Abstract
Post-translational modifications of histones are critical not only for local regulation of gene expression, but also for higher-order structure of the chromosome and genome organization in general. These modifications enable a preset state to be maintained over subsequent generations and thus provide an epigenetic level of regulation. Heterochromatic regions of the genome are epigenetically regulated to maintain a “silent state” and protein coding genes inserted into these regions are subject to the same epigenetic silencing. The fission yeast Schizosaccharomyces pombe has well characterized regions of heterochromatin and has proven to be a powerful model for elucidation of epigenetic silencing mechanisms. Research in S. pombe led to the breakthrough discovery that epigenetic silencing is not solely a chromatin-driven transcriptional repression and that RNA interference of nascent transcripts can guide epigenetic silencing and associated histone modifications. Over the last 10 years, an eloquent integration of genetic and biochemical studies have greatly propelled our understanding of major players and effector complexes for regulation of RNAi-mediated epigenetic silencing in S. pombe. Here, we review recent research related to regulation of the epigenetic state in S. pombe heterochromatin, focusing specifically on the mechanisms by which transcription and RNA processing interact with the chromatin modification machinery to maintain the epigenetically silent state.
Keywords: epigenetic silencing, fission yeast, heterochromatin, histone methyltransferase, RNA interference
Introduction
The organization of chromosomal DNA within the nucleus is achieved through direct interaction with a wide range of proteins. The basic unit of the DNA-protein complex, known as chromatin, is the nucleosome that contains approximately 147 bp of DNA wrapped around a core of eight histone proteins: two copies each of the H2A, H2B, H3 and H4 histone proteins. Post-translational modification of core histone proteins and further interactions with nuclear proteins regulates the nature of chromatin and its subsequent activity. In general, the majority of active protein-coding genes are located within euchromatin, regions of chromatin in which the nucleosome particles have an “open” packaging structure amenable to access by regulatory proteins and the transcriptional machinery. In contrast, inactive genes and structural regions of the genome, such as the centromeres and telomeres, are packaged with the nucleosome in a more tight “closed” state known as heterochromatin. The post-translational modifications of chromatin that define its state enable a preset gene activity to be maintained even in the absence of a regulatory signal. This is also a heritable state that can be transferred to subsequent generations without requiring change of DNA sequence, and is thus referred to as epigenetic regulation.
The N-terminal regions of histone proteins, commonly called the “histone tails” as they extend from the nucleosomes, are highly conserved among eukaryotic organisms and represent the target site for most common epigenetic modifications. The major post-translation modifications involved in epigenetic regulation are found in the histone H3 tails, particularly at the lysine 4 (H3K4) and lysine 9 (H3K9) residues. The chromatin at actively transcribed genes and associated euchromatic regions is marked by methylation of lysine 4 (H3K4me) and acetylation of lysine 9 (H3K9ac). The opposite is found in heterochromatin, where methylation of lysine 9 (H3K9me) and subsequent coating by Heterochromatin Protein 1 (HP1) proteins is a hallmark of epigenetically silent chromatin (Fig. 1). It is important to note that these are not the only modifications found in histone tails. A large number of modifications can exist depending on the specific context or DNA process, and positive and negative interactions between different modifications also occur (for review see Kouzarides 2007). Phosphorylation of histone H3 serine 10 (H3S10) contributes to disruption of heterochromatin structure during the cell cycle (Dormann et al. 2006), whereas methylation of H3K27 by the Polycomb machinery is associated with epigenetic suppression of developmental genes in multicellular organisms (Schuettengruber & Cavalli 2009). In the case of plants and vertebrates, methylation of DNA cytosine residues also plays a significant role in epigenetic control of gene expression.
Fig. 1.
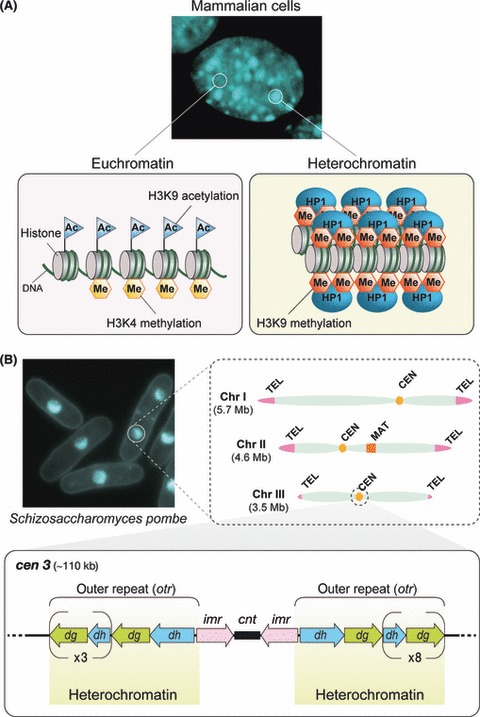
Heterochromatin and epigenetically silent regions of the genome. (A) Active protein coding genes are generally contained within euchromatin, typically modified by acetylation (Ac) of histone H3 lysine 9 (H3K9) and methylation (Me) of histone H3 lysine 4 (H3K4). In contrast, heterochromatin is generally considered to have a compact structure and can be observed as densely stained regions in nuclei. Heterochromatin is characterized by H3K9 methylation, which is recognized by Heterochromatin Protein 1 (HP1). The upper panel shows one complete 4′6′-diamidino-2-phenylindole dihydrochloride (DAPI)-stained nucleus (center) of a mouse NIH3T3 cell. (B) Schizosaccharomyces pombe chromosomes and heterochromatin. The S. pombe genome is organized across three chromosomes, with heterochromatin found at the telomeres (TEL), centromeres (CEN) and mating type region (MAT). Centromeric regions (lower panel) are arranged with a unique core centromere region (cnt and imr) flanked by heterochromatin covering the pericentromeric outer repeat regions (otr). The otr contains multiple copies of dg and dh repeat sequences that vary in number depending on the chromosome. The upper left panel shows S. pombe cells stained with Hoechst33342.
Although heterochromatic regions of the genome contain few protein-coding genes, the mechanisms for epigenetic silencing are directly relevant to that occurring at protein-coding genes and for the control of mobile transposon elements that can strongly influence neighboring genes. Epigenetically silent heterochromatin also serves an essential function in chromosome dynamics, supporting kinetochore formation to ensure correct chromosome segregation and for maintenance of telomere integrity at chromosome ends. Mutations that disrupt any aspect of epigenetic silencing, such as those that impair H3K9me or reduce HP1 association with chromatin, often display phenotypes of lagging chromosomes and missegregation, and in severe cases cell lethality (Ekwall et al. 1995; Peters et al. 2001).
The fission yeast Schizosaccharomyces pombe serves as a highly effective model for the study of basic cellular processes. Research in S. pombe has provided significant insight on the regulation of the cell cycle, and it was discoveries in this area that led to Sir Paul Nurse being awarded the Nobel Prize in Physiology or Medicine 2001. S. pombe also shares many of the chromatin modifications with higher organisms. Chromatin research in S. pombe benefits from the fact that the structural organization of the centromeres and associated histone modifications are more complex than that in budding yeast and similar to that of multicellular organisms, yet it contains only three chromosomes and in many cases only one copy of key regulatory proteins. The centromeres in S. pombe are arranged with a core centromere region containing a central sequence (cnt) flanked by two inverted inner repeats (imr) that share exact identity, which is further flanked by the pericentromeric outer repeat region (otr) that contains multiple copies of dg and dh repeat sequences (Fig. 1). The core region (cnt and imr) is unique to each chromosome whereas the pericentromeric repeats of each chromosome otr are similar and vary in number depending on the chromosome (Takahashi et al. 1992; Rhind et al. 2011). The cnt acts to form the kinetochore for spindle attachment during mitosis and has a chromatin structure specific to this purpose (for review see Allshire & Karpen 2008). On the other hand, the pericentromeric repeats function as sites of typical heterochromatin formation and are essential for higher order structure and correct function of the centromere itself.
Due to the above characteristics, S. pombe is a powerful tool to elucidate epigenetic silencing mechanisms of heterochromatin (Martienssen et al. 2005) and to infer the relationship of heterochromatin formation with centromere function (Pidoux & Allshire 2004). In 2002, research with S. pombe provided the breakthrough discovery that RNAi-mediated processing of native RNA transcripts can guide epigenetic silencing and histone modification (Volpe et al. 2002). This prompted a new field of research and parallels have now been found in multicellular organisms including plants, flies, and vertebrate cells (Huisinga & Elgin 2009; Matzke et al. 2009; Bourc’his & Voinnet 2010; Malecova & Morris 2010). Although the requirement for transcription and RNAi in heterochromatic silencing was immediately apparent, the molecular mechanisms behind this regulation and the nature of interactions between different protein components remained elusive. An eloquent combination of genetic and biochemical studies by different laboratories is now enabling a clearer picture of how this epigenetic silencing is formed at heterochromatic regions and inherited through mitotic division.
The breakthrough: a role for RNA in epigenetic silencing
In addition to the centromeric and pericentromeric regions, the S. pombe genome contains the mating type (mat) region that is another well studied region of heterochromatin. The mating type region contains an upstream active mat1 locus and two downstream donor loci, mat2 and mat3, within a silent heterochromatin domain. Switching of the upstream mat1 gene with one of two downstream donor loci determines the mating type of the cell. Correct regulation of heterochromatin silencing is essential for efficient mating type switching. This region long served as a target site for heterochromatin studies in S. pombe (Klar 2007), and it was the results from early genetic screens using this locus that enabled identification of what are still considered to be the major key players for chromatin modification in S. pombe, particularly swi6, rik1, and clr4 (Ekwall et al. 1995, 1996). Early analyses of mutants resulting from these genetic screens all pointed to a mechanism for epigenetic silencing based on transcriptional gene silencing (TGS). However, it is now clear that redundant mechanisms operate at the mating type locus (Jia et al. 2004). This redundancy had been continually masking the presence of another unexpected mechanism for heterochromatin silencing that is readily observable at the pericentromeric repeats, one that requires nascent RNA transcripts and involves a post-transcriptional gene silencing (PTGS) activity.
RNA interference (RNAi) is a PTGS regulation that either represses translation of target transcripts or results in their cleavage and rapid degradation. The RNAi process is mediated by small RNAs, microRNAs or short interfering RNAs (siRNAs), that have complementary sequence to the target transcript and determine the site of action for the RNA machinery (Hannon 2002; Mello & Conte 2004; Ghildiyal & Zamore 2009). Small RNAs are generated from longer double stranded RNAs by the ribonuclease III enzyme Dicer and then loaded into the Argonaute protein, which functions in a silencing complex and binds target transcripts using the small RNA as a guide (Hammond et al. 2000; Song et al. 2003). PTGS that involves cleavage of the target is achieved by an endonuclease “slicing” activity contained within the Argonaute protein itself (Song et al. 2004; Irvine et al. 2006). An additional activity associated with RNAi is that of RNA-dependent RNA polymerase (RdRP). Found in plants, fungi and invertebrate animals, this is considered an amplification or enhancement step where RdRP synthesizes a complementary strand of the target transcript to form dsRNA that serves as a template for Dicer and additional small RNA production (Cogoni & Macino 1999; Dalmay et al. 2000; Smardon et al. 2000; Sijen et al. 2001). Prior to 2002, a role for PTGS in the nucleus was unknown and RNAi was thought of as a cytoplasmic regulation of protein-coding transcripts that was unrelated to chromatin state.
The discovery that RNAi may participate in epigenetic silencing was the result of clever foresight originating from research into control of plant development. Argonaute genes were suggested to have an important role in cell differentiation in plants (Kidner & Martienssen 2004), and to gain further insight on how these proteins function, this research was extended into S. pombe due to the fact that its genome only contained a single copy of Argonaute (ago1+). Deletion of ago1+ and the related single Dicer (dcr1+) and RdRP (rdp1+) genes led to the surprising finding that these core components of the RNAi machinery were required for maintaining the silent state of pericentromeric regions (Volpe et al. 2002). Further analysis revealed that, contrary to common belief, one strand of the heterochromatin DNA was actually being transcribed in wild-type cells and this was being turned-over by the RNAi machinery. RNAi processing of this strand was required to maintain TGS of the opposite strand that was enforced by typical chromatin modifications. Loss of the RNAi machinery resulted in transcription from both strands of the pericentromeric repeats and a reduction of H3K9me at transgenes inserted within heterochromatin, demonstrating the requirement for RNAi in maintaining the epigenetic state (Volpe et al. 2002). This seminal discovery revealed that the two seemingly unrelated fields of RNA processing and chromatin modification could be tightly integrated in key developmental processes, and was accordingly recognized as the “Breakthrough of the Year” for 2002 (Couzin 2002).
While the above discovery demonstrated a role for RNAi in epigenetic silencing, the nature of this role and mechanisms for interaction between RNAi and chromatin machineries were less clear. Intensive biochemical and genetic experiments by several groups over the following years have revealed that many of the core RNAi and chromatin regulatory proteins function in concert with each other as larger complexes. Each of these complexes has a specific function in RNAi-mediated epigenetic silencing as described in the section below.
Silencing by not being silent
Due to its highly compact structure, heterochromatin was long considered to be inaccessible to the transcriptional machinery and thus completely devoid of transcription activity. It is now known that this is not necessarily the case, and that nascent RNA polymerase II-dependent transcripts from low levels of strand-specific transcription are required to maintain the epigenetically silent state (Volpe et al. 2002; Djupedal et al. 2005; Kato et al. 2005). Although this appears to be a paradox at first glance, the contradiction arises by a definition of silencing as simply whether transcription takes place or not. This can easily be resolved by a clearer definition of epigenetic silencing. Indeed, although low levels of strand-specific transcription may occur across transgenes located within heterochromatic regions (Volpe et al. 2002; Irvine et al. 2006), these protein-coding genes are not transcribed in a way that facilitates translation and thus remain functionally silent (Allshire et al. 1995). Epigenetic silencing may therefore better be defined as an epigenetic regulation that maintains a functionally silent state, irrespective of whether it is achieved through a transcription-independent (TGS) or transcription-dependent (PTGS) activity, or a combination of both.
Current major complexes and their activities
The list of proteins known to be involved in RNAi-mediated epigenetic silencing has increased considerably in the last few years, and biochemical characterization is providing an insight into their functions. At present, four major complexes have been identified that are required for maintaining epigenetic silencing at heterochromatin. When considering the biological functions of these complexes, it is important to keep in context the regions of heterochromatin studied. Epigenetic silencing may be maintained differently within a heterochromatin region depending on the nature of specific sequence being examined. When a transgene is inserted within the pericentromeric region, heterochromatin spreads over the gene and it becomes epigenetically silenced similar to that for the repeats. The presence of H3K9me and epigenetic silencing at inserted transgenes is almost entirely dependent on the RNAi machinery, consistent with RNAi having an upstream role and recruiting the chromatin modification machinery to these sequences (Volpe et al. 2002; Sadaie et al. 2004; Verdel et al. 2004; Irvine et al. 2006). However, at endogenous pericentromeric repeat sequences, H3K9me and heterochromatin can still be maintained at specific sites by the action of the H3K9 methyltransferase Clr4 even when silencing of both strands is released by mutation of RNAi (Noma et al. 2004; Sadaie et al. 2004; Li et al. 2008). In this context, RNAi appears to have a role to enforce silencing and spread heterochromatin to neighboring regions after initial recruitment by chromatin modification machinery (Kagansky et al. 2009). Both observations were supported by an analysis of Ago1 slicing activity, which demonstrated that catalytic inactive Ago1 could still associate with repeat sequences, whereas Ago1-dependent processing of read-through transcripts from the repeats was required for its localization to inserted transgenes and to maintain H3K9me at these sequences (Irvine et al. 2006).
The RNAi-induced transcriptional silencing complex
In S. pombe, four of the nine chromodomain proteins contained within its genome (Wood et al. 2002) have been confirmed to have a functional role in heterochromatin modification: Clr4 H3K9 methyltransferase, HP1 proteins Chp2 and Swi6, and a unique chromodomain protein Chp1 (Ekwall et al. 1995; Doe et al. 1998; Ivanova et al. 1998; Partridge et al. 2000, 2002; Thon & Verhein-Hansen 2000; Bannister et al. 2001; Nakayama et al. 2001; Sadaie et al. 2004, 2008; Motamedi et al. 2008). A critical link in understanding how RNAi activity interacts with heterochromatin was provided by a biochemical analysis of tagged Chp1 purified from S. pombe cells. This revealed that Chp1 directly interacts with the RNAi Ago1 protein and forms a complex that includes a third protein named Tas3 (Verdel et al. 2004). The Chp1-Tas3-Ago1 complex interacts with siRNAs derived from pericentromeric sequences in a Dcr1-dependent manner, and is required for epigenetic silencing and H3K9me at transgenes within the pericentromeric region. Based on these features, this complex was named as RNAi-induced transcriptional silencing (RITS) complex (Fig. 2) (Verdel et al. 2004). The association of RITS with heterochromatin depends on H3K9me through the Chp1 subunit, although in the absence of dcr1+, siRNA loading or Ago1 catalytic activity, the inactive RITS does not spread H3K9me into transgenes or repress accumulation of heterochromatin transcripts and epigenetic silencing is released (Noma et al. 2004; Verdel et al. 2004; Irvine et al. 2006). RITS therefore appears to function to recruit the RNAi machinery to heterochromatin for reinforcement of the silencing and guide the spread of epigenetic silencing across the entire region.
Fig. 2.
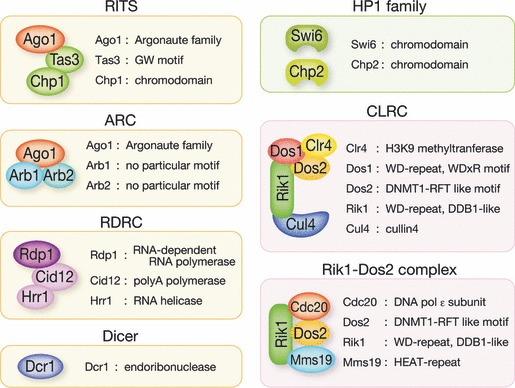
Proteins and effector complexes linked to RNAi-mediated epigenetic silencing in Schizosaccharomyces pombe. Complexes containing known RNAi components are shaded in yellow, whereas complexes interacting directly with chromatin are shaded in pink. Heterochromatin Protein 1 (HP1) proteins are also a key component of heterochromatin structure and are included for consistency.
The RNA-dependent RNA polymerase complex
The RNAi process depends on dsRNA that is cleaved by Dcr1 to provide siRNAs for loading into Ago1. Although one strand of the pericentromeric repeats is weakly transcribed, TGS of the opposite strand in wild-type cells means an absence of a direct complementary strand for these transcripts. It could be argued that the repetitive nature of pericentromeres may inadvertently generate an appropriate source of dsRNA; however, this would not explain how RITS and slicing activity of Ago1 is able to expand into non-repetitive transgene sequences. The S. pombe genome encodes an RNA-dependent RNA polymerase, Rdp1, that functions in a complex with a RNA helicase, Hrr1, and a polyA polymerase family protein, Cid12, to synthesize dsRNA complementary strands from target transcripts (Fig. 2) (Motamedi et al. 2004). The RNA-dependent RNA polymerase complex (RDRC) is not tightly associated with chromatin like RITS; however, an association between the two complexes can be detected and loss of siRNAs from RITS in RDRC mutants is consistent with this complex functioning to provide the Dcr1-dependent siRNA substrate for RITS (Motamedi et al. 2004; Iida et al. 2008). As it is being synthesized by RDRC, it is likely that the dsRNA substrate is provided directly to Dcr1 for siRNA production in a coupled process. Dcr1 can associate with RDRC through a domain independent from its RNaseIII activity and this interaction does not require dsRNAs or heterochromatin assembly (Colmenares et al. 2007). As suggested by the presence of Cid12 within RDRC, select components of the spliceosome also functionally contribute to RDRC function and epigenetic silencing at the pericentromeric region, although this is not coupled to splicing activity and likely represents a structural support for RNA processing by the RNAi machinery (Bayne et al. 2008; Chinen et al. 2010).
The Argonaute siRNA chaperone complex
Epigenetic silencing by the RITS complex at heterochromatin requires that it is loaded with relevant siRNAs. The Argonaute siRNA chaperone (ARC) complex is a second Ago1-containing complex that is proposed to mediate the loading of Ago1 with mature single-stranded siRNA for its activity within the RITS complex (Fig. 2) (Buker et al. 2007). The ARC complex contains two other conserved proteins, Arb1 and Arb2, and differs from RITS in that it is not localized to chromatin and shows diffuse distribution in both the cytoplasm and nucleus. Duplex siRNAs are predominantly found in the ARC complex and the maturation and conversion of these to single-stranded siRNAs requires the slicer activity of Ago1. The Arb1 subunit shows suppression activity against Ago1 slicing catalytic activity, suggesting that the ARC complex functions to obtain duplex siRNAs from Dcr1 and prevent their autodegradation until they are required as single-stranded siRNAs in the RITS complex (Buker et al. 2007).
The CLR4 complex
The accumulation of high levels of siRNA derived from the heterochromatin repeats requires functional Clr4, the sole H3K9 methyltransferase in S. pombe (Motamedi et al. 2004; Noma et al. 2004; Hong et al. 2005; Buhler et al. 2006; Halic & Moazed 2010). Clr4 is present in a complex containing the Rik1 protein, Rik1-associated factors Dos1/Raf1 and Dos2/Raf2, and the ubiquitin ligase scaffold family protein cullin 4 (Cul4) (Fig. 2; Sadaie et al. 2004; Horn et al. 2005; Li et al. 2005). Deletion of the CLR4 complex (CLRC) subunits releases epigenetic silencing of both strands of pericentromeric repeats and abolishes H3K9me, confirming its upstream role in heterochromatin formation and epigenetic silencing (Sadaie et al. 2004; Hong et al. 2005; Horn et al. 2005; Jia et al. 2005; Li et al. 2005; Thon et al. 2005). Consistent with this, siRNAs derived from the pericentromeric repeats fail to accumulate in a CLRC mutant background (Hong et al. 2005; Li et al. 2005).
Although Clr4 was identified as the namesake component of CLRC, stoichiometrical validation will be necessary to understand exactly how the complex functions to support Clr4 activity and H3K9me. In several purification studies, Rik1, Cul4, and other CLRC components were stably identified by mass spectrometric analyses, but Clr4 was missing (Horn et al. 2005; Bayne et al. 2010). Based on amino-acid similarity, Rik1 is thought to have an intertwined three β-propeller cluster and to be a functional homologue of DNA-damage binding protein 1 (DDB1). DDB1 also interacts with Cul4 to function in nucleotide excision repair (Angers et al. 2006; Li et al. 2006; Scrima et al. 2008), and binds to a family of WD40-repeats containing proteins that are thought to act as receptors for substrates of the DDB1-CUL4 E3 machinery (Angers et al. 2006). Dos1 is a WD40 protein containing conserved signatures for DDB1-interacting proteins and may serve a similar function with Rik1-Cul4 as part of CLRC. In filamentous fungus Neurospora crassa, a H3K9 methyltransferase DIM-5 has been shown to form a multiprotein complex with DDB1-CUL4. Moreover, this interaction is also mediated by proteins related to Dos1 and Dos2 (Lewis et al. 2010). It is thus conceivable that CLRC may represent a scenario in which modification of Clr4 by Rik1-Cul4-Dos1 promotes its recruitment to DNA or H3K9me activity at target regions.
Rik1-Dos1 can directly interact with a JmjC-domain H3K4 demethylase, Lid2, that is thought to bring CLRC to the pericentromeric repeats. Hypomethylation of H3K4 by Lid2 is a required step that precedes H3K9me by Clr4 for heterochromatin formation (Li et al. 2008). The subsequent recruitment of RITS/RNAi to these regions can then be explained by the association of Chp1 with H3K9me. Although RNAi is not absolutely required to recruit CLRC to the pericentromeric repeats or for CLRC-mediated heterochromatin formation (Jia et al. 2004; Kagansky et al. 2009), RNAi is required for spreading of CLRC activity across heterochromatin and transgenes within these regions, suggesting an additional interaction exists for RNAi recruitment of CLRC within the centromere (Volpe et al. 2002; Bayne et al. 2010).
A complex interaction
The relationship between CLRC and RITS/RNAi exemplifies the presence of a self-enforcing loop for maintenance of heterochromatin and epigenetic silencing at S. pombe centromeres. CLRC is required upstream of RNAi, however, slicing activity of Ago1 at transcripts is also required to recruit CLRC and H3K9me into non-repeat sequences. The maintenance of epigenetic silencing at heterochromatin therefore requires cross-talk between complexes with specific functions. These can be direct interactions between complex subunits, or involve additional proteins that play important regulatory roles. A schematic model of the self-enforcing loop based on current knowledge of the relevant complexes is shown in Figure 3.
Fig. 3.
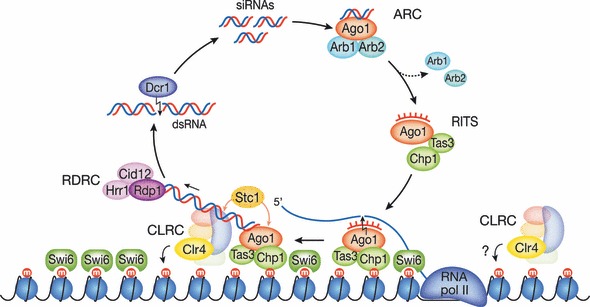
Schematic model of the self-enforcing loop for RNAi-mediated epigenetic silencing in Schizosaccharomyces pombe. Heterochromatin is shown as DNA (black line) wrapped around nucleosomes (blue circles) modified by histone H3 lysine 9 methylation (H3K9me; red circles “m”). Nascent transcripts generated by RNA polymerase II are targeted by an activated RNAi-induced transcriptional silencing (RITS) complex that also interacts with H3K9me through the Chp1 subunit. RDRC is then recruited for synthesis of dsRNA from the transcripts, which is cleaved into duplex siRNAs by Dcr1 and bound by Argonaute siRNA chaperone (ARC). These siRNAs are then processed into single-stranded siRNAs and loaded back into RITS completing the loop. Catalytically active RITS recruits the CLR4 complex (CLRC) for spreading of epigenetic silencing into adjacent regions through interaction with Stc1, although CLRC can also associate with these regions independent of RNAi and recruit inactive RITS.
Interactions within the self-enforcing loop
Of the four main complexes described above, RITS and CLRC are thought to be tightly associated with chromatin, whereas RDRC appears to have a more peripheral association involving nascent transcripts and ARC has a wider distribution throughout the cell. In a wild-type cell, one strand of the pericentromeric repeats is transcribed and turned over when all complexes are functioning normally. These nascent transcripts serve as a basal point for the self-enforcing loop and as substrate for siRNAs that participate in silencing. Transcription of pericentromeric repeats is carried out by RNA polymerase II (RNAPII) (Djupedal et al. 2005; Kato et al. 2005), although the contribution of RNAPII in epigenetic silencing appears to be more than just providing the transcript substrate. A mutation in the largest subunit of RNAPII resulted in loss of siRNAs corresponding to the repeats and disrupted heterochromatin at transgenes within the pericentromeric region. However, this mutation did not affect RNAPII localization to the pericentromeric region and transcripts from the repeats also accumulated similar to that observed in RNAi mutants (Kato et al. 2005). This suggested that loss of epigenetic silencing was not due to absence of transcript template but rather that RNAPII interacts with RITS to enable coupling of transcription with RNAi-mediated epigenetic silencing.
The RITS complex interacts with nascent transcripts from the pericentromeric regions, and the activity of RITS in processing these transcripts is thought to be guided by complementary single-stranded siRNAs loaded into Ago1 (Buker et al. 2007). RITS also directly interacts with Clr4-methylated H3K9me through the Chp1 subunit (Noma et al. 2004; Sadaie et al. 2004; Verdel et al. 2004). It is expected that competition exists between Swi6 (HP1) and Chp1 (RITS) for binding the H3K9me target site. Although the number of Chp1 proteins is lower than that of Swi6, the affinity of Chp1 for H3K9me is approximately 10-fold greater than that of Swi6 (Schalch et al. 2009) and thus RITS can likely access H3K9me when necessary due to dynamic association of Swi6.
In the next step of the loop, RITS interacts with both RDRC and CLRC through its Ago1 subunit (Motamedi et al. 2004; Bayne et al. 2010). When RITS is activated, the interaction with RDRC promotes synthesis of dsRNA based on the target transcript, whereas the interaction with CLRC promotes further H3K9me of the surrounding chromatin. Both of these interactions are dependent on Ago1 catalytic activity and siRNAs, raising the question of how specific interactions with RNA targets and two distinct protein complexes can be achieved by a single Ago1 protein (Motamedi et al. 2004). It can be expected that additional factors are supporting the complex interactions. Evidence for this was recently provided by the discovery of the LIM domain-containing protein, Stc1, that can bind both CLRC and Ago1 but not RDRC (Bayne et al. 2010). The interaction between CLRC and Ago1 was dependent on the presence of Stc1, suggesting it functions as a mediator for RITS recruitment of CLRC. Deletion of Stc1 also caused a strong reduction in centromeric siRNA accumulation and disrupted the association between RITS and RDRC, consistent with it having a function to also support RITS activity at the repeats.
The dsRNA products provided by RDRC activity then proceed through the loop to function as substrates for Dcr1 and are processed into siRNAs. The mechanism by which dsRNA is released from RDRC and interacts with Dcr1 remains unclear, although it may have parallels to that for transcription as suggested by the presence of polyA polymerase subunit in RDRC and contribution of splicing components (Motamedi et al. 2004; Bayne et al. 2008; Chinen et al. 2010). Dcr1 has also been found to associate with RDRC (Colmenares et al. 2007), suggesting that the synthesis and transfer of dsRNA may be partially coupled. However, similar to that for interactions between RITS, CLRC and RDRC, it can be assumed that additional proteins yet to be identified are also mediating the interaction between RDRC and Dcr1. The duplex siRNAs produced by Dcr1 are then fed into ARC, which mediates processing of the siRNA into a single-strand of the correct length for loading into RITS (Buker et al. 2007). At present it is not known if the same Ago1 molecule shuttles between ARC and RITS permitting siRNA maturation by Ago1 slicing activity as the complex composition changes, or whether a mechanism is in place to transfer siRNAs between the two separate Ago1-containing complexes and remove the siRNA passenger strand. Whichever the mechanism, this final step results in a chromatin-associated active RITS complex and a return to the beginning of the self-enforcing loop.
It is important to emphasize that the complexes described here should not be considered as fixed, stable complexes. One example of this is the ability of CLRC subunits Rik1 and Dos1 to associate and interact with chromatin even in the absence of Clr4 (Bayne et al. 2010). Indeed, it has been proposed that this complex may function in a stepwise fashion with the Rik1-Dos1 subunits first interacting with a H3K4 demethylase and then bringing Clr4 to the chromatin for H3K9me (Li et al. 2008). Both the complexes themselves and the supporting interactions between them are thought to be highly dynamic with constant exchange of subunit components and in various states of assembly.
Interactions activating the self-enforcing loop
The concept of the self-enforcing loop is based on the interdependent relationship between Clr4 activity and the RNAi pathway. Clr4 is required for the accumulation of high siRNA levels and the mutual interaction between RNAi complexes (Motamedi et al. 2004; Noma et al. 2004; Hong et al. 2005; Buhler et al. 2006; Halic & Moazed 2010). On the other hand, the RNAi pathway and siRNA products are required for H3K9me at centromeric repeats and inserted marker loci (Volpe et al. 2002). However, the recruitment or targeting of complex components to chromatin does not exclusively depend on this self-enforcing loop. CLRC can associate with chromatin independent of the RITS complex or RNAi (Jia et al. 2004). In addition, RITS can still associate with heterochromatin independent of Ago1 catalytic activity (Irvine et al. 2006). More recently, it was reported that Dcr1 and low levels of the RDRC component Rdp1 could also bind to centromeric repeats independent of Clr4 activity (Woolcock et al. 2011).
The process for de novo formation of heterochromatin that leads into the self-enforcing loop is one aspect that requires further investigation. Low levels of H3K9me persist at the centromeric repeat regions in RNAi-deficient cells (Sadaie et al. 2004) and play an essential role in the initial step of heterochromatin formation (Partridge et al. 2007). These results suggest that an RNAi/siRNA-independent mechanism provides an initial H3K9me mark at the centromeric repeat repeats and this drives the self-enforcing loop. However, this idea was challenged by the discovery of Dicer- and Rdp1-independent small RNAs, called primal RNAs (priRNAs), that associated with Ago1 in S. pombe (Halic & Moazed 2010). Based on a comparison of H3K9me levels in mutants lacking Dcr1, Ago1 and Clr4, it was proposed that these priRNAs play a role promoting H3K9me, and that RNAi-dependent factors initiate de novo heterochromatin assembly. However, another report argues against an upstream role for siRNAs. Using a genetic approach to transiently deplete CLRC or RNAi components, it was shown that Ago1 was not required to initiate de novo H3K9me at centromeres by Clr4 (Shanker et al. 2010). Although it is seemingly difficult to assemble these contradictory results, a plausible idea would be that different heterochromatic regions use distinct pathway to establish primal marks that drive the self-enforcing loop. This is supported by the finding that Clr4 has activity against substrates other than H3K9 and that this alternative activity is able to promote siRNA production from one of the two types of endogenous pericentromeric repeats (Gerace et al. 2010).
Interactions with the cell cycle and DNA replication
Similar to the fact that epigenetic silencing complexes are thought to be highly dynamic, consideration also needs to be made of the dynamic nature of chromatin and how RNAi-mediated epigenetic silencing interacts with other molecular processes that occur along the chromosome. One chromatin remodeling process expected to conflict with the self-enforcing loop is that of DNA replication. Centromeric transcripts and siRNAs accumulate predominantly during S-phase, and it is at this point that H3K9me needs to be transferred or spread along the newly synthesized DNA (Chen et al. 2008; Kloc et al. 2008). This suggests that both RNAi-mediated silencing and DNA replication machineries are required to function within the repeats at the same time. Surprisingly, a direct interaction between DNA replication factors and the epigenetic silencing factor Rik1 has recently been found (Li et al. 2011; Zaratiegui et al. 2011).
Rik1 and Dos2 have been identified as part of a second complex separate from CLRC, which contains the transcriptional activator Mms19 and DNA polymerase subunit Cdc20 (Li et al. 2011). This complex, referred to as the Rik1-Dos2 complex (Fig. 2), promotes the transcription of pericentromeric repeats during S-phase. The presence of Rik1 in this complex is presumed to result in subsequent recruitment of CLRC for spreading of H3K9me across both chromatids of the newly replicated DNA. However, spreading of H3K9me via the self-enforcing loop is dependent on transcription of the repeats by RNAPII and it was unclear how this could be achieved since transcription and replication at the same location would conflict and cause the replication fork to stall. It has now been shown that this conflict is overcome by the RNAi-dependent processing of nascent transcripts that, while recruiting heterochromatin formation through the self-enforcing loop, promotes degradation of the transcripts and releases RNAPII (Zaratiegui et al. 2011). Removal of RNAPII enables the replication fork to continue and the chromatin remodeling machinery to spread H3K9me into the replication fork. Without this RNAi processing of the transcripts, the stalled replication forks are interpreted as sites of DNA damage and enter into a homologous recombination-mediated repair pathway that causes both the Rik1 complexes and epigenetic silencing to be lost.
Perspectives
Our understanding of the major players and effector complexes for RNAi-mediated epigenetic silencing and heterochromatin assembly has advanced considerably over the last 10 years. However, as new components and functions are discovered, it becomes apparent that that there are still many questions that remain unanswered. Examples of this are Ago1 and Rik1, which are two core proteins that are located in multiple complexes. The identification of ARC provided a concept for siRNA loading of Ago1 in RITS, but it remains to be determined how siRNA transfers between the two complexes, whether the same Ago1 molecule shuttles between the two complexes, and the extent that separate functions for Ago1 are controlled by its other subunits. Rik1 was one of the first genes to be implicated in heterochromatin formation and is now known to form two distinct complexes. These complexes explain the critical role for Rik1 and how multiple processes of transcription, chromatin modification and replication are intertwined. However, the nature of Rik1 contribution to these complexes still requires clarification and the dynamics behind subunit interactions remain vague. The HP1 protein Swi6 gained prominence as a signal for completion of typical heterochromatin formation irrespective of RNAi involvement and its role was thought to be clear. However, Swi6 has now been shown to participate in the initiation of replication within pericentromeric repeats (Hayashi et al. 2009) and to be required for efficient RNAi processing of centromeric transcripts (Motamedi et al. 2004, 2008; Buhler et al. 2006; Halic & Moazed 2010). Much research is still required before a complete picture of heterochromatin assembly can be drawn.
A central question for future research to understand heterochromatin formation is what exactly defines the region to be targeted for epigenetic silencing. It has been eloquently demonstrated that tethering of Clr4 is sufficient to trigger formation of heterochromatin, and importantly, that the RNAi pathway is dispensable for assembling of this silent heterochromatin into a functional centromere (Kagansky et al. 2009; Bayne et al. 2010). This appears to suggest that the primary purpose of RNAi processing in heterochromatin is to recruit Clr4 methyltransferase, which then raises the question of what type of chromatin signature is destined to be targeted by Clr4. Several lines of evidence suggest that genes with convergent transcripts are marked by H3K9me and Swi6 (Gullerova & Proudfoot 2008), whereas another study showed that Clr4 also plays a critical role in degradation of read-through transcripts from non-heterochromatic coding sequences (Zofall et al. 2009; Zhang et al. 2011). While it is difficult to determine whether these typically euchromatic regions have formed heterochromatin, these observations imply that unfavorable transcriptional states are targeted by Clr4 activity. The simplest conclusion from these observations is that double stranded RNA formed by read-through transcripts are processed and targeted by the RNAi pathway to recruit Clr4. Considering that Rik1 shows structural similarity to DDB1 that functions in DNA repair and to CPSF-160 that functions in pre-mRNA 3′ processing (Mandel et al. 2008), the Rik1-containing CLRC complex may also target chromatin by recognizing structural DNA or RNA signatures representative of unfavorable transcription.
Acknowledgments
Research in the authors laboratories is supported in-part by Grants-in-aid for Scientific Research on Innovative Areas (No. 22115501 to DBG; No. 23114005 to JN), for Scientific Research (B) (No. 23370004 to JN), and for Challenging Exploratory Research (No. 23651209 to DBG) from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT). DBG also wishes to acknowledge the support of the start-up fund of the Leader Development System in the Basic Interdisciplinary Research Areas at Hokkaido University, Japan.
References
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, Maccoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe K-L, Kim D-U, Park H-O, Ponting CP, Rappsilber J, Allshire RC. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Buker SM, Iida T, Buhler M, Villén J, Gygi SP, Nakayama J, Moazed D. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Chinen M, Morita M, Fukumura K, Tani T. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J. Biol. Chem. 2010;285:5630–5638. doi: 10.1074/jbc.M109.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Couzin J. Breakthrough of the year. Small RNAs make big splash. Science. 2002;298:2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Wang G, Chow C, Fricker MD, Singh PB, Mellor EJ. The fission yeast chromo domain encoding gene chp1+ is required for chromosome segregation and shows a genetic interaction with alpha-tubulin. Nucleic Acids Res. 1998;26:4222–4229. doi: 10.1093/nar/26.18.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 2006;5:2842–2851. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrøm B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol. Cell. 2010;39:360–372. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat. Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA. Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Elgin SC. Small RNA-directed heterochromatin formation in the context of development: what flies might learn from fission yeast. Biochim. Biophys. Acta. 2009;1789:3–16. doi: 10.1016/j.bbagrm.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu. Rev. Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, Selker EU. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010;6:e1001196. doi: 10.1371/journal.pgen.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr. Opin. Mol. Ther. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Debeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol. Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Peters AH, O’carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, Fitzgerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO. J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol. Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thomä NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S, Job G, George OL, Creamer KM, Shaban A, Partridge JF. Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex. PLoS Genet. 2010;6:e1001174. doi: 10.1371/journal.pgen.1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klar AJ. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2011;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schäfer M, Müller-Auer S, Gabel C, Fuchs M, Düsterhöft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dréano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sánchez M, del Rey F, Benito J, Domínguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Castel S, Irvine D, Kloc A, Ren J, Li F, de Castro E, Marin L, Antequera F, Goto D, Cande Z, Arcangioli B, Martienssen R. RNA interference promotes heterochromatic silencing through replication-coupled release of RNA polymerase II. Nature. 2011 doi: 10.1038/nature10501. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]


