Abstract
The objective of this work was to isolate and identify fungi associated with R. reniformis in cotton roots. Soil samples were collected in cotton fields naturally infested with R. reniformis and from cotton stock plants cultured in the greenhouse. Nematodes extracted from the soil were observed under the stereoscope, and discolored eggs and vermiform stages colonized with mycelia were cultured on 1.5% water agar supplemented with antibiotics, and incubated at 27°C. Identification of the nematophagous fungi was based on the morphological characters, and the ITS regions and 5.8S rDNA amplified by PCR using the primers ITS1 and ITS4. The parasitism percentage on vermiform nematodes from greenhouse samples was 21.2%, and the percentages from cotton fields in Limestone, Henry, and Baldwin counties in Alabama were 3%, 23.2%, and 5.6%, respectively. A total of 12 fungi were identified from R. reniformis vermiform stages and eggs. The most frequently isolated fungi were Arthrobotrys dactyloides (46%) and Paecilomyces lilacinus (14%), followed by Phoma exigua (4.8%), Penicillium waksmanii and Dactylaria brochophaga (3.6%), Aspergillus glaucus group (2.4%). Cladosporium herbarum, Cladosporium cladiosporioides, Fusarium oxysporum, Torula herbarum, Aspergillus fumigatus, and an unidentified basidiomycete were less frequent (1.2%). A high percentage (16.8%) of fungi from colonized nematodes was not cultivable on our media. Out of those 12 fungi, only four have been previously reported as nematophagous fungi: three isolates of Arthrobotrys dactyloides, and one isolate of Dactylaria brochopaga, Paecilomyces lilacinus, and Fusarium oxysporum. Molecular identification of Arthrobotrys dactyloides and Dactylaria brochopaga was consistent with the morphological identification, placing these two fungi in the new genus Drechslerella as proposed in the new Orbilaceae classification.
Keywords: Arthrobotrys dactyloides, Dactylaria brochopaga, Paecilomyces lilacinus, reniform nematode, Rotylenchulus reniformis
Rotylenchulus reniformis is a serious problem in cotton production in the southeastern United States, causing an economic loss of $98 million per year (Blasingame et al., 2007). There are no commercial cotton varieties resistant to R. reniformis (Weaver et al., 2007; Starr et al., 2007; Usery et al., 2005); consequently, management is based on the application of nematicides, such as aldicarb, oxamyl, 1,3-dichloropropene, thiodicarb, and metam sodium (Koenning et al., 2007; Lawrence et al., 2005; Kinloch and Rich, 2001; Lawrence et al., 1990), and crop rotation with corn, peanut, sorghum, and resistant cultivars of soybean (Davis et al., 2003; Gazaway et al., 2000). The economic cost of managing this nematode pest indicates the need to explore new management options that can be integrated in sustainable systems. Biological control options for R. reniformis nematode management are not currently available in the United States (Robinson, 2007).
Numerous studies of fungi colonizing plant-parasitic nematodes have been reported on soybean cyst (Heterodera glycines) and root-knot nematodes (Meloidogyne spp.) (Kim and Riggs, 1991; Carris and Glawe, 1989; Jatala, 1986). In the case of R. reniformis, suppressive soils from five fields in the Lower Rio Grande Valley (LRGV) of Texas and five fields in Louisiana have been reported. In the LRGV the suppression of R. reniformis populations was as great as 95%, but the organisms involved in this suppression were not identified (Robinson et al., 2008). In other studies, nematophagous fungi have been isolated and evaluated on different life stages of R. reniformis. Paecilomyces lilacinus significantly reduced numbers of R. reniformis eggs under greenhouse conditions (Walters and Barker, 1994). Pochonia chlamydospora reduced numbers of R. reniformis on cotton roots or in soil after a single application of 5,000 chlamydospores per gram of soil (Wang et al., 2005). Furthermore, a fungus known as Arkansas Fungus (ARF) parasitized 17-21% of R. reniformis eggs (Wang et al., 2004). For the juvenile stages, Arthrographis sp., Pseudorobillarda sp., and Fusarium equiseti significantly reduced R. reniformis population development on cotton (McLean et al., 2000).
Taxonomy of nematophagous fungi is being redefined with the development of new molecular techniques. In the past, identification of ring-trapping fungi species was based on the morphology of the conidia and conidiophores (Drechsler, 1937), however Scholler et al. (1999) reclassified the Orbiliaceae genera based on the ITS1, ITS2, and 5.8S rDNA sequences, demonstrating that trapping devices are more informative than other morphological structures.
Based on the life cycle of R. reniformis (semi-endoparasitic), we hypothesize that the nematodes are constantly exposed to many antagonists and other organisms in the rhizosphere. The objective of this study was to isolate and identify nematophagous fungi which parasitize R. reniformis vermiform life stages and eggs. Fungal isolates were identified by morphological characters and molecular techniques based on the sequence of the ITS and 5.8S rDNA regions.
Materials and Methods
Collection of nematode samples: Rotylenchulus reniformis nematodes were collected from roots of cotton plants grown to maintain stock cultures of nematodes at the Plant Science Research Center of the Alabama Agricultural Experiment Station on the campus of Auburn University. Continuous long-term culture of cotton with R. reniformis without repotting increased populations of antagonistic fungi against this nematode in our greenhouse cultures. These nematodes were cultured in 15-cm-diam polystyrene pots which had each been inoculated with 2,000 eggs/500 cm3. The cotton was grown in the greenhouse for 6-18 months. Rotylenchulus reniformis nematode samples were also collected from R. reniformis infested cotton fields from three cotton production counties of Alabama (Limestone, Henry, and Baldwin). Approximately 1,000 cm3 of soil were collected to a depth of 15 cm in the root zone of the cotton plants during the growing season from June through August. Soils were sealed in plastic bags, labeled, and stored in a cooler for transport to the greenhouse. Vermiform adults and nematode juveniles were extracted from the soil by a modified gravity screening and centrifugation-flotation method (Jenkins, 1964). Eggs were extracted from cotton roots by shaking the roots in 1% NaOCl for 4 minutes at 120 rpm and sieving the solution (Hussey and Baker, 1973).
Nematode samples were observed at 6.3× total magnification under the stereoscope (Nikon SMZ800), where the vermiform stages with trapping-rings and healthy one were counted to determine the fungal parasitism percentage. Discolored eggs of R. reniformis and vermiform life stages colonized with fungal mycelia were carefully removed from the counting dish, placed in a syracuse dish, and rinsed with sterile water. Colonized nematodes and eggs were aseptically cultured on 1.5% water agar supplemented with 12.5 mg of chlortetracycline HCl and 300 mg of streptomycin sulfate per liter (Kim and Riggs, 1994). Cultures were incubated at 27°C and identified after 5 days. Hyphal tips from the fungi growing from the eggs and vermiform life stages of the nematodes were aseptically transferred to a Potato Dextrose Agar (PDA/SA) (Sigma Chemical Co.) culture plates supplemented with 300 mg/liter of streptomycin sulfate (Sigma Chemical Co.) to establish pure cultures.
Morphological identification: Identification of the nematophagous fungi was based on the morphological characteristics of conidiophores and conidia (Nelson et al., 1983, Gerlach and Nirenberg, 1982, Domsch et al., 1980, Barron, 1977 and Drechsler, 1937). For some fungi, it was necessary to induce sporulation on water agar media (WA) by adding 20 R. reniformis vermiform stages to each plate. Sporulation was also induced in some cultures by exposing fungal mycelium to a black light lamp (Model X-15B 115 volts 60Hz).
Molecular identification of fungi: Identification of fungi isolated from R. reniformis and previously reported as nematophagous fungi was confirmed by sequencing of the internal transcribed spacer (ITS) regions 1 and 2, including the 5.8 rDNA. Fungal isolates were grown on PDA/SA at 27°C for 7 days. After this period, mycelia were harvested, placed in a mortar, frozen with liquid nitrogen, and transferred to an eppendorf tube without letting the sample thaw. DNA was extracted using the Dneasy Plant Mini Kit® (Quiagen Inc.). PCR reactions of 50 μL were prepared using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al.,1998). PCR conditions included an initial denaturation for 3 min at 95°C, 35 cycles of 1 min at 95°C, annealing for 40 s at 54°C, extension for 40 s at 72°C, and final extension for 10 min at 72°C. PCR amplification products were purified with QIAquick columns (Quiagen Inc.) according to manufacturer's instructions. The resulting amplified products were sequenced at the Auburn University Genomics facility using the same ITS1 and ITS4 primers.
Sequences were manually edited using Chromas Lite 2.01 software (http://www.technelsyum.com.au). Alignments were done with ClustalW software (Tamura et al., 2007), and then consensus sequences were subjected to Blast analysis in National Center for Biotechnology Information (NCBI) using blastn option. Phylogenetic analyses were performed using Neighbor joining algorithm in Mega 4.1 software (Tamura et al., 2007).
Results
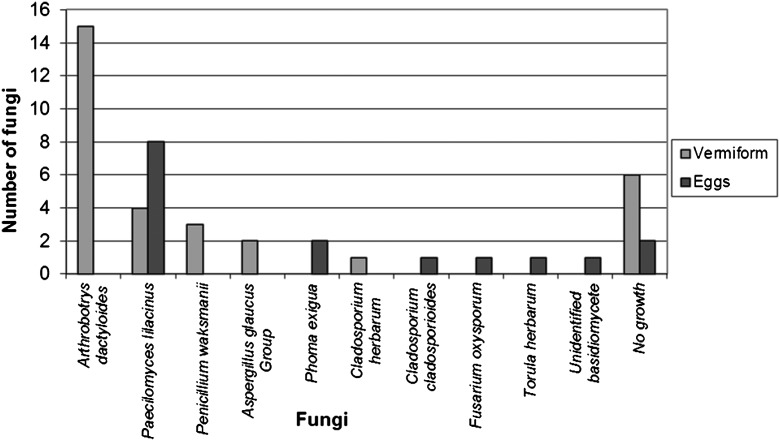
Morphological identification: The percentage of parasitized vermiform nematodes from greenhouse samples was 21.2%, and the percentages from Limestone, Henry, and Baldwin counties were 3%, 23.2%, and 5.6%, respectively. In the greenhouse samples, number of fungi associated with vermiform life stages were Arthrobotrys dactyloides (15 isolates), Paecilomyces lilacinus (4 isolates), Penicillium waksmanii (3 isolates), Aspergillus glaucus Group (2 isolates), and Cladosporium herbarum (1 isolate) (Fig. 1). No fungal growth was observed from 6 vermiforms visually colonized, and cultured. Numbers of fungi associated with eggs were Paecilomyces lilacinus (8 isolates), Phoma exigua (2 isolates), Cladosporium cladosporioides (1 isolate), Fusarium oxysporum (1 isolate), Torula herbarum (1 isolate), and an unidentified basidiomycete (1 isolate). No fungal cultures were found in 2 discolored eggs plated (Fig. 1).
Fig. 1.
Number of fungi isolated from symptomatic Rotylenchulus reniformis greenhouse samples.
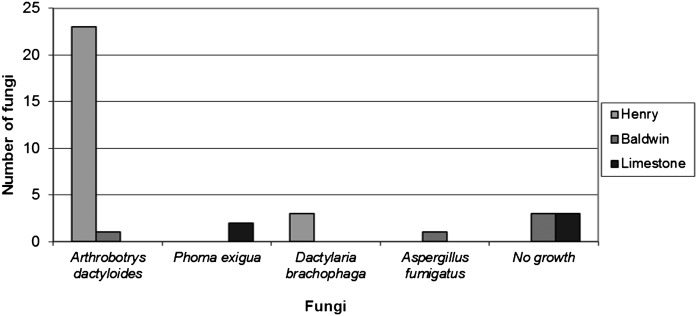
From the Limestone field location, only Phoma exigua (2 isolates) was associated with vermiform stages of R. reniformis; however, three of the colonized vermiform nematodes did not produce any fungal growth in PDA media with antibiotics (Fig. 2). The most frequent fungi founded in the colonized nematodes from Henry field were Arthrobotrys dactyloides (23 isolates), and Dactylaria brochophaga (3 isolates). In the Baldwin location Arthrobotrys dactyloides was also isolated along with Aspergillus fumigatus (1 isolate). However, in the Baldwin location, three of the cultured nematodes did not produce fungal growth on culture medium (Fig. 2).
Fig. 2.
Fungi isolated from symptomatic Rotylenchulus reniformis vermiform life stages from the field locations.
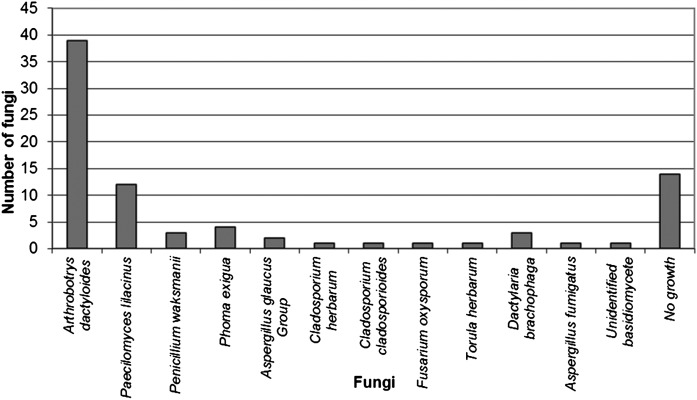
From all the samples taken, the most frequently isolated fungi from R. reniformis were A. dactyloides (39 isolates) and P. lilacinus (12 isolates) (Fig. 3). Followed by Phoma exigua (4 isolates), Penicillium waksmanii and D. brachophaga (3 isolates), Aspergillus glaucus group (2 isolates), and C. cladiosporioides, C. herbarum, F. oxysporum, T. herbarum, D. brachophaga, A. fumigatus, and unidentified basidiomycete were less frequent (1 isolate). None the less, 14 of of the fungal colonized nematodes cultured did not produce fungal culture on the media (Fig. 3).
Fig. 3.
Percent frequency of occurrence of the total fungi isolated from symptomatic Rotylenchulus reniformis from all samples.
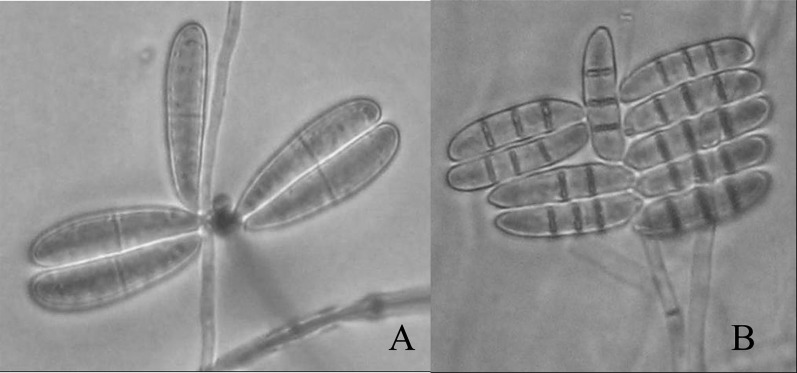
Morphological characteristics: Arthrobotrys dactyloides is a nematode-trapping fungus that produces constricting rings to capture nematodes. The rings were 20 - 32 μm diameter, composed of 3 individual arcuate cells. Conidia are ellipsoidal, 40 to 48 μm long, 8 to 9 μm wide, with 2 cells equally divided by one septum (Fig. 4A), which is the main morphological characteristics that differentiated it from A. anchonia. Chlamydospores have been reported on this fungus (Haard, 1968), but they were not observed in any of our strains. Dactylaria brochophaga is another nematode-trapping fungus producing constricting rings of 20 to 35μm in diameter with 3 arcuate cells. Conidia are ellipsoidal, slightly curved, 30 to 40 μm long, 5 to 8 μm wide, and were divided into 4 cells by 3 septa (Fig. 4B).
Fig. 4.
Conidia of Arthrobotrys dactyloides (A), and Dactylaria brachophaga (B) (x100).
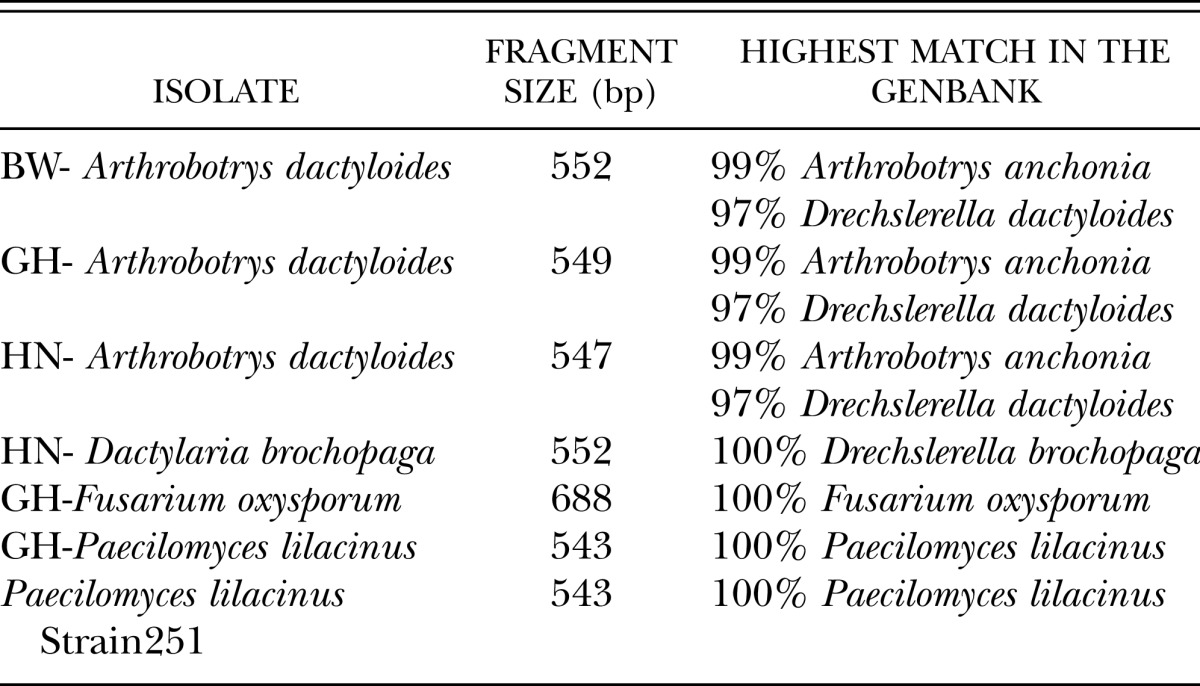
Molecular identification of fungi: The morphological identification of the fungi reported above as nematophagous fungi was confirmed by sequencing of the ITS1 – 5.8 rDNA – ITS2 region. The three strains of Arthrobotrys dactyloides (GH-A. dactyloides, HN-A. dactyloides, BW-A. dactyloides) displayed 99% similarity with Arthrobotrys anchonia (AY965753.1) and 97% with Drechslerella dactyloides (AY773463.1). Paecilomyces lilacinus and Fusarium oxysporum isolates presented a 100% coverage of the sequences and a 100% identity with P. lilacinus (EU306174.1) and F. oxysporum (FJ605247.1) when blast in the GenBank (Table 1).
Table 1.
Relationship of fungal isolates colonizing Rotylenchulus reniformis with other fungi from the blast analysis in GenBank.
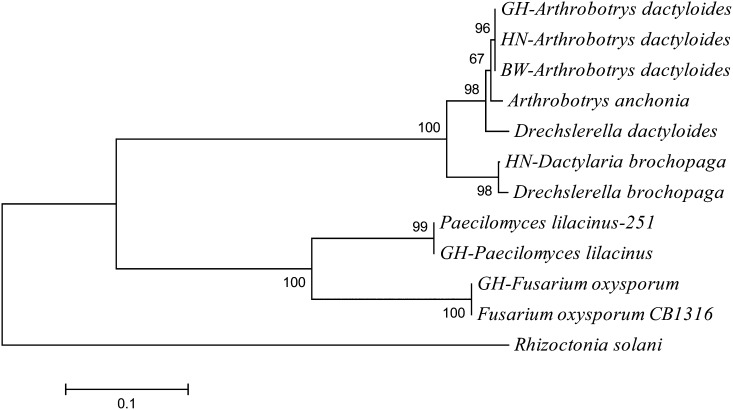
All of the isolates of Arthrobotrys dactyloides group were in a clade in the phylogram, and grouped closely to Arthrobotrys anchonia, and Drechslerella dactyloides. HN-Dactylaria brochopaga grouped with Drechslerella brochopaga. The isolates of P. lilacinus grouped together in a monophyletic clade. Also, GH-Fusarium oxysporum grouped together with a biocontrol strain. Rhizoctonia solani was used in this graph as an outgroup (Fig. 5).
Fig. 5.
Phylogram from the fungi isolated from Rotylenchulus reniformis nematodes based on ITS and 5.8s rDNA (Neighbor-joining algorithm).
Discussion
Long-term cultures of R. reniformis maintained in cotton plants in the greenhouse increased the population of fungi antagonistic to reniform nematode. Parasitism of the vermiform stages of reniform nematodes was higher on plants in the greenhouse than in the field. Wang et al. (2004), also obtained nematode-antagonisitic fungal isolates from cotton field soil maintained under greenhouse conditions. Of all the fungi isolated from all locations, only Arthrobotrys dactyloides, D. brachophaga, P. lilacinus, and F. oxysporum have been reported as pathogens on other nematode species (Kiewnick and Sikora, 2006; Stirling and Smith, 1998; Freitas et al., 1995). The most predominant nematode-trapping fungus was A. dactyloides, which was isolated from the greenhouse and two counties (Henry and Baldwin). Dactylaria brachophaga is closely related to A. dactyloides and was only identified in Henry County. Both fungi present white mycelium on PDA, however, the most abundant septation in D. brochopaga is the feature most decisively distinguishing it as species (Drechsler, 1937; Haard, 1968; Barron, 1977). Arthrobotrys dactyloides and D. brachophaga have been reported to reduce populations of Meloidogyne graminicola on rice (Singh et al., 2007). Additionally, Stirling and Smith (1998a) reported reductions in populations of Meloidogyne javanica on tomato roots using isolates of A. dactyloides. These two fungi have potential as biological control agents of juvenile stages, and commercial formulations of A. dactyloides have been attempted (Stirling et al., 1998b).
Paecilomyces lilacinus is typically a soilborne fungus and is common in the rhizosphere of a number of plants (Domsch et al., 1980). This fungus has been reported on R. reniformis on tomato plants, where it reduced the populations of the nematode in greenhouse and microplot trials (Walters and Barker, 1994). This fungus was also very predominant on the cotton plants used for culturing R. reniformis in the greenhouse.
Fusarium oxysporum has been previously reported to be destructive on eggs of the soybean cyst nematode in Alabama soybean fields (Morgan-Jones and Rodriguez-Kabana, 1981). There are some biotypes capable of penetrating eggs and causing disorders on the embryonic development through enzymatic and/or toxic effects (Morgan-Jones and Rodriguez- Kabana, 1988). This fungus has never been reported on R. reniformis so it is necessary to evaluate the biocontrol potential for this nematode.
Morphological and molecular fungal identification are in agreement for HN-D. brochopaga, GH-P. lilacinus, and GH-Fusarium oxysporum. Only the three strains of Arthrobotrys dactyloides showed differences between the two types of identification. Molecular identification indicated 99% of similarity to A. anchonia and 97% identity with A. dactyloides. Arthrobotrys dactyloides is closely related to Arthrobotrys anchonia, both are ring-trapping fungi, differing in the shape and size of the conidia. Arthrobotrys anchonia produce elongated ovoid conidia with unequal cells within each of the conidia, while the Arthrobotrys dactyloides conidia are ellipsoid with equal cell lengths (Haard, 1968). Therefore, our morphological observations of the three isolates showed ellipsoid conidia with equal cell size. This allowed us to confirm that the fungi we isolated are A. dactyloides. Thus, our isolates will be named Drechslerella dactyloides, based on the classification proposed by Scholler et al. 1999, which also renames A. brochopaga as Drechslerella brochopaga.
Overall, our results suggest that Arthrobotrys dactyloides, D. brochopaga, Fusarium oxysporum, and Paecilomyces lilacinus are pathogens of R. reniformis. Evaluations of these fungi under in vitro and greenhouse conditions will be performed to evaluate their biocontrol potential. Additionally, is it important to develop and evaluate different media and techniques for isolation and identification of fungi associated with colonized nematodes that did not grow on artificial media. There is a possibility that these fungi are endoparasitic fungi that complete their life cycle inside the nematode host, or bacteria parasitized on nematodes. The current research had identified several natural enemies of R. reniformis in different life stages of this nematode. The outcome of this research will provide a foundation to develop a biocontrol program that could be added to the actual management practices and improve the nematode control.
Literature Cited
- Barron GL. Canada: Lancaster Press; 1977. The nematode-destroying fungi. [Google Scholar]
- Blasingame D, Patel MV, Gazaway W, Olsen M, Kirkpatrick T, Davis M, Sprenkel RK, Kemerait B, Colyer P, Wrather A, Goldberg N, Koenning S, Banks JC, Muller J, Newman M, Woodward J, Phipps P. Cotton disease loss estimate committee report; Proceedings of the Beltwide Cotton Conferences of the National Cotton Council of America; 2007. www.cotton.org/beltwide/proceedings/2007. Confirmed on Jan. 4, 2008. [Google Scholar]
- Carris LM, Glawe DA. Bulletin 786. University of Illinois at Urbana-Chamapaig. College of Agriculture. Agricultural Experiment Station. U.S. Department of Agriculture; 1989. Fungi colonizing cyst of Heterodera glycines. [Google Scholar]
- Davis RF, Koenning SR, Kemerait RC, Cummings TD, Shurley WD. Rotylenchulus reniformis management in cotton with crop rotation. Journal of Nematology. 2003;35(1):58–64. [PMC free article] [PubMed] [Google Scholar]
- Domsch KH, Gams W, Traute-Heidi A. Compendium of Soil fungi – Volumen 1. London: Academic Press; 1980. [Google Scholar]
- Drechsler C. Some Hyphomycetes that prey on free-living terricolous nematodes. Mycologia. 1937;29:447–552. [Google Scholar]
- Freitas LG, Ferraz S, Muchovej JJ. Effectiveness of different isolates of Paecilomyces lilacinus and an isolate of Cylindrocarpon destructans on the control of Meloidogyne javanica. Nematropica. 1995;25:109–115. [Google Scholar]
- Gazaway WS, Akridge JR, McLean K. Proceedings Beltwide Conference National Cotton Council America. San Antonio, TX: 2000. Impact of various crop rotations and various winter cover crops on reniform nematode in cotton; pp. 162–163. [Google Scholar]
- Gerlach W, Nirenberg H. Biologische Bundesanstalt für Land – und Forstwirtschalt. Berlin-Dahlem: Institut für Mikrobiologie; 1982. The genus Fusarium – A Pictorial Atlas. [Google Scholar]
- Haard K. Taxonomic studies on the genus Arthrobotrys Corda. Mycologya. 1968;60:1140–1159. [Google Scholar]
- Hussey RS, Barker KR. A comparision of methods of collecting inocula for Meloidogyne spp. including a new technique. Plant Disease Report. 1973;57:1025–1028. [Google Scholar]
- Jatala P. Biological control of plant parasitic nematodes. Annual Review of Phytopathology. 1986;24:453–489. [Google Scholar]
- Jenkins WR. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Disease Reporter. 1964;48:692. [Google Scholar]
- Kiewnick S, Sikora RA. Biological control of the root-knot nematode Meloidogyne incognita by Paecilomyces lilacinus strain 251. Biological Control. 2006;38:179–187. [Google Scholar]
- Kim DG, Rigs D. Techniques for isolation and evaluation of fungal parasites of Heterodera glycines. Journal of Nematology. 1994;26(4S):592–595. [PMC free article] [PubMed] [Google Scholar]
- Kim DG, Rigs D. Characteristics and efficacy of the sterile hyphomycete (ARF18), a new biocontrol agent for Heterodera glycines and other nematodes. Journal of Nematology. 1991;23(3):275–282. [PMC free article] [PubMed] [Google Scholar]
- Kinloch RA, Rich JR. Management of Root-knot and reniform nematode in Ultra-narrow row cotton with 1,3-Dichloropropene. Supplement to the Journal of Nematology. 2001;33(4S):311–313. [PMC free article] [PubMed] [Google Scholar]
- Koenning SR, Morrison DE, Edmisten KL. Relative efficacy of selected nematicides for management of Rotylenchulus reniformis in cotton. Nematropica. 2007;37:227–235. [Google Scholar]
- Lawrence KS, Feng Y, Lawrence GW, Burmester CH, Norwood SH. Accelerated degradation of Aldicarb and its metabolites in cotton field soils. Journal of Nematology. 2005;37(2):190–197. [PMC free article] [PubMed] [Google Scholar]
- Lawrence GW, McLean KS, Batson WE, Miller D, Borbon JC. Response of Rotylenchulus reniformis to nematicide applications on cotton. Supplement to Journal of Nematology. 1990;22(4S):707–711. [PMC free article] [PubMed] [Google Scholar]
- McLean KS, Palmateer AJ, Morgan-Jones G. Fungal antagonists of Rotylenchulus reniformis. Proceedings of the Beltwide Cotton Conference. 2000;1:145–146. [Google Scholar]
- Morgan-Jones G, Rodriguez-Kabana R. Diseases of nematodes Volumen II. 1988. 1988. Fungi colonizing cyst and eggs. CRC Press. Edited by Poinar, G. O., Jansson, H. B. [Google Scholar]
- Morgan-Jones G, Rodriguez-Kabana R. Fungi associated with cyst of Heterodera glycines in Alabama soil. Nematropica. 1981;11:69–74. [Google Scholar]
- Nelson PE, Toussoun TA, Marasas WFO. The Pensilvania State University Press: University Park and London; 1983. Fusarium species – An illustrated manual for identification. [Google Scholar]
- Robinson AF. Nematode management in cotton. In: Ciancio A, Mukerji K, editors. Integrated management and biological control of vegetable and grain crops nematodes. Berlin: Springer; 2008. pp. 149–182. [Google Scholar]
- Robinson AF, Westphal A, Overstreet Ch., Padgett GB, Greenberg SM, Wheeler TA, Stetina SR. Detection of suppressiveness against Rotylenchulus reniformis in soil from cotton (Gossypium hirsutum) fields in Texas and Louisiana. Journal of Nematology. 2008;40(1):35–38. [PMC free article] [PubMed] [Google Scholar]
- Robinson AF. Reniform in the U.S. cotton: When, where, why, and some remedies. Annual Review of Phytopathology. 2007;45:263–288. doi: 10.1146/annurev.phyto.45.011107.143949. [DOI] [PubMed] [Google Scholar]
- Scholler M, Hagedorn G, Rubner A. A reevaluation of predatory orbilaceous fungi. II. A new generic concept. Sydowia. 1999;51(1):89–113. [Google Scholar]
- Singh KP, Jaiswal RK, Kumar K, Kumar D. Nematophagous fungi associated with root galls of rice caused by Meloidogyne graminicola and its control by Arthrobotrys dactyloides and Dactylaria brochopaga. Journal of Phytopathology. 2007;155:193–197. [Google Scholar]
- Starr JL, Koenning SR, Kirkpatrick TL, Robinson AF, Roberts PA, Nichols RL. The future of nematode management in cotton. Journal of Nematology. 2007;39(4):283–294. [PMC free article] [PubMed] [Google Scholar]
- Stirling GR, Smith LJ. Field test of formulated products containing either Verticillium chlamydosporium or Arthrobotrys dactyloides for biological control of the Root-knot nematodes. Biological Control. 1998a;11:231–239. [Google Scholar]
- Stirling GR, Smith LJ, Licastro KA, Eden LM. Control of Root-knot nematode with formulations of the nematode-trapping fungus Arthrobotrys dactyloides. Biological Control. 1998b;11:224–230. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Usery SR, Lawrence KS, Lawrence GW, Burmester CH. Evaluation of cotton cultivars and tolerance to Rotylenchulus reniformis. Nematropica. 2005;35:121–133. [Google Scholar]
- Walters SA, Barker KR. Efficacy of Paecilomyces lilacinus in suppressing Rotylenchulus reniformis on tomato. Supplement to Journal of Nematology. 1994;26(4S):600–605. [PMC free article] [PubMed] [Google Scholar]
- Wang K, Riggs RD, Crippen D. Isolation, selection, and efficacy of Pochonia chlamydosporia for control of Rotylenchulus reniformis on cotton. Phytopathology. 2005;95:890–893. doi: 10.1094/PHYTO-95-0890. [DOI] [PubMed] [Google Scholar]
- Wang K, Riggs RD, Crippen D. Suppression of Rotylenchulus reniformis on cotton by the nematophagous fungus ARF. Journal of Nematology. 2004;36(2):186–191. [PMC free article] [PubMed] [Google Scholar]
- Weaver DB, Lawrence KS, Van Santen E. Reniform nematode in Upland cotton germplasm. Crop Science. 2007;47:19–24. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1998. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp. 315–322. In: Innis, M. A., Gelfand, D. H., Sninsky, J. J., and White, T. J. eds. PCR protocols A guide to methods and applications. Academic Press, Inc. [Google Scholar]