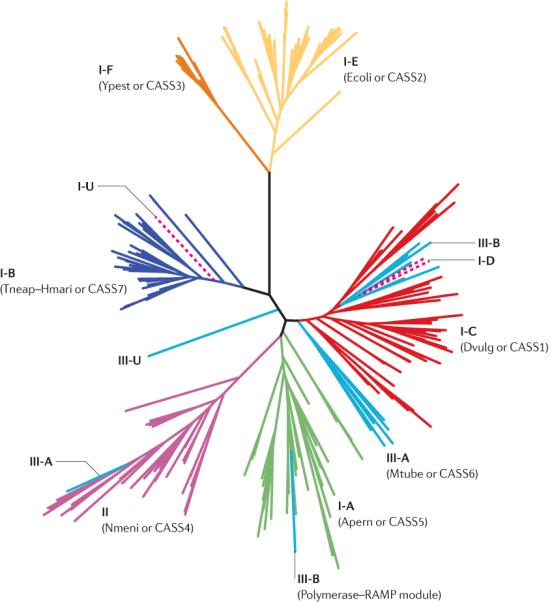
The BLASTCLUST program was used to cluster the sequences of CRISPR (clustered regularly interspaced short palindromic repeats)-associated protein 1 (Cas1) by similarity (parameters: the sequence length to be covered was 75%, and the score identity threshold was 0.9), and one representative from each cluster was chosen (see the list in Supplementary information S4 (table)). Six major subtypes of type I CRISPR–Cas system (I-A to I-F), as well as type II and type III systems, are colour coded. Dashed lines show cas1 genes that are found in `hybrid' CRISPR loci containing genes from both type I and type III CRISPR–Cas systems (see main text for details). Subtypes I-U and III-U (U for unclassified) denote CRISPR–Cas systems that lack currently defined subtype-specific signature genes (see main text for details). The maximum likelihood tree was constructed using the PHYML program46, from 182 informative positions in the multiple alignment of a representative set of 228 Cas1 proteins from 442 complete genomes (those that encode Cas1 from the set of 703 genomes listed in Supplementary information S1 (table)). For each CRISPRCas subtype (except for the newly identified subtype I-D), the old names from REFS 13,14 are indicated in parentheses.
The CRISPR repeats can be classified into at least 12 groups on the basis of sequence similarity40. Four groups of CRISPR repeats clearly correspond to distinct CRISPR–Cas subtypes: group 2 corresponds to sub-type I-E systems, group 3 corresponds to subtype I-C systems, group 4 corresponds to subtype I-F systems and group 10 corresponds to type II systems. These four variants of CRISPR–Cas systems have the most stable operon organizations; by contrast, subtypes I-A, I-B and I-D and type III systems seem to be prone to recombination between different types and subtypes. Structural characteristics of the CRISPR repeats of these four groups could potentially be used for classification, in addition to phylogenetic data and signature genes. The other eight groups of repeats cannot be unequivocally associated with particular CRISPR–Cas system subtypes.
Integration of all the above considerations into a dendrogram reflects our present understanding of the evolutionary history of CRISPR–Cas systems (FIG. 2). Subtypes of the type I system are grouped according to their operon organizations and the phylogeny of the respective Cas1 proteins.
A new CRISPR–Cas nomenclature
We propose to retain the well-established names for core genes of the CRISPR–Cas systems: the ubiquitous cas1 and cas2 (found in all three types), cas3 (type I), cas4 (types I and II), cas5 (type I) and cas6 (types I and III). In the cases for which orthology can be confidently traced, we extend the usage of these six cas gene names; for example, cmx5 of subtype I-C is renamed cas5, and cmx6 is renamed cas6. In cases for which significant sequence similarity between Cas proteins is observed but orthologous relationships cannot be definitively assigned, a letter derived from the subtype label is added; hence, cse3 and csy4 in the former nomenclature become cas6e and cas6f, respectively, as they are likely to be extremely divergent derivatives of cas6 (TABLE 2).
In type I systems, there are two additional genes for which orthology is readily detectable between different subtypes. We refer to these genes as cas7 and cas8 (which can be further divided into cas8a, cas8b and cas8c); both encode subunits of the Cascade complex (TABLE 2). The cas8a, cas8b and cas8c genes are the signature genes for subtypes I-A, I-B and I-C, respectively. In type II and type III systems, the respective signature genes are designated cas9, and cas10 (formerly cmr2, csm1 and csx11).
When a gene is clearly a fusion or fission of established genes, we propose an ad hoc nomenclature indicating the relationship of this variant to the `canonical' forms. Thus, cas2–cas3 in subtype I-F systems is a fusion of cas2 and cas3, whereas cas3′ and cas3″ denote the genes that encode only the helicase domain or only the HD domain of Cas3, respectively.
For less common genes that have been named previously13, the `legacy' nomenclature can be retained. As the Cas protein sequences are highly diverged, it is expected that, with the increasing representation of sequences and structures, many of these genes will eventually be incorporated into existing families. We propose to continue assigning further `numerical' names to newly merged cas gene families in the future (such as cas11, cas12, and so on).
For the remaining CRISPR-associated genes, we propose to assign interim gene names (csx1, in which `x' indicates an unclassified family), with an indication of the family or superfamily where known (such as csx1, COG1517 family, or csx10, RAMP superfamily).
Outstanding problems
Subtype assignment
The phylogenetic tree of Cas1 reproduces most of the previously established groups fairly well, with the exception of the type III systems (FIG. 3). However, for the deep branches, assigning a subtype can be problematic. In many cases, detailed analysis of the gene orders reveals a more complicated picture with different arrangements of cas genes in the operons, potentially owing to frequent horizontal gene transfer and recombination involving the CRISPR–cas loci. In particular, a notable recombinant CRISPR–Cas system is present in approximately 30 archaeal and bacterial genomes, including cyanobacteria (such as the region spanning the loci slr7010–ssr7072 in Synechocystis sp. PCC 6803). In this CRISPR–Cas system, the type I-C system has combined with a distinct type III gene arrangement encoding the polymerase–RAMP module, containing cas3, cas10 (which is predicted to be an inactivated polymerase with an HD domain), csc2 (from the COG1337 family, and the RAMP super-family), csc1 (from the RAMP superfamily), cas6, cas4, cas1 and cas2. This hybrid system containing signature genes for both type I and type III systems is represented in approximately 30 archaeal and bacterial genomes. As this system is likely to be functional, we have classified it as subtype I-D (FIG. 2).
Another interesting CRISPR–Cas system, typified by A. ferrooxidans str. ATCC 23270 (loci AFE_1037-AFE_1040), has been detected in only four genomes to date. This CRISPR–cas locus seems to possess a distinct gene content and could potentially contribute to our understanding of the functions and evolution of CRISPR–Cas systems in general. This system contains neither of the two ubiquitous core genes (cas1 or cas2) nor any other signature genes of the three CRISPR–Cas types or the ten subtypes. The A. ferrooxidans system consists of four genes denoted csf1, csf2, csf3 and csf4 (TIGRFAMs entries TIGR03114, TIGR03115, TIGR03116 and TIGR03117, respectively), which encode a Zn-finger domain-containing protein, a protein containing two RAMP domains, another distinct RAMP protein and a DinG-like helicase of the XPD family, respectively39. According to the CRISPRdb database41, a CRISPR array is present in the vicinity of these four genes in all of the respective genomes, although the architecture of these arrays is unique in each genome. Thus, this system might function in conjunction with different CRISPR arrays and does not require a distinct repeat signature. Indeed, three of the four genomes containing this system possess cas1 and cas2 genes that are located in other parts of the genome and are associated with type I CRISPR–Cas systems. It remains unclear whether this is a self-sufficient system or rather a defective system that captures and utilizes pre-existing CRISPR arrays that are generated by other, Cas1-containing CRISPR–Cas systems. More data are needed to classify this novel system as a separate CRISPR–Cas type, but this finding illustrates the diversity of CRISPR–Cas systems and the challenges that are associated with their classification.
Gene name assignments
Many cas genes, in particular genes that encode RAMP proteins, seem to evolve at exceptionally high rates. CRISPR–Cas systems can contain genes that encode highly divergent proteins which may not fall into a known Cas protein family after the structure is solved. For such genes and proteins, family assignment is extremely complicated. For example, a CRISPR system very similar to subtype I-F, as determined by Cas1 similarity, is present in Photobacterium profundum and several other bacteria. This system includes two proteins, PBPRB1992 and PBPRB1993, that show no significant sequence similarity to any Cas proteins. However, analyses of the sequence motifs that are conserved in these proteins, the predicted secondary structure of the proteins, and the length and position of the corresponding genes in the operon strongly suggest that they belong to the Cas7 and Cas5 families of RAMPs, respectively. Another example is the CRISPR–Cas system of Geobacter sulfurreducens: according to the phylogeny of Cas1, this system should be assigned to subtype I-C. The operon for this system encodes three uncharacterized proteins, GSU0052, GSU0053 and GSU0054; the last two of these proteins contain several motifs that are similar to the characteristic motifs of the RAMP superfamily and thus might be RAMP homologues (TABLE 2). However, none of these proteins could be linked to known Cas families, even using the most sensitive of the available methods for the detection of remote sequence similarity36,42,43. Therefore, only a comparison of the solved structures might shed light on the relationships of these and other highly diverged Cas proteins with known Cas families. In such cases, assignment of new gene names seems to be premature because these proteins are likely to eventually assume already existing names. Therefore, it is proposed that these genes are given temporary csx names.
Many CRISPR–cas loci belong to `islands' that contain various `high-mobility' genes such as toxins–antitoxins, transposases and components of other defence systems44. Some of these genes can be erroneously linked to CRISPR–Cas systems, so caution should be exercised in the classification and naming of genes as cas or even csx before functional connections with CRISPR–Cas systems are convincingly established.
An additional challenge to the nomenclature is presented by the variable domain architectures of some of the Cas proteins, including the domain fusions and fissions discussed above for Cas3. Other notable fusions include the fusion of cas2 and cas3 (in subtype I-F systems), of cas1 and cas4 (such as is found in GSU0057 from G. sulfurreducens), of cas1 and a DEDDh family exonuclease (for example, LBUL_0800 from Lactobacillus delbrueckii subsp. bulgaricus) and of cas1 and a reverse transcriptase (for example, VVA1544 from Vibrio vulnificus).
In several genomes, homologues of some cas genes also appear in contexts other than CRISPR–Cas systems. These proteins might represent distinct antivirus defence systems or components thereof, or they could be involved in other functions such as DNA repair. The latter possibility is emphasized by the recent demonstration that cas1 mutants of E. coli have DNA repair-deficient phenotypes45. Homologues of Cas proteins that probably function in processes other than adaptive immunity include RAMPs of the COG5551 sub-family and the COG1517, COG1468 and COG3513 families. In cases such as these, classification and labelling of the genes as cas should be avoided.
The CRISPR arrays contain few stop codons and, accordingly, are often erroneously translated into hypothetical proteins. Unfortunately, these artefacts then enter the databases and tend to be amplified during the analysis of new genomes, so there are currently at least two Pfam entries that consist of non-existent `pseudo-Cas proteins' (PF11194 and PF11664). Care should be taken during the annotation of new genome sequences to avoid further proliferation of such errors.
Conclusion
Given the complexity and the highly dynamic mode of evolution of the CRISPR–Cas systems, it would be counterproductive to attempt classification on the basis of any single criterion — for instance, the phylogeny of Cas1. Thus, we propose a polythetic classification that integrates the phylogenies of the conserved cas genes, the sequences of and structural similarities between other Cas proteins, and the composition and organization of the known and putative operons. It should be emphasized that a robust family classification of the Cas proteins, many of which diverge rapidly, is not only a matter of convenient description but also a basis for experimental validation of the respective functional predictions. Therefore, it is important that this classification be continuously updated and revised when necessary, using new sequence and structure information combined with state-of-the-art computational methods. The classification described here is available at the NCBI CRISPR/Cas website, along with tools for the identification of Cas proteins. In the future, a fine-grained classification of the CRISPR–Cas systems should become feasible on the basis of phylogenies and structures of Cas proteins, the operon organizations of cas genes and the architectures of CRISPR repeats.
Supplementary Material
Acknowledgements
The authors thank M. Terns for critical reading of the manuscript and useful discussions. K.S.M., Y.I.W. and E.V.K. are supported by the intramural funds of the US Department of Health and Human Services (National Library of Medicine); D.H.H. is supported by a US National Institutes of Health grant (1 R01 HG004881); E.C. acknowledges funding from Umeå University, Sweden, and the Swedish Research Council. S.M. acknowledges funding from the National Sciences and Engineering Research Council of Canada (the Discovery programme); F.J.M.M. acknowledges support from the University of Alicante, Spain, (Vicerrectorado de Investigacion, and Desarrollo e Innovacion) for the use of its research technical services; A.F.Y. is supported by the Government of Canada through Genome Canada and the Ontario Genomics Institute (grant 2009-OGI-ABC-1405). S.J.B. and J.O. are supported by Veni and TOP grants from the Netherlands Organization for Scientific Research (NWO).
Footnotes
Competing interests statement The authors declare no competing financial interests.
References
- 1.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 2.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol. Rep. 2009;1:95. doi: 10.3410/B1-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorek R, Kunin V, Hugenholtz P. CRISPR — a widespread system that provides acquired resistance against phages in bacteria and archaea. Nature Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 6.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 9.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 11.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 12.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 13.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Garrett RA, et al. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem. Soc. Trans. 2011;39:51–57. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 18.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 19.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 2011;392:277–289. doi: 10.1515/BC.2011.042. [DOI] [PubMed] [Google Scholar]
- 20.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 21.Sontheimer EJ, Marraffini LA. Slicer for DNA. Nature. 2010;468:45–46. doi: 10.1038/468045a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 23.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedenheft B, et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Beloglazova N, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/ Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soding J, Remmert M, Biegert A, Lupas AN. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006;34:W374–W378. doi: 10.1093/nar/gkl195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleanthous C, et al. Structural and mechanistic basis of immunity toward endonuclease colicins. Nature Struct. Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 38.Jakubauskas A, Giedriene J, Bujnicki JM, Janulaitis A. Identification of a single HNH active site in type IIS restriction endonuclease Eco31I. J. Mol. Biol. 2007;370:157–169. doi: 10.1016/j.jmb.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White MF. Structure, function and evolution of the XPD family of iron–sulfur-containing 5′→3′ DNA helicases. Biochem. Soc. Trans. 2009;37:547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 40.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Koonin EV. PSI-BLAST — a tool for making discoveries in sequence databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babu M, et al. A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 47.Han D, Lehmann K, Krauss G. SSO1450 – a CAS1 protein from Sulfolobussolfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009;583:1928–1932. doi: 10.1016/j.febslet.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 48.Han D, Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 49.Guy CP, Majernik AI, Chong JP, Bolt EL. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186. doi: 10.1093/nar/gkh960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selengut JD, et al. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.