Abstract
In the androdioecious nematode Caenorhabditis elegans, self-fertilization is the predominant mode of reproduction. Nevertheless, males do occur, and it is still unclear if these represent a selective advantage or merely an evolutionary relict. In this study, we first tested the hypothesis that the production of males might benefit invaders to resident populations. We added single, GFP-marked worms to established laboratory populations and followed GFP frequencies over time. Mated hermaphrodites and also males were more successful in invading resident populations if compared to single, unmated hermaphrodites. The observed higher frequencies should increase the likelihood that any of the associated invading alleles persist. Second, we tested the hypothesis that males and, thus, higher outcrossing rates, are specifically favored under changing environmental conditions. After an outbred population was subjected to changing stress or to control laboratory conditions, we measured the male maintenance of the resulting populations. Interestingly all populations, experimental and control alike, showed high male maintenance, suggesting that persistence of males is also favored under standard laboratory conditions.
Keywords: Caenorhabditis elegans, ecology, hermaphrodite, male, mode of reproduction
Androdioecious reproductive systems consisting primarily of hermaphrodites, instead of females and males, exist in multiple phyla (for an overview see Stewart and Phillips, 2002). From an evolutionary perspective, androdioecy is puzzling, and the replacement of females with hermaphrodites appears only possible under certain rather strict conditions (Charnov et al., 1976). Further, once hermaphrodites are present and can reproduce by self-fertilization, males appear superfluous. In analogy with the two-fold cost of males in theories on the evolution of sex (Maynard-Smith, 1978; Bell, 1982), males might even represent a burden, decreasing individual fitness.
The arguably best-characterized androdioecious species is Caenorhabditis elegans. C. elegans hermaphrodites reproduce either by self-fertilization or by cross-breeding with a male [for a general introduction see Wood (1988) and Hope (1999)]. Cross-fertilization between hermaphrodites does not occur. Hermaphrodites are essentially females that initially produce a limited quantity of sperm and then switch to the production of eggs for the rest of their reproductive life. The sperm is stored in the spermatheca and can be used to fertilize the eggs after the sex switch of the germ line. Sex is determined genetically by the presence of two (hermaphrodites) or one (males) X chromosomes along with five pairs of autosomes. Consequentially, almost the entire self-progeny is hermaphroditic. The very few males that arise spontaneously in the self-progeny of hermaphrodites (around 0.2% in the standard laboratory strain N2) are the result of X chromosome non-disjunction events. Half of the sperm produced by males contain no X chromosome, and as a consequence, 50% of the progeny sired by males is male. Upon mating, male-derived sperm is also stored in the spermatheca along with the hermaphrodite's own sperm. Usually, male sperm has a competitive advantage over the hermaphrodite's sperm, at least in part due to its larger size (LaMunyon and Ward, 1998, 1999, 2002).
It has been suggested that C. elegans males might merely be evolutionary relicts with no particular function, which are still present after a relatively recent evolution of females into self-fertilizing hermaphrodites (Chasnov and Chow, 2002). Indeed, hermaphroditism in all three contemporary hermaphroditic species within the genus Caenorhabditis has likely arisen independently, and all of these species have close gonochoristic relatives, indicating that the transition to self-fertilization happened relatively recently (Kiontke et al., 2004; K. Kiontke and D. H. Fitch, NYU, pers. com.). A similar picture emerges in Pristionchus, another well-studied nematode genus, where hermaphroditism arose at least six times independently (Mayer et al., 2007; W. E. Mayer, M. Herrmann and R. J. Sommer, MPI Dev. Biol., pers. com.). However, given that “relatively recently” in this context still means up to tens of millions of years (Kiontke et al., 2004; Mayer et al., 2007), it is striking that males still exist in all known hermaphroditic species of Caenorhabditis (K. Kiontke and D. H. Fitch, NYU, pers. com.) and Pristionchus (M. Herrmann, W. E. Mayer and R. J. Sommer, MPI Dev. Biol., pers. com.). Indeed, androdioecy may be maintained by as yet unknown selective forces, which prevent the complete loss of males as well as a switch back to a dioecous reproduction system (Stewart and Phillips, 2002). It has been estimated that a purely selfing C. elegans population would be driven to extinction within less than a million year by the accumulation of slightly deleterious mutations (Loewe and Cutter, 2008).
A first prerequisite for males to play a role in populations is that they, and consequentially out-crossing, exist at a level that significantly influences population genetics. In cultures of the standard laboratory strain N2 that are initiated with high numbers of males, the male frequency declines rapidly, and most of the time males disappear from the populations within less than 20 generations (Chasnov and Chow, 2002; Stewart and Phillips, 2002; Cutter et al., 2003; Cutter, 2006; Teotonio et al., 2006; Wegewitz et al., 2008). However, other natural isolates behave differently if assayed under the same standard laboratory conditions and maintain males over longer periods of time (Teotonio et al., 2006; Wegewitz et al., 2008). Recently, several authors have attempted to infer the outcrossing frequencies in natural populations by measuring linkage disequilibrium, heterozygosity, or genetic diversity (Sivasundar and Hey, 2003; Barriere and Felix, 2005; Haber et al., 2005; Sivasundar and Hey, 2005; Cutter, 2006; Barriere and Felix, 2007). All these studies support the notion that outcrossing does occur in wild populations, and that males do leave an appreciable genetic footprint in natural populations. Except for Sivasundar and Hey (2005), who estimated an outcrossing rate of 0.2, it is broadly accepted that outcrossing is rare in natural populations, ranging in between 10-5 and 0.02. Nevertheless, even these rare outcrossing events may be sufficient to reduce the mutational load and/or maintain sufficient genetic diversity required for rapid adaptation to fluctuating environments (Charlesworth and Charlesworth, 1998; Agrawal and Lively, 2001; Pannell, 2002). Laboratory evolution experiments suggested that elevated mutation rates induced either by chemical mutagens (Manoel et al., 2007) or by a deficient DNA repair mechanism (Cutter, 2005) represent a selective force for higher male frequencies. Very recently, it has been shown that outcrossing was not only favored under conditions of increased mutation rate but also during the adaptation to the presence of a pathogen (Morran et al., 2009b). These observations are in agreement with the expectation based on theoretical considerations, according to which males and frequent outcrossing are beneficial under variable environmental conditions and/or high deleterious mutation rates (Fischer, 1930; Muller, 1932; Muller, 1964; Kondrashov, 1988; Hamilton et al., 1990; Agrawal and Lively, 2001). In this context, it is interesting to note that C. elegans also appears to plastically increase the outcrossing rate in response to stressful conditions (Morran et al., 2009a).
In a previous study, we showed that in a situation with virtually unlimited access to hermaphrodites a male can produce a considerably higher number of progeny than a hermaphrodite, which reproduces by self-fertilization (Wegewitz et al., 2008). This effect was much more pronounced in the strain CB4856 where males sired more than 10 times as many progeny as unmated hermaphrodites produced. Even the “poorly” mating N2 males still gave rise to more than three times as many progeny as unmated hermaphrodites of the same strain. However, males needed to mate with multiple hermaphrodites to reach this reproductive success. From these numbers, one would expect that a male that arose spontaneously or invaded into a population of hermaphrodites would contribute more to the gene pool of the next generation than any of the hermaphrodites, provided the population density is high enough that the male finds multiple mates during its life. In order to test this prediction, we asked if particular phenotypes may spread and persist more easily if they invade a resident, largely hermaphrodite population as male individuals or mated hermaphrodites rather than virgin (and thus exclusively selfing) hermaphrodites. We performed experiments where we added single individuals that were marked with a transgene to stable populations of unmarked worms and followed the frequency of the transgenic phenotype.
In a second experiment we asked if varying non-mutagenic stress conditions also act as selective pressure in favor of higher male frequencies, because these could possibly enhance the spread of novel advantageous phenotypes. To create a starting population with the genetic potential to achieve various levels of male maintenance, we interbred N2 and CB4856, two strains on the low and the high ends, respectively, of the spectrum of male maintenance found in natural isolates (Wegewitz et al., 2008). The resulting hybrid populations were subjected to varying environmental conditions (high salt, low and high temperatures, pathogenic bacteria, and standard laboratory conditions) or continuous standard laboratory conditions.
Materials and Methods
C. elegans cultures: C. elegans was cultured on NGM plates with Escherichia coli strain OP50 as food (Stiernagle, 1999). Mating plates consisted of 6 cm NGM plates seeded in the center with 30 μl of an E. coli (OP50) culture. Cultures were incubated in an air-conditioned room at a temperature of 21±1°C and 40% humidity. Cultures were kept in boxes, randomized in piles that were evenly distributed within the boxes.
Strains used: N2: Standard laboratory wild type strain, isolated in Bristol, UK CB4856: Standard polymorphic mapping strain, isolated in Hawaii. PD4792: mIs11[myo-2::gfp + pes-10::gfp + gut::gfp] IV All three strains were requested from the Caenorhabditis Genetics Center at the University of Minnesota (http://biosci.umn.edu/CGC/). QA351: ytIs3[sur-5::gfp] (created by microinjection and UV induced integration (Jin, 1999) of pTG96 (Gu et al., 1998) followed by five back crosses with N2). QA353: mIs11[myo-2::gfp + pes-10::gfp + gut::gfp] IV (created by backcrossing PD4792 to N2 five times) QA354: mIs11[myo-2::gfp + pes-10::gfp + gut::gfp] IV (created by backcrossing PD4792 to CB4856 five times).
Male maintenance assays: Male maintenance assays were done with a population size of 150 as described by Wegewitz et al., (2008).
Invasion experiments: “Stable populations” were started with N2 hermaphrodites and maintained by transferring a fixed number (population size) of worms to a new plate every three days, without paying attention to developmental stage, sex, or GFP fluorescence. To transfer the worms, they were washed off the old plate with M9 solution, and the number of animals in an aliquot were counted. Based on this count, the total number of worms on the plate was estimated, and the appropriate number of worms was pipetted onto a new plate. To initiate the experiment, one individual that was genetically marked with myo-2::gfp (invader) was added to each “stable population” immediately after a transfer. The invaders were either virgin hermaphrodites or mated hermaphrodites or males. The populations were maintained as described above for eight transfers. Prior to each transfer and at the end of the experiment, the number of GFP positive worms on each plate was counted. As invaders we used QA351 and QA353 (essentially the genetic background of N2) and QA354 (essentially the genetic background of CB4856).
For Invasion Experiment 1, the population size was 500. Four replicates for each type of invader were done in parallel, and the experiment was repeated twice with QA351 and twice with QA353, resulting in two times eight replicates per type of invader. For Invasion Experiment 2, population sizes of 100 and 500 and with QA353 and QA354 as invaders were used, resulting in 12 different treatments. One replicate for each of the treatments were carried out simultaneously, and the experiment was repeated six times.
Experimental Evolution Experiment: The different environmental conditions were:
High Salt: NGM/OP50 plates containing 20 g/l NaCl at 20°C.
Low temperature: NGM/OP50 plates at 15°C.
High temperature: NGM/OP50 plates at 25°C
Pathogen: NGM plates seeded four days prior to use with 1 ml of a 5:2 mixture of CBX102 (Microbacterium nematophilum) and E. coli OP50 overnight cultures at 20°C.
Control: NGM/OP50 plates at 20°C.
Populations were initiated by placing 10 N2 and 10 CB4856 hermaphrodites on mating plates together with 30 males of the other strain for 24 h. Then the hermaphrodites from each cross were transferred to a 10 cm NGM/OP50 plate and allowed to reproduce for three days. The worms were washed from the plates with M9 buffer (Stiernagle, 1999), and from each cross 60 individuals were placed on five high salt or five control plates without paying attention to developmental stage or sex, resulting in five replicates for selection (series B) and five control replicates (series A) with starting populations of 120 individuals. Another five selection (series D) and control (series C) replicates were initiated one day later.
After four days 120 individuals from each plate were transferred to new plates without paying attention to developmental stage or sex and subjected to low temperature (selection) or control conditions.
After four days, 120 individuals from each plate were transferred to new plates and subjected to high temperature (selection) or control conditions.
After four days, 120 individuals from each plate were transferred to plates containing pathogenic bacteria (selection) or control plates. The reminder of the cultures was frozen as described by (Stiernagle, 1999).
After four days, all cultures were treated with hypochloride (Stiernagle, 1999) to remove the pathogenic bacteria, and the isolated embryos were allowed to hatch on plates without food for one day. Then, 120 larvae were placed on NGM/OP50 plates. Two days later, three N2 and three CB4856 males were added to each culture to avoid statistical loss of genotypes. One day later, 120 individuals per culture were placed on NaCl (selection) or control plates to start the second round of selection. In total, five rounds were performed. The experiment was terminated with the freezing step of the fifth round. For the further analysis, aliquots of the cultures were thawed, and the male maintenance test started within two generations.
“Chunking” Experiment: The whole experiment was performed on 10 cm NGM/OP50 plates. The cultures were started by combining a total of 10 hermaphrodites and, if applicable, 10 males on one plate as specified below.
Mixed: hermaphrodites: Five progeny of a cross N2 x CB4856 plus 5 progeny of a cross CB4856 x N2; males: Five progeny of a cross N2 x CB4856 plus 5 progeny of a cross CB4856 x N2.
N2 with males: hermaphrodites: 10 N2; males: 10 N2.
CB4856 with males: hermaphrodites: 10 CB4856; males: 10 CB4856.
N2 no males: hermaphrodites: 10 N2; males: none.
CB4856 no males: hermaphrodites: 10 CB4856; males: none.
For each combination, three replicates were performed.
The cultures were incubated at 20°C. After seven days, a square (chunk) of 16 x 16 mm was cut out from the agar about one third of a plate radius away from the center and transferred upside down onto a new plate (chunking). The chunking step was repeated every seven days until a total of 12 transfers were completed. At the end, the worms were frozen. Aliquots of the cultures were thawed for further analysis, and the male maintenance test started within two generations.
Statistical analysis: The data were analysed using generalized linear models, based on logistic regression analysis. The analyses were performed with the program JMP 8.0 (SAS Institute Inc.), and all graphs were produced with Sigmaplot 11.0 (Systat Software Inc.).
For the invasion experiments, the model included the following factors: treatment (i.e. addition of hermaphrodites, males, or mated hermaphrodites), day of measurement, the interaction between day and treatment, and block (i.e. date when a particular combination of experiments was started). The response variable was the proportion of GFP-positive nematodes within the population. We assessed the overall model and then separately several models, which specifically evaluated the difference between two of the treatment alternatives (e.g., addition of hermaphrodites versus addition of males). In all cases, likelihood ratio effect tests were used to evaluate the impact of a particular factor on the variance of the data. The significance level was adjusted using the false-discovery rate (FDR) to account for multiple testing. For Invasion Experiment 1, we performed separate analyses for two types of crosses (i.e. QA351 vs. QA353). For Invasion Experiment 2, we performed separate analyses for the two population sizes (i.e. 100 vs. 500) and the two types of crosses (i.e. QA354 vs. QA353).
For the Experimental Evolution and the Chunking experiments, the models included the following factors: treatment (i.e. the different types of crosses and strains), day of measurement, interaction between treatment and day, and parental population (from which the nematodes were taken). We again assessed an overall model and then several separate models, which particularly addressed the difference between two treatments. Likelihood ratio effect tests were used to test the impact of a factor and FDR to account for multiple testing.
Results and Discussion
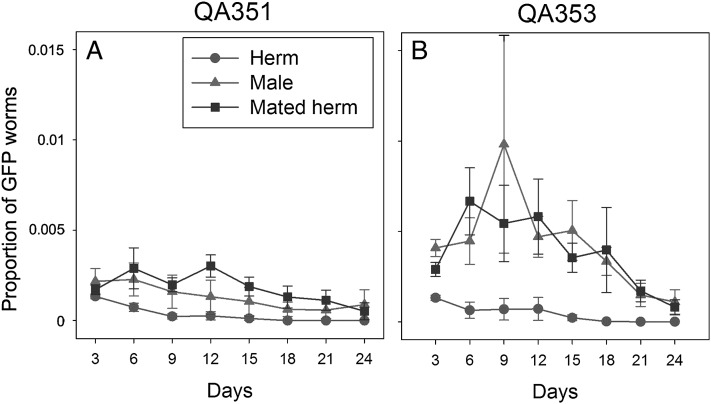
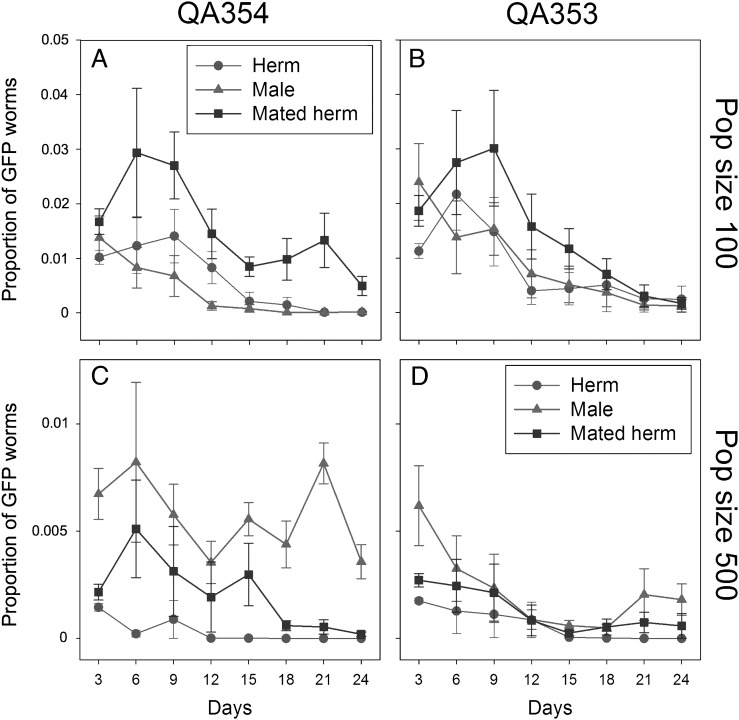
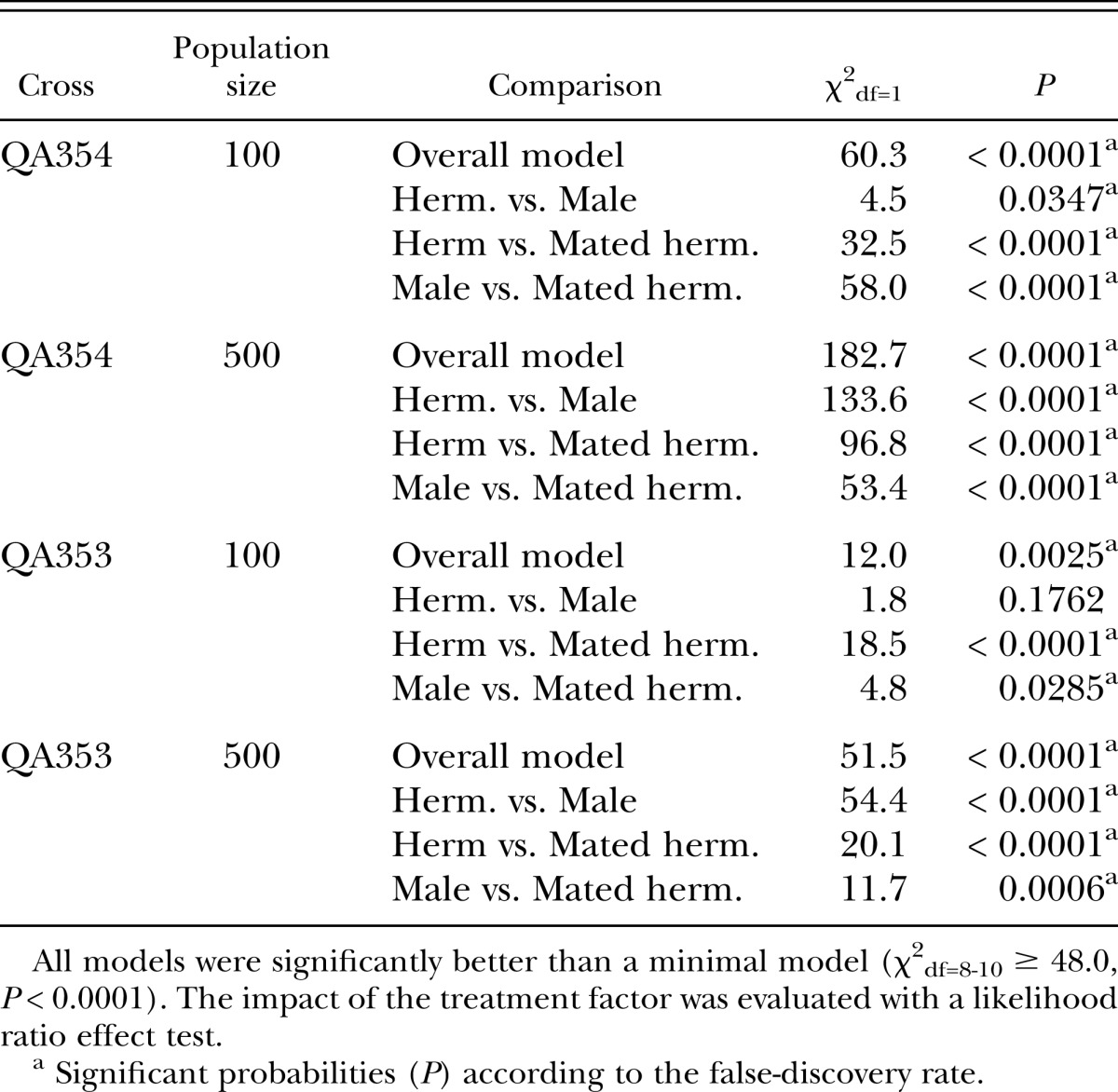
We added individual worms marked with a gfp reporter gene (strains QA351 and QA353) to plates with 500 hermaphrodites to mimic an event of an invasion or the occurrence of a spontaneous mutation (Invasion Experiment 1). Consistent with the expectation, gfp positive worms reached higher frequencies when the reporter gene was brought to the population through a male or a mated hermaphrodite than through a non-mated hermaphrodite (Fig. 1, Table 1). Next, we repeated the experiment with QA353 with two different population sizes (100 and 500), and we also included QA354, a strain with essentially the genetic background of the more efficiently mating strain CB4856 (Wegewitz et al., 2008) as invader (Fig. 2, Table 2). In all four treatments, mated hermaphrodites did significantly better than non-mated hermaphrodites. Interestingly, males did better in the larger populations of 500 than in the smaller populations of 100 individuals. A possible explanation for this is that the reproductive success of the males might have been limited by a low rate of finding suitable mates (young adult hermaphrodites) in the 100-individuals mixed-stage populations. Males must mate with multiple hermaphrodites to maximize their reproductive potential (Wegewitz et al., 2008), and it has been suggested that mate encounter rates are an important factor for male reproductive success (Lopes et al., 2008). In contrast, hermaphrodites – mated or unmated – could immediately reproduce and thus contribute to the next generation of these small populations.
Fig. 1.
Invasion Experiment 1: Proportion of GFP-positive worms (Y-axis) over time (X-axis) after the addition of a single GFP marked (QA351 [A] or QA353 [B]) virgin hermaphrodite (circles), male (triangles) or mated hermaphrodite (squares) to a population of 500 N2 worms on day 0. Every three days the populations were reduced to 500. GFP-positive worms were counted immediately before the reduction of the population. The error bars designate standard errors. Each point is the average of 8 independent measurements. For the statistical analysis see Table 1.
Table 1.
Statistical analysis of the effect of treatment on the proportion of GFP-positive offspring in Invasion Experiment 1.
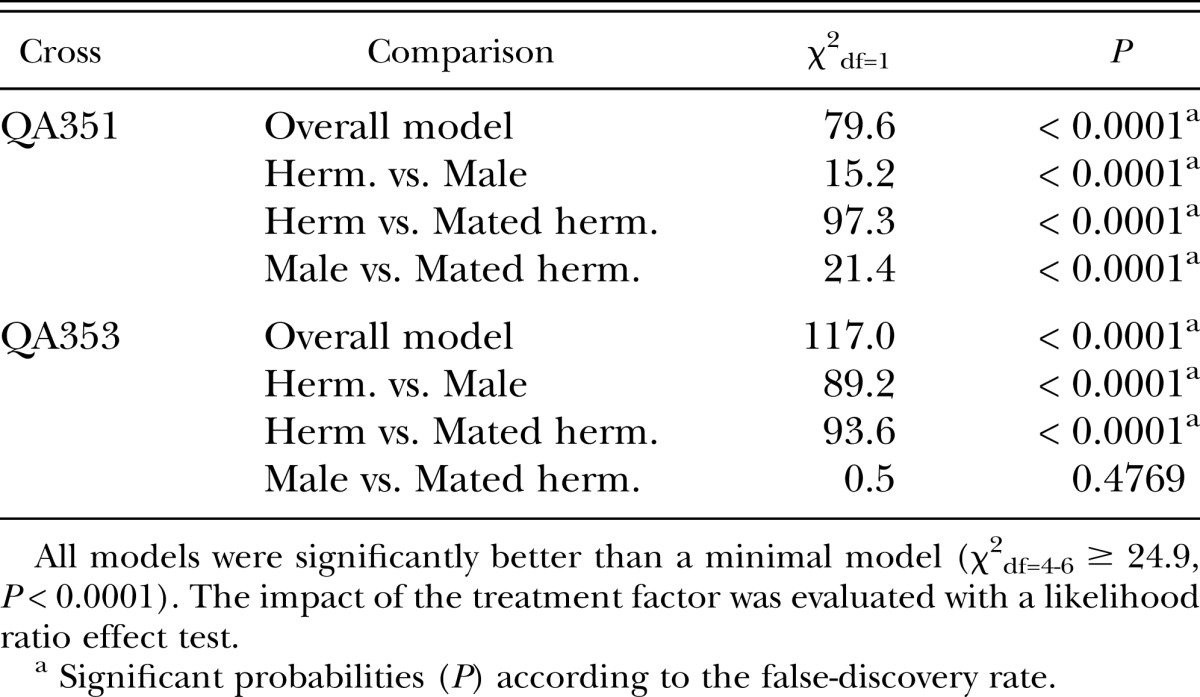
Fig. 2.
Invasion Experiment 2: Proportion of GFP-positive worms (Y-axis) over time (X-axis) after the addition of a single GFP marked virgin hermaphrodite (circles), male (triangles) or mated hermaphrodite (squares) to a population of 500 (A, B) or 100 (C, D) N2 worms on day 0. The added worms were of strain QA354 (essentially the genetic background of CB4856; A, C) or QA353 (essentially the genetic background of N2; B, D). The error bars designate standard errors. Each point is the average of 6 independent measurements. For the statistical analysis see Table 2.
Table 2.
Statistical analysis of the effect of treatment on the proportion of GFP-positive offspring in Invasion Experiment 2.
For the purpose of this experiment, we considered the myo-2::gfp a neutral genetic marker. Although we did not observe any obvious deleterious effects of the transgene, we cannot exclude a slight selective disadvantage for worms that carry the marker. However, this effect would have been the same for all worms that were compared directly (i.e. the transgenic males, the transgenic unmated hermaphrodite, and the mated hermaphrodite). Therefore, while the transgene might have slightly affected the absolute numbers, it would not have compromised the comparison. As expected after the addition of a single invader, the frequency of the introduced gene remained rather low, and in several cases it disappeared across time through genetic drift, especially if the invader was a virgin hermaphrodite. In particular, the GFP marker disappeared by the end of the experiment in all of the 32 replicated 500-individual populations with labeled, virgin hermaphrodites of any strain, whereas with labeled males the marker was lost in only 8 out of 32 and with labeled mated hermaphrodites in 6 out of 32 cases. For the population size of 100, the marker was completely lost in 9 out of 12 replicated populations with virgin hermaphrodites, 6 out of 12 with males and 1 out of 12 with mated hermaphrodites. Nevertheless, our results illustrate that rare males in a hermaphroditic population cause an increase in frequency of their alleles in the next generations. This is obviously also the case for genes involved in male formation and development, which are necessarily functional if they occur in a male. If the occasional boost of frequency of functional alleles of these genes caused by the sporadic males is large enough to offset the loss of functional alleles by mutational degradation and drift (which is expected to happen in hermaphrodites), this might be sufficient to maintain the genetic machinery for the production of males, even if there is no fitness advantage of out-crossing for hermaphrodites as has been proposed by Chasnov (2002). However, it might also be advantageous for an individual hermaphrodite to produce males, by allowing X-chromosome non-disjunctions or by mating, as long as the hermaphrodite density is high and the frequency of males is low, because this should indirectly lead to an increase of the frequency of the hermaphrodite's alleles. This advantage is expected to be strongly enhanced if novel environmental conditions (i.e. new selective constraints) can be expected to favor new phenotypes, because such new phenotypes are more rapidly produced through out-crossing and recombination than a series of mutations (Maynard-Smith 1978; Bell 1982). This notion recently was supported experimentally (Morran et al., 2009b).
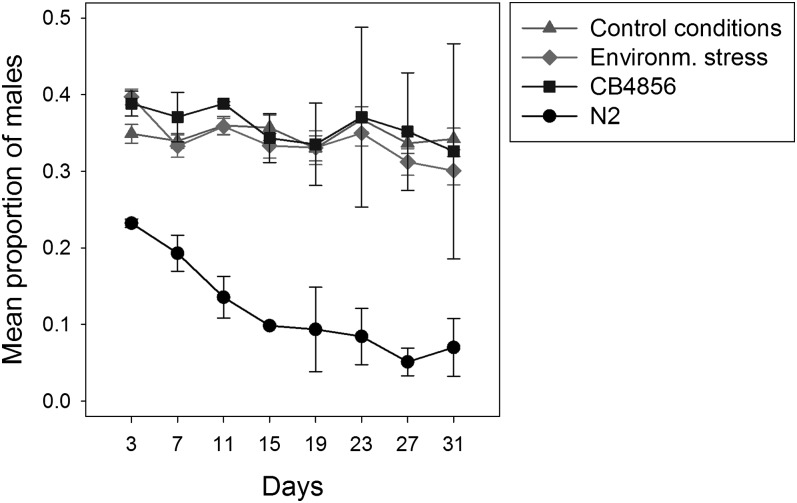
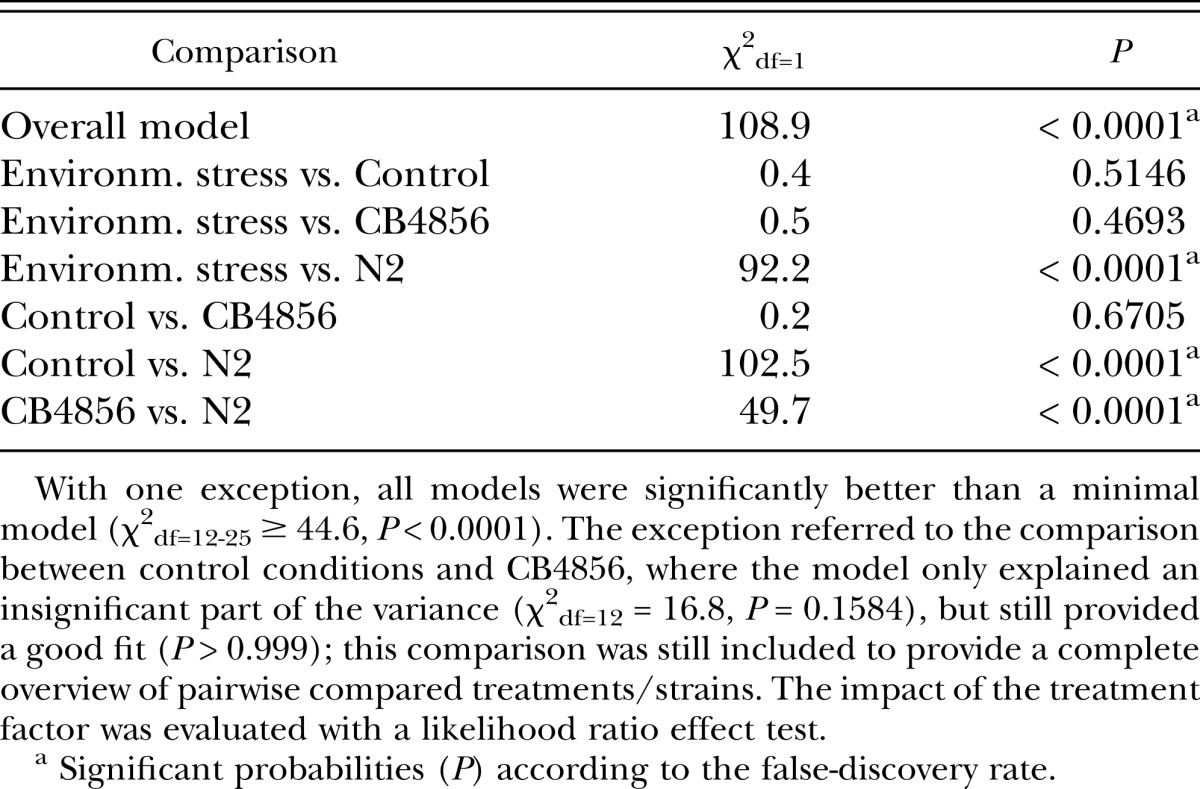
Our prior work has shown that under standard laboratory conditions, different strains of C. elegans lose or maintain males at very different rates and levels (Wegewitz et al., 2008). This indicates that male maintenance is, at least in part, genetically determined and is therefore a selectable trait. In order to address the question if changing environmental stress conditions represent a selective pressure in favor of more outcrossing, we subjected worm populations to alternating conditions of high salt, low temperature, high temperature, and pathogenic bacteria or permanent standard laboratory conditions as control (Experimental Evolution Experiment). The conditions were changed every four days, which is slightly longer than one generation-time under standard laboratory conditions. Genetically heterogeneous starting populations were generated by interbreeding the strains N2 (low male maintenance) and CB4856 (high male maintenance). After five cycles of selection (corresponding to about 20 generations), we analyzed the male maintenance of the resulting populations under standard laboratory conditions (Fig. 3, Table 3). There was no difference between the selection and the control treatments. However, both control and selection populations had adopted a high male maintenance, which was significantly different from N2 but indistinguishable from CB4856.
Fig. 3.
Persistence of males over time in populations after the Experimental Evolution Experiment. The mean proportion of males (Y-axis) over time (X-axis) is given. Every four days the populations were reduced to 150 individuals and transferred to new plates. The error bars indicate standard errors. All experiments were started with populations containing approximately 50% males. The first actual measurement was done after the first generation at day 3. For the experimental (diamonds) and the control (triangles) treatments each point is the average of two independent measurements for each of the 10 replicates of the selection experiment (total of 20 data points per treatment and time point). Two independent, male maintenance assays for each of N2 (circles) and CB4856 (squares) were done in parallel as experimental controls. The average of these two measurements is shown. For the statistical analysis see Table 3.
Table 3.
Statistical analysis of the effect of treatment/strains on the proportion of males in the Experimental Evolution Experiment.
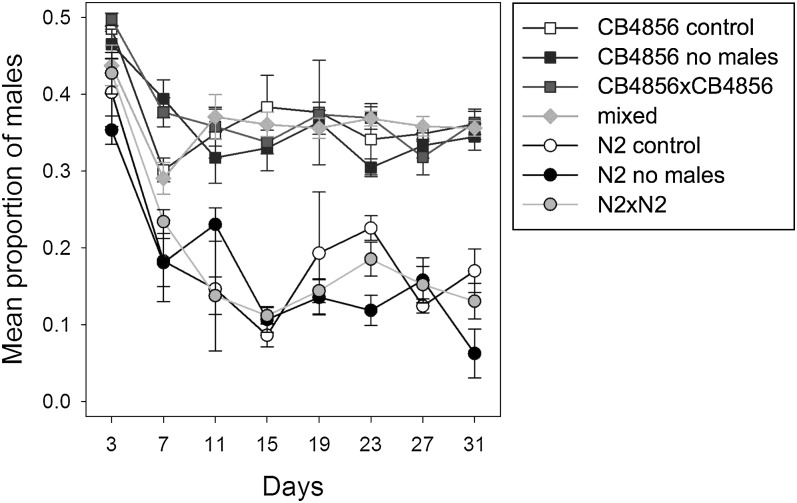
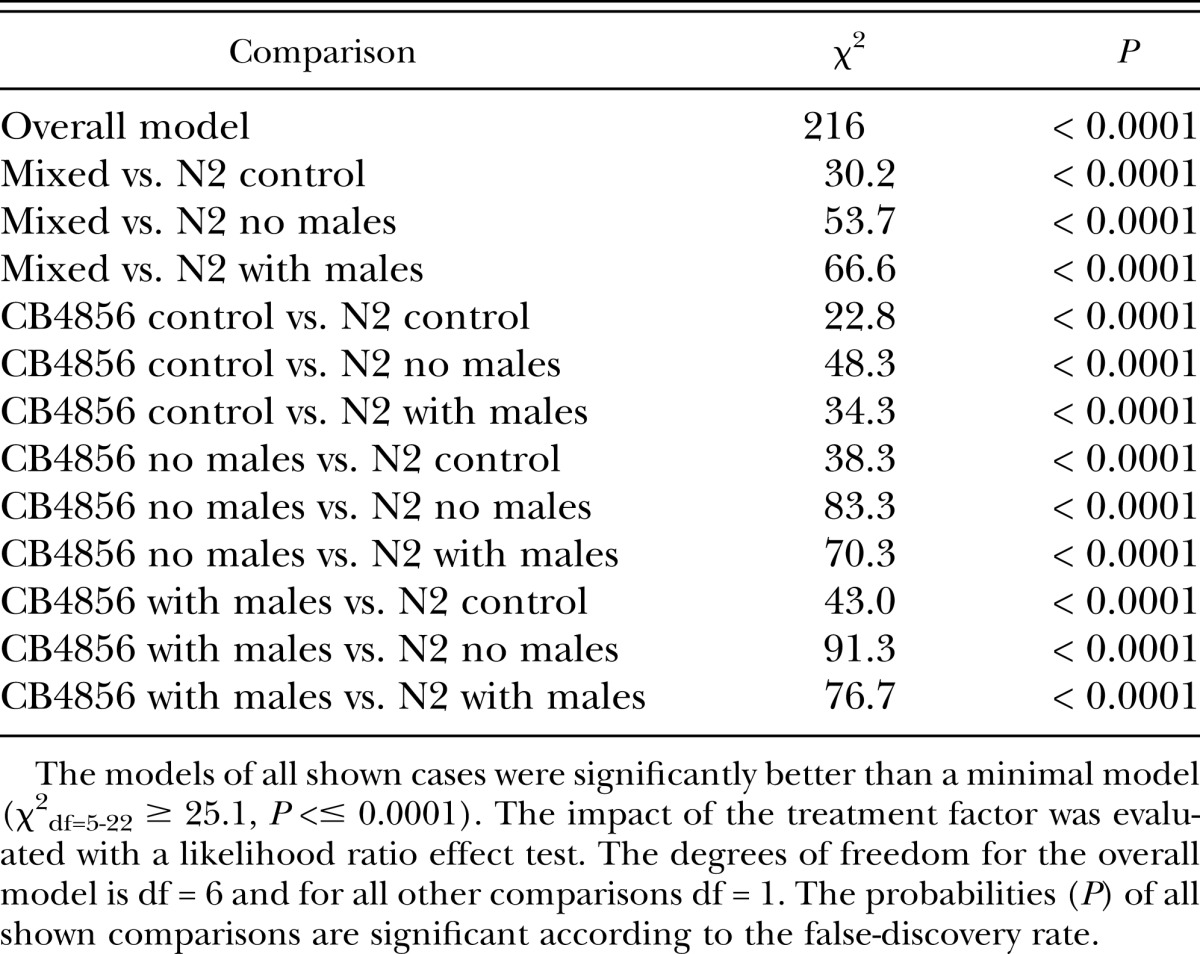
Since during the Experimental Evolution Experiment small numbers of males of both parental strains were added to prevent stochastic loss of genotypes, we suspected that this might have led to the dominance of the CB4856-like phenotype. To address this, we repeated the control experiment in a simplified form, without the periodic addition of males during the experiment (Chunking Experiment). Again, all heterogeneous populations assumed a high male maintenance significantly different from N2 and undistinguishable from CB4856 (Fig. 4, Table 4). This result indicates that under standard laboratory conditions, subpopulations that behave like CB4856 with respect to male maintenance are selected for from N2 x CB4856 hybrid populations, at least if males are present in the cultures. The higher maintenance of males itself does not need to be the selected trait but it may be the consequence of selection for the CB4856 variant at closely linked loci. The high male maintenance might also be independent of the environment and result from intrinsic factors. Male maintenance may be influenced by a fairly large number of loci, and only if all or most of them are N2-derived, low male maintenance occurs. If this is the case, the likelihood of recreating this situation from a mixed population by chance is very small. The low male maintenance in the standard laboratory strain N2 might then be the result of decade-long selection by geneticists, who prefer their strains to self-reproduce, unless mated deliberately. Alternatively, the relevant locus might reside in a region of genetic incompatibility between the two strains, such that preferentially individuals survive, which are homozygous for CB4856 derived genetic material in the particular region. We consider this explanation rather unlikely. These two strains are used extensively in the C. elegans field for genetic mapping and QTL analysis. Nevertheles, only one single region of partial genetic incompatibility between these two strains was found (Seidel et al., 2008). It affects an interval on chromosome I and clearly favors the N2-derived genetic material in this region.
Fig. 4.
Persistence of males over time in populations after the Chunking Experiment. The mean proportion of males (Y-axis) over time (X-axis) is given. Every four days the populations were reduced to 150 individuals and transferred to new plates. The error bars indicate standard errors. All experiments were started with populations containing approximately 50% males. The first actual measurement was done after the first generation at day 3. For the experimental and the control treatments each point is the average of two independent measurements for each of the 3 replicates of the Chunking Experiment. Two independent, male maintenance assays for each of N2 and CB4856 were done in parallel as experimental controls. The average of these two measurements is shown. CB4856 control (white squares); treatment CB4856 no males, (black squares); treatment CB4856 with males (grey squares); treatment mixed (grey diamonds); N2 control (white circles); treatment N2 no males (black circles); treatment N2 with males (grey circles).
Table 4.
Significant results for the statistical analysis of treatment effects on the proportion of males during the Chunking Experiment.
It still remains to be addressed in the future whether higher outcrossing rates generally allow a population to adapt to new selective constraints more rapidly, as the recent findings suggest for pathogens (Morran et al., 2009b), and if this effect is indeed the decisive driving force behind the continuous existence of males in C. elegans.
Footnotes
Some of the C. elegans strains were supplied by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. The contributions of HS and AS to this manuscript should be considered equal. This work was funded by the Max Planck Society and grant SCHU1415/5-1 from the Deutsche Forschungsgesellschaft to HS.
This paper was edited by Amy Treonis.
Literature Cited
- Agrawal AF, Lively CM. Parasites and the evolution of self-fertilization. Evolution: International Journal of Organic Evolution. 2001;55:869–879. doi: 10.1554/0014-3820(2001)055[0869:pateos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Current Biology. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics. 2007;176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. Berkeley: University of California Press; 1982. Masterpiece of Nature: The evolution of sexuality. [Google Scholar]
- Charlesworth B, Charlesworth D. Some evolutionary consequences of deleterious mutations. Genetica. 1998;102–103:3–19. [PubMed] [Google Scholar]
- Charnov EL, Smith JM, Bull JJ. Why be an hermaphrodite? Nature. 1976;263:125–126. [Google Scholar]
- Chasnov JR, Chow KL. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics. 2002;160:983–994. doi: 10.1093/genetics/160.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. Mutation and the experimental evolution of outcrossing in Caenorhabditis elegans. Journal of Evolutionary Biology. 2005;18:27–34. doi: 10.1111/j.1420-9101.2004.00804.x. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Aviles L, Ward S. The proximate determinants of sex ratio in C. elegans populations. Genetics Research. 2003;81:91–102. doi: 10.1017/s001667230300613x. [DOI] [PubMed] [Google Scholar]
- Fischer RA. Oxford: Clarendon Press; 1930. The genetical theory of natural selection. [Google Scholar]
- Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Molecular and Cellular Biology. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Schungel M, Putz A, Muller S, Hasert B, Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Molecular biology and evolution. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA. Background on Caenorhabditis elegans. In: Hope IA, editor. C. elegans a practical approach. Oxford: Oxford University Press; 1999. pp. 1–15. [Google Scholar]
- Jin Y. Transformation. In: Hope IA, editor. C. elegans a practical approach. Oxford: Oxford University Press; 1999. pp. 69–96. [Google Scholar]
- Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proceedings of the Royal Society - Biological Sciences. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proceedings of the Royal Society - Biological Sciences. 1999;266:263–267. doi: 10.1098/rspb.1999.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proceedings of the Royal Society - Biological Sciences. 2002;269:1125–1128. doi: 10.1098/rspb.2002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L, Cutter AD. On the potential for extinction by Muller's ratchet in Caenorhabditis elegans. BMC Evolutionary Biology. 2008;8:125. doi: 10.1186/1471-2148-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes PC, Sucena E, Santos ME, Magalhaes S. Rapid experimental evolution of pesticide resistance in C. elegans entails no costs and affects the mating system. PLoS ONE. 2008;3:e3741. doi: 10.1371/journal.pone.0003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel D, Carvalho S, Phillips PC, Teotonio H. Selection against males in Caenorhabditis elegans under two mutational treatments. Proceedings of the Royal Society - Biological Sciences. 2007;274:417–424. doi: 10.1098/rspb.2006.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer WE, Herrmann M, Sommer RJ. Phylogeny of the nematode genus Pristionchus and implications for biodiversity, biogeography and the evolution of hermaphroditism. BMC Evolutionary Biology. 2007;7:104. doi: 10.1186/1471-2148-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J. Cambridge: Cambridge University Press; 1978. The evolution of sex. [Google Scholar]
- Morran LT, Cappy BJ, Anderson JL, Phillips PC. Sexual Partners for the Stressed: Facultative Outcrossing in the Self-Fertilizing Nematode C. elegans. Evolution. international journal of organic evolution. 2009a;63:1473–1482. doi: 10.1111/j.1558-5646.2009.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009b;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Some genetic aspects of sex. American Naturalist. 1932;66:118–138. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advantage. Mutation Research. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Pannell JR. The evolution and maintenance of Androdioecy. Annual Review of Ecology and Systematics. 2002;33:397–425. [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A, Hey J. Population genetics of Caenorhabditis elegans: The paradox of low polymorphism in a widespread species. Genetics. 2003;163:147–157. doi: 10.1093/genetics/163.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A, Hey J. Sampling from natural populations with RNAI reveals high outcrossing and population structure in Caenorhabditis elegans. Current Biology. 2005;15:1598–1602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Stewart AD, Phillips PC. Selection and maintenance of androdioecy in Caenorhabditis elegans. Genetics. 2002;160:975–982. doi: 10.1093/genetics/160.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. In: Hope IA, editor. C. elegans a practical approach. Oxford: Oxford University Press; 1999. pp. 51–67. [Google Scholar]
- Teotonio H, Manoel D, Phillips PC. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution: International Journal of Organic Evolution. 2006;60:1300–1305. [PubMed] [Google Scholar]
- Wegewitz V, Schulenburg H, Streit A. Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda) BMC Ecology. 2008;8:12. doi: 10.1186/1472-6785-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. Introduction to C. elegans Biology. In: Wood WB, editor. The Nematode Caenorhabditis elegans. New York: Cold Spring Harbor Laboratory Press; 1988. pp. 1–16. [Google Scholar]