Abstract
Purpose
The adenosine A2A receptor (A2AR) modulates collagen synthesis and extracellular matrix production in ocular tissues that contribute to eye growth and the development of myopia. We aimed to determine if single nucleotide polymorphisms (SNPs) in A2AR exons associates with high myopia found in Chinese subjects.
Methods
DNA samples were prepared from venous lymphocytes of 175 Chinese subjects with high myopia of less than –8.00 diopters (D) correction and 101 ethnically similar controls with between –1.00 D and +1.00 D correction. The coding region sequences of A2AR were amplified by PCR and analyzed by Sanger sequencing. The detected variations were confirmed by reverse sequencing. Allelic frequencies of all detected common SNPs were assessed for Hardy–Weinberg equilibrium.
Results
Five variations in A2AR exons, 5675 A>G, 5765 C>T, 13325 G>A, 13448 C>T, and 14000 T>A, were detected in controls at a low frequency (<1%). However, one SNP, 13772 T>C (rs5751876), showed its polymorphism in 53.3% of the total study population. The rs5751876 is a synonymous substitution located in a tyrosine codon of exon 2. Despite no significant difference in genotype distribution between cases and controls, the frequency of heterozygotes with the rs5751876 genotype was significantly lower in subjects with high myopia.
Conclusions
The reduced frequency of the heterozygote rs5751876 genotype in subjects suggests a possible association of A2AR with high myopia in a Chinese population.
Introduction
Human high myopia is an extreme form of myopia with an excessive increase in the ocular axial length and with pathological structural changes in the sclera. These changes include thinning of sclera due to reduced collagen production and tissue loss in the posterior segment [1-3]. The exact mechanism that causes such abnormal eye development is unknown. It is now widely recognized that in addition to a series of biologic factors and neurotransmitter pathways [4-11], genetic factors affecting transcription and other regulatory activities contribute to the complex process of ocular development and the occurrence of high myopia [12-15].
The role of adenosine in vision development and myopia formation has recently been reported. In the tiger salamander, the adenosine A2A receptor (A2AR) regulates vision formation by mediating inhibition of rod opsin mRNA expression [16]. In form deprivation myopic guinea pigs, adenosine A2AR is increased in the retina, choroid, and sclera [17]. In addition, treatment with the adenosine receptor antagonist 7-methylxanthine decreases myopia progression and eye elongation in myopic children and in form deprivation myopic knockout guinea pigs [18,19]. In previous findings we detected a greater myopic shift with longer axial length, increased vitreous chamber depth, and altered scleral collagen fiber structure in A2AR knockout mice compared to wild-type littermates [20]. We also found that the A2AR agonist CGS21680 increases the expression of collagen I, III, and V mRNAs in cultured human scleral fibroblasts and promotes production of soluble collagen in a concentration-dependent manner [20].
Adenosine is the product of ATP metabolism present in all cells and body fluids, where it also acts as an important neurotransmitter and an effective vasodilator [21]. It elicits biologic responses by binding to adenosine receptors, including A1, A2A, A2B, and A3, that are distributed throughout the body. Adenosine A2AR is expressed in ocular tissues of vertebrates [22-25]. Based on previous functional studies, we hypothesized that A2AR plays an important role in postnatal refractive development and thus is also a candidate susceptibility gene for high myopia.
By using single nucleotide polymorphisms (SNPs) as markers, association analysis is a powerful tool to find disease-related genes. Recently, various susceptibility genes have been identified by myopia- or high-myopia-association studies, including transforming growth factor-1 [26], catenin delta 2 [27], BH3-like motif containing cell death inducer [28], gap junction protein delta 2, actin alpha cardiac muscle 1 [29], and Ras protein-specific guanine nucleotide-releasing factor 1 [30]. However, most association analyses at either candidate gene or whole genome level have focused on tag SNPs; few functional SNPs (coding SNPs) have been tested. The purpose of this study was to determine if there are any significant associations between high myopia and SNPs in the A2AR exons in a Chinese population.
Methods
Subjects
We matched 175 unrelated subjects with high myopia (spherical power of less than −8.00 diopters [D]) in both eyes with 101 ethnically and socially similar, unrelated control subjects with refractive errors within ±1.00 D in each eye. All the subjects were from the Optometry Eye Hospital of Wenzhou Medical College, Wenzhou, China. Each of the participating subjects received a complete ocular examination including visual acuity (Topcon RM-8800; Topcon Corp., Tokyo, Japan), slit-lamp evaluation of the anterior segment (Topcon SL-1E Slit Lamp; Topcon Corp.), dilated fundus examination (Heine Omega 180 Binocular Indirect Ophthalmoscope; Heine Optotechnik, Herrsching, Germany) and axial-length measurements (Zeiss IOL Master; Carl Zeiss Meditec, Jena, Germany) [31]. Exclusion criteria included subjects with any known ocular, genetic, or systemic connective tissue disorders associated with myopia. Written consent was obtained from every subject after being fully informed of the purpose and procedures of the study. This study adhered to the tenets of the Declaration of Helsinki with subsequent revisions and was approved by the Human Subjects Ethics Committees of Wenzhou Medical College and the Eye Hospital, Wenzhou, China.
DNA extraction and amplification
Genomic DNA for polymerase chain reaction (PCR) was extracted from 2 to 5 ml of peripheral venous blood from all participants. DNA was purified from lymphocytes according to the manufacturer’s instructions using a kit (BBI, Toronto, ON, Canada) [31]. The accession number for the A2AR sequence used for the construction of our primers was NT_011520. Four pairs of primers (Table 1) were used to amplify exon regions of A2AR. PCR was performed in a 50-μl volume with 31.25 μl double distilled H2O, 5 μl 10× PCR buffer (100 mM Tris-HCl pH 8.8, 500 mM KCl, 15 mM MgCl2, 0.8% Nonidet P40), 3.5 μl 25 mmol/l MgCl2, 4 μl 2.5 mmol/l deoxy-ribonucleoside triphosphate, 2 μl template DNA, 2 μl sense primer (all these ingredients were from Invitrogen, Carlsbad, CA), 2 μl antisense primer (Invitrogen, Shanghai, China), and 0.25 μl Ex Taq polymerase (Takara BIO Inc., Tokyo, Japan). The PCR reaction was initiated at 94 °C for 2 min, followed by 35 cycles (94 °C for 30 s, 65 °C for 30 s, 72 °C for 45 s), and ended at 72 °C for 10 min. PCR amplification products were purified using the PCR Cleanup Kit (Axygen Biosciences, Union City, CA).
Table 1. Primers used for the amplification and sequencing of A2AR.
| Fragment | Direction | Primer sequence | PCR product (bp) | Exon |
|---|---|---|---|---|
| A2A1 |
S |
5′- CGCCCTCTGCAGATGGTTCAGCT-3′ |
428 |
1 |
| |
AS |
5′- GTTGCTGTTGAGCCACACGGCC −3′ |
|
|
| A2A2 |
S |
5′- CCGTGCTGAGCCTGCCTGTCGTC −3′ |
434 |
2 |
| |
AS |
5′- CCTGGCTCCGGGCACAGACCAA −3′ |
|
|
| A2A3 |
S |
5′- CCCTGGCTTCTCAGATCTCTGAT −3′ |
577 |
2 |
| |
AS |
5′- GGCGTAGATGAAGGGATTCACA −3′ |
|
|
| A2A4 |
S |
5′- CTCATGTACCTGGCCATCGTCCTC −3′ |
611 |
2 |
| AS | 5′- CTTCTTGCTGGGCCTCATGCTG −3′ |
S, sense strand; AS, antisense strand.
Single nucleotide polymorphism genotyping by sequencing
After the PCR amplification products were purified, nucleotide sequence analysis was performed with an ABI 3700 sequencer (Applied Biosystems Inc., Foster City, CA). Using Clustal W (version 3.0; European Bioinformatics Institute, Hinxton, Cambridgeshire, Great Britain), we compared all sequencing results to identify variations and to check the sequence map of mutable points to confirm the genotype. Positive results were further confirmed by reverse sequencing.
Analysis of common single nucleotide polymorphisms
Allelic frequencies of all detected common SNPs were assessed for Hardy–Weinberg equilibrium (HWE). The frequency (P) of allele A was calculated as
| , |
where NAA is the number of wild types, NAB is the number of heterozygotes, and N is the sample size of the group. The frequency of allele B was calculated as
| . |
The theoretical value of the three genotypes was calculated according to the following allele frequencies:
| , |
| , |
| , |
where (E) is the expected value and NBB is the number of homozygous mutations. The HWE for the genotype distributions was examined by the χ2 test in each group, where
| . |
The level of HWE was set at p>0.05. All samples were assumed to be from the same Mendelian population. Differences in the observed genotype, allelic frequencies, and heterozygous frequency (AB versus AA+BB) between the subjects with high myopia and the control subjects were also examined by the χ2 test. Statistical analyses were performed with the Statistical Package for the Social Sciences software (SPSS, version 13.0 for Windows; SPSS Science Inc., Chicago, IL). The power analysis for the χ2 test was run using Statistical Analysis Software (SAS, version 9.0; SAS Institute, Cary, NC).
Results
For the high myopia group, the spherical power (OD; OS, mean±SD, D) was -15.44±6.06 D; -15.18±6.28 D; the astigmatism (OD; OS, mean±SD, D) was -1.60±1.21 D; -1.85±1.26 D; the axial length (OD; OS, mean±SD, mm) was 29.58±2.64 mm; 29.52±2.86 mm [31].
Common and rare single nucleotide polymorphisms in A2AR exons
Purified PCR products specific to A2AR exons were sequenced to detect functional SNPs correlating to high myopia (Table 2). In total, six exonic substitutions were found (Table 3). Five of these, 5675 A>G, 5765 C>T, 13325 G>A, 13448 C>T, and 14000 T>A, were seen only in the control panel and had minor allele frequencies lower than 1%; as a consequence, no further analysis was conducted for these rare variants. In contrast, SNP 13772 T>C was common, with a frequency of 53.3% in all subjects (Figure 1, Table 3). Listed as rs5751876 (National Center for Biotechnology Information, Entrez SNP database, dbSNP Build 129), this is a synonymous substitution in codon Y361, which is located in the second exon of A2AR (Figure 2). The rs5751876 was also reported to be associated with anxiety in various investigations [32-34].
Table 2. Quantities of effective sequence results.
| Fragment | High myopia group (n) | Control group (n) | Total (n) |
|---|---|---|---|
| A2A1 |
170 |
98 |
268 |
| A2A2 |
163 |
101 |
264 |
| A2A3 |
158 |
97 |
255 |
| A2A4 | 161 | 96 | 257 |
Table 3. Observed frequency of the variations in high myopia and control groups.
| Variations | Polymorphism | High myopia group | Control group | Total |
|---|---|---|---|---|
| 5675 |
A→G |
0/196 (0.00%) |
1/340 (0.30%) |
1/536 (0.19%) |
| 5765 |
C→T |
0/196 (0.00%) |
2/340 (0.59%) |
2/536 (0.37%) |
| 13325 |
G→A |
0/194 (0.00%) |
1/316 (0.32%) |
1/510 (0.20%) |
| 13448 |
C→T |
0/194 (0.00%) |
1/316 (0.32%) |
1/510 (0.20%) |
| 14000 |
T→A |
0/192 (0.00%) |
2/322 (0.62%) |
2/514 (0.39%) |
| 13772 (rs5751876) | T→C | 104/192(54.17%) | 170/322 (52.80%) | 274/514 (53.31%) |
Figure 1.
The common SNP 13772 T>C (rs5751876) was identified in A2AR. Although rs5751876, which codes for tyrosine, was located in coding regions, it did not affect the amino acid sequence.
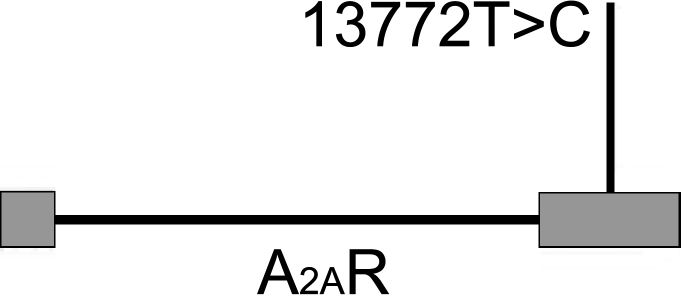
Figure 2.
Schematic diagram of the single nucleotide polymorphism rs5751876 within A2AR. The gray rectangles represent exons, and the lines represent introns.
Lower heterozygous frequency of rs5751876 in subjects with high myopia
To examine the association between rs5751876 and high myopia, we compared the variation frequency of this SNP in subjects with high myopia and control subjects by the χ2 test. No significant difference was observed between the case and control groups by either genotype frequency test (p=0.11) or allele frequency test (p=0.76).
However, the genotype distribution of rs5751876 in subjects with high myopia deviated significantly from HWE (p=0.0016), while no such deviation was seen in control samples. Further testing on the distribution of homozygotes and heterozygotes demonstrated a significantly lower frequency of the heterozygous genotype in subjects with high myopia compared to that of the control subjects (p=0.033, Table 4). This result suggests a possible association of A2AR with high myopia in Chinese population.
Table 4. Genotype frequency of 13772 T>C (rs5751876).
| Genotype* | High myopia group | Control group | Total |
|---|---|---|---|
| TT |
26/96 (27.08%) |
35/161 (21.74%) |
61/257 (23.74%) |
| TC |
36/96 (37.50%) |
82/161 (50.93%) |
118/257 (45.91%) |
| CC | 34/96 (35.42%) | 44/161 (27.33%) | 78/257 (30.35%) |
TT, genotype with homozygous normal allele; TC, genotype with heterozygous sequence alterations; CC, genotype with homozygous sequence alterations. *Based on association test using χ2 statistics, Poverall=0.11 and PTC versus TT+CC=0.03.
Discussion
Two facts prompted us to conduct this investigation to determine if coding SNPs in the A2AR gene contributes to the development of high myopia. First, previous association studies on high myopia mainly focused on tag SNPs [35]; few functional variants (e.g, coding SNPs) were identified for their association with high myopia [36]. Furthermore, myopic changes observed in knockout mice suggest that certain genes may cause or contribute to refraction error in humans. In particular, myopic changes occur in early growth response 1 knockout mice [37] and in A2AR knockout mice [20]. Although these pathologic abnormalities are similar to those in humans with high myopia [1-3], no mutation was found in early growth response 1 exons in Chinese subjects with high myopia [37], and correlation between A2AR and high myopia remains unknown.
In our sequencing results only one SNP (rs5751876) shows a statistical power for association analysis (minor allele frequency=46.7%). Previous association studies have linked this locus to susceptibility of anxiety [32-34]. Despite no observed correlation in the genotype and allele frequency test, the significant deviation from HWE and the lower heterozygote genotype frequency in case but not in control subjects suggest an association between this SNP and high myopia, at least in Chinese population. Moreover, the C allele frequency of rs5751876 in our individuals (53.3%) was lower than that in Europeans and Africans (72.2% and 60%, respectively, dbSNP), further implying different signals of A2AR among populations. Expanding the sample size may provide stronger signals for the association of this SNP to high myopia.
Being a synonymous substitution, the association signal of rs5751876 to high myopia could be explained in two ways. First, this substitution may change the codon of Y361 (TAC to TAT), which in turn affects the translation efficiency by tRNA-dependent codon usage bias [38]. Another possibility may be a high linkage disequilibrium of this SNP with rs2298383 (D’=1) in an upstream transcription factor binding site [33]. Therefore, rs5751876 is perhaps a marker for surrounding SNPs corresponding to A2AR expression.
Adenosine A2AR is expressed in ocular tissues and participates in eye growth. As described above the relevance of A2AR to high myopia is supported by a line of evidence from cellular and animal studies [20]. These findings as well as our genetic results indicate that adenosine and adenosine receptors contribute to the development of high myopia. The mechanism(s) by which A2AR and/or other adenosine receptors contribute to myopia is currently unknown. Functional analysis will help elucidate the molecular pathways of A2AR and determine if pharmaceutical intervention targeting adenosine receptors may inhibit development of myopia.
Acknowledgments
We thank Dr. Britt Bromberg, Xenofile Editing, for providing editing services for early version of manuscript. This study was sponsored by the National Basic Research Program of China (973 project, 2011CB504604) to X.Z.; the National Natural Science Foundation of China (81070751, 30830107) and the Research Fund for the Doctoral Program of Higher Education of China (20060343002) to J.Q.; the Zhejiang Provincial Natural Science Foundation of China (R205739, Z2100065) to X.Z.; the Key Research and Development Program project grant 2008C14097 from Zhejiang Province to X.Z.; and the Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents to X.Z.
References
- 1.Norton TT. Experimental myopia in tree shrews. Ciba Found Symp. 1990;155:178–94. [PubMed] [Google Scholar]
- 2.Norton TT, Rada JA. Reduced extracellular matrix in mammalian sclera with induced myopia. Vision Res. 1995;35:1271–81. doi: 10.1016/0042-6989(94)00243-f. [DOI] [PubMed] [Google Scholar]
- 3.Rada JA, Shelton S, Norton TT. The sclera and myopia. Exp Eye Res. 2006;82:185–200. doi: 10.1016/j.exer.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Rohrer B, Spira AW, Stell WK. Apomorphine blocks form-deprivation myopia in chickens by a dopamine D2-receptor mechanism acting in retina or pigmented epithelium. Vis Neurosci. 1993;10:447–53. doi: 10.1017/s0952523800004673. [DOI] [PubMed] [Google Scholar]
- 5.Rohrer B, Stell WK. Basic fibroblast growth factor (bFGF) and transforming growth factor beta (TGF-beta) act as stop and go signals to modulate postnatal ocular growth in the chick. Exp Eye Res. 1994;58:553–61. doi: 10.1006/exer.1994.1049. [DOI] [PubMed] [Google Scholar]
- 6.Pendrak K, Nguyen T, Lin T, Capehart C, Zhu X, Stone RA. Retinal dopamine in the recovery from experimental myopia. Curr Eye Res. 1997;16:152–7. doi: 10.1076/ceyr.16.2.152.5090. [DOI] [PubMed] [Google Scholar]
- 7.Ohngemach S, Feldkaemper M, Schaeffel F. Pineal control of the dopamine D2-receptor gene and dopamine release in the retina of the chicken and their possible relation to growth rhythms of the eye. J Pineal Res. 2001;31:145–54. doi: 10.1034/j.1600-079x.2001.310208.x. [DOI] [PubMed] [Google Scholar]
- 8.McFadden SA, Howlett MH, Mertz JR, Wallman J. Acute effects of dietary retinoic acid on ocular components in the growing chick. Exp Eye Res. 2006;83:949–61. doi: 10.1016/j.exer.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Yan DS, Zhou XT, Chen XY, Lu F, Wang J, Hu DN, Qu J. Expression of retinoid acid receptors in human scleral fibroblasts and regulation of growth of fibroblasts by retinoic acid. Zhonghua Yan Ke Za Zhi. 2007;43:750–3. [PubMed] [Google Scholar]
- 10.Fan DS, Lam DS, Chan CK, Fan AH, Cheung EY, Rao SK. Topical atropine in retarding myopic progression and axial length growth in children with moderate to severe myopia: a pilot study. Jpn J Ophthalmol. 2007;51:27–33. doi: 10.1007/s10384-006-0380-7. [DOI] [PubMed] [Google Scholar]
- 11.Barathi VA, Weon SR, Beuerman RW. Expression of muscarinic receptors in human and mouse sclera and their role in the regulation of scleral fibroblasts proliferation. Mol Vis. 2009;15:1277–93. [PMC free article] [PubMed] [Google Scholar]
- 12.Saw SM, Chua WH, Wu HM, Yap E, Chia KS, Stone RA. Myopia: gene-environment interaction. Ann Acad Med Singapore. 2000;29:290–7. [PubMed] [Google Scholar]
- 13.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–6. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–6. [PubMed] [Google Scholar]
- 15.Liang CL, Yen E, Su JY, Liu C, Chang TY, Park N, Wu MJ, Lee S, Flynn JT, Juo SH. Impact of family history of high myopia on level and onset of myopia. Invest Ophthalmol Vis Sci. 2004;45:3446–52. doi: 10.1167/iovs.03-1058. [DOI] [PubMed] [Google Scholar]
- 16.Alfinito PD, Alli R, Townes-Anderson E. Adenosine A(2a) receptor-mediated inhibition of rod opsin mRNA expression in tiger salamander. J Neurochem. 2002;83:665–72. doi: 10.1046/j.1471-4159.2002.01162.x. [DOI] [PubMed] [Google Scholar]
- 17.Cui D, Trier K, Zeng J, Wu K, Yu M, Ge J. Adenosine receptor protein changes in guinea pigs with form deprivation myopia. Acta Ophthalmol. 2010;88:759–65. doi: 10.1111/j.1755-3768.2009.01559.x. [DOI] [PubMed] [Google Scholar]
- 18.Trier K, Munk Ribel-Madsen S, Cui D, Brogger Christensen S. Systemic 7-methylxanthine in retarding axial eye growth and myopia progression: a 36-month pilot study. J Ocul Biol Dis Infor. 2008;1:85–93. doi: 10.1007/s12177-008-9013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui D, Trier K, Zeng J, Wu K, Yu M, Hu J, Chen X, Ge J. Effects of 7-methylxanthine on the sclera in form deprivation myopia in guinea pigs. Acta Ophthalmol. 2009 doi: 10.1111/j.1755-3768.2009.01688.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Huang Q, An J, Lu R, Qin X, Jiang L, Li Y, Wang J, Chen J, Qu J. Genetic Deletion of the Adenosine A2A Receptor Confers Postnatal Development of Relative Myopia in Mice. Invest Ophthalmol Vis Sci. 2010;51:4362–70. doi: 10.1167/iovs.09-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaar R, Jones MR, Chen JF, Ravid K. Animal models for the study of adenosine receptor function. J Cell Physiol. 2005;202:9–20. doi: 10.1002/jcp.20138. [DOI] [PubMed] [Google Scholar]
- 22.Blazynski C. Characterization of adenosine A2 receptors in bovine retinal pigment epithelial membranes. Exp Eye Res. 1993;56:595–9. doi: 10.1006/exer.1993.1073. [DOI] [PubMed] [Google Scholar]
- 23.Kvanta A, Seregard S, Sejersen S, Kull B, Fredholm BB. Localization of adenosine receptor messenger RNAs in the rat eye. Exp Eye Res. 1997;65:595–602. doi: 10.1006/exer.1996.0352. [DOI] [PubMed] [Google Scholar]
- 24.Taomoto M, McLeod DS, Merges C, Lutty GA. Localization of adenosine A2a receptor in retinal development and oxygen-induced retinopathy. Invest Ophthalmol Vis Sci. 2000;41:230–43. [PubMed] [Google Scholar]
- 25.Dong F, An JH, Ren YP, Yan DS, Zhou XT, Lu F, Hu DN, Chen JF, Qu J. Expression of dopamine receptor D2 and adenosine receptor A2A in human retinal pigment epithelium. Zhonghua Yan Ke Za Zhi. 2007;43:1110–3. [PubMed] [Google Scholar]
- 26.Metlapally R, Ki CS, Li YJ, Tran-Viet KN, Abbott D, Malecaze F, Calvas P, Mackey DA, Rosenberg T, Paget S, Guggenheim JA, Young TL. Genetic association of insulin-like growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci. 2010;51:4476–9. doi: 10.1167/iovs.09-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YJ, Goh L, Khor CC, Fan Q, Yu M, Han S, Sim X, Ong RT, Wong TY, Vithana EN, Yap E, Nakanishi H, Matsuda F, Ohno-Matsui K, Yoshimura N, Seielstad M, Tai ES, Young TL, Saw SM. Genome-Wide Association Studies Reveal Genetic Variants in CTNND2 for High Myopia in Singapore Chinese. Ophthalmology. 2011 doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Chen X, Yan D, Xue A, Lu F, Qu J, Zhou XT. Association analysis of retinoic acid receptor beta (RARbeta) gene with high myopia in Chinese subjects. Mol Vis. 2010;16:855–61. [PMC free article] [PubMed] [Google Scholar]
- 32.Childs E, Hohoff C, Deckert J, Xu K, Badner J, de Wit H. Association between ADORA2A and DRD2 polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology. 2008;33:2791–800. doi: 10.1038/npp.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohoff C, Mullings EL, Heatherley SV, Freitag CM, Neumann LC, Domschke K, Krakowitzky P, Rothermundt M, Keck ME, Erhardt A, Unschuld PG, Jacob C, Fritze J, Bandelow B, Maier W, Holsboer F, Rogers PJ, Deckert J. Adenosine A(2A) receptor gene: evidence for association of risk variants with panic disorder and anxious personality. J Psychiatr Res. 2010;44:930–7. doi: 10.1016/j.jpsychires.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Rogers PJ, Hohoff C, Heatherley SV, Mullings EL, Maxfield PJ, Evershed RP, Deckert J, Nutt DJ. Association of the anxiogenic and alerting effects of caffeine with ADORA2A and ADORA1 polymorphisms and habitual level of caffeine consumption. Neuropsychopharmacology. 2010;35:1973–83. doi: 10.1038/npp.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 36.Young TL. Molecular genetics of human myopia: an update. Optom Vis Sci. 2009;86:E8–22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Xiao X, Li S, Xing Y, Guo X, Zhang Q. Evaluation of EGR1 as a candidate gene for high myopia. Mol Vis. 2008;14:1309–12. [PMC free article] [PubMed] [Google Scholar]
- 38.Comeron JM, Aguade M. An evaluation of measures of synonymous codon usage bias. J Mol Evol. 1998;47:268–74. doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]