Abstract
Background
Agrobacterium-mediated transformation is widely used to produce insertions into plant genomes. There are a number of well-developed Agrobacterium-mediated transformation methods for dicotyledonous plants, but there are few for monocotyledonous plants.
Methods
Three hydrolase genes were transiently expressed in Brachypodium distachyon plants using specially designed vectors that express the gene product of interest and target it to the plant cell wall. Expression of functional hydrolases in genotyped plants was confirmed using western blotting, activity assays, cell wall compositional analysis and digestibility tests.
Key Results
An efficient, new, Agrobacterium-mediated approach was developed for transient gene expression in the grass B. distachyon, using co-cultivation of mature seeds with bacterial cells. This method allows transformed tissues to be obtained rapidly, within 3–4 weeks after co-cultivation. Also, the plants carried transgenic tissue and maintained transgenic protein expression throughout plant maturation. The efficiency of transformation was estimated at around 5 % of initially co-cultivated seeds. Application of this approach to express three Aspergillus nidulans hydrolases in the Brachypodium cell wall successfully confirmed its utility and resulted in the expected expression of active microbial proteins and alterations of cell wall composition. Cell wall modifications caused by expression of A. nidulans α-arabinofuranosidase and α-galactosidase increased the biodegradability of plant biomass.
Conclusions
This newly developed approach is a quick and efficient technique for expressing genes of interest in Brachypodium plants, which express the gene product throughout development. In the future, this could be used for broad functional genomics studies of monocots and for biotechnological applications, such as plant biomass modification for biofuel production.
Keywords: Transient expression, Agrobacterium, Brachypodium distachyon, hydrolases, plant cell wall, biodegradability
INTRODUCTION
Monocotyledonous plants, particularly grass crop plants, are the major sources of human nutrition and biofuels. The four most agriculturally important grasses are maize, rice, wheat and sugarcane, which are staple food crops; prominent bioenergy crops include perennial grasses, such as switchgrass, rye grass and Miscanthus species. Improvements to crop sustainability, nutrition and biomass properties are important for meeting the challenges of creating sustainable agricultural systems that can produce both adequate food supplies and efficient bioenergy. Studies of the model plant arabidopsis have successfully advanced our understanding of dicotyledonous crops. Development of a model system for grasses with similar potential would permit investigations comparable with those possible in arabidopsis, and allow cereal crop improvements. Recently, Brachypodium distachyon has emerged as a new model system for grass crop genomics research because it is self-pollinating, fast growing, has a small genome and is closely related to a large and diverse group of temperate cereal crops, giving Brachypodium several advantages over other model crops such as rice (Oryza sativa) or sorghum (Sorghum bicolor) (Garvin et al., 2008; Thole et al., 2009). Molecular resources, such as community standard genetic stocks, expressed sequence tag collections, bacterial artificial chromosome (BAC) libraries, genetic linkage maps and segregating populations, have all been developed for Brachypodium (Garvin et al., 2008). Indeed, many areas of plant biology will directly benefit from using Brachypodium as a model system, e.g. studies of development (Olsen et al., 2006; Faricelli et al., 2009; Schwartz et al., 2010), seed storage proteins (Laudencia-Chingcuanco and Vensel, 2008; Charles et al., 2009; Gu et al., 2010; Xu and Messing, 2009), fatty acid turnover (Yang and Ohlrogge, 2009), plant–pathogen interactions (Parker et al., 2008) and wounding/insect responses (Mur et al., 2004; Azhaguvel et al., 2009).
Rhizogenesis is another research area where Brachypodium provides an advantage because, unlike arabidopsis, it readily forms mycorrhizal associations. These associations are important for many crops, particularly for phosphorus uptake, which is especially important for low-input biomass crop production, as well as for agriculture in developing countries. Brachypodium root development is very similar to that of wheat, making Brachypodium an excellent model for studies of rhizogenesis (Bevan et al., 2010).
Transient gene expression and stable transformation are essential tools for analysing gene function in plants. Thus far, several methods have been developed for transformation of monocotyledonous plants. One method is biolistic transformation, or microparticle bombardment, of Brachypodium embryogenic calluses (Draper et al., 2001; Christiansen et al., 2005). Biolistic transformation frequently results in complex insertions containing many copies of the inserted DNA, often along with rearrangements of the native DNA. For many applications, particle bombardment is not suitable due to probable gene silencing in subsequent generations (Taylor and Fauquet, 2002). A second method is Agrobacterium-mediated transformation using Brachypodium excised immature embryos. Published protocols for the transformation of Brachypodium are based on tissue culture methods (Vogel and Hill, 2008). Cereal transformation using tissue culture has been successful, but has several limitations. For example, a significant number of rice mutants among the hundreds of thousands of T-DNA lines produced via tissue culture are somaclonal variants (An et al., 2005). The transformation method is laborious, involves extensive tissue culture and requires a minimum of 8 months before obtaining seeds of the next generation. There are two additional methods of Agrobacterium-mediated transformation described for monocotyledonous plants. One is floral transformation of allohexaploid wheat (Triticum aestivum), which was published (Zale et al., 2009) prior to the development of the floral dip method in arabidopsis by Hess et al. (1990). In this method, target tissue, pollen and a basal medium containing Agrobacterium carrying genetic constructs is pipetted into open wheat florets during anthesis. Another method described for wheat is Agrobacterium-mediated transformation of germinating seeds. The embryo of each soaked seed is inoculated by piercing a region of the embryonic apical meristem with a needle that has been dipped in an Agrobacterium inoculum (Supartana et al., 2006).
Transient gene expression methods are widely used to analyse the functions of selected gene products in vivo and, depending on the part of the plant used for inoculation, may lead to stable transformation of the seeds developed from the transiently transformed tissues. Recently, an Agrobacterium-mediated transient gene expression system has been proposed that uses inoculation of germinating switchgrass (Panicum virgatum) seedlings (Chen et al., 2010).
Here, we propose and demonstrate an alternative method for Brachypodium transient gene expression that does not require the use of tissue culture and is based on Agrobacterium-mediated gene transfer into the seeds. This new technique is a faster, less laborious, more efficient and more predictable approach to generating transgenic monocotyledonous plants. To confirm the robustness of our approach, we used it to express three Aspergillus nidulans hydrolases: rhamnogalacturonan acetylesterase (AN2528·2; EC 3·1·1·6), α-l-arabinofuranosidase (AN7908·2; EC 3·2·1·55) and α-galactosidase (AN8138·2; EC 3·2·1·22). These proteins were expressed in Brachypodium plants, localized to the apoplast in an active form, and acted to modify cell wall composition. We believe that this technique will allow us to perform stable transformations, and will facilitate monocot biology research, including for biotechnology applications of agriculturally important cereal crops.
MATERIALS AND METHODS
Plant growth conditions
Seeds of Brachypodium distachyon (genotype Bd21) were obtained from the USDA/ARS National Germplasm Resources Information Network (GRIN, http://www.ars-grin.gov/). Plants were grown in controlled conditions in a growth chamber, at 170–180 µmol m−2 s−1 light intensity; 20 h light/4 h dark; 21 °C during the day and 18 °C during the night as recommended by Garvin et al. (2008).
Creation of vectors and expression cassettes
The pMono_T-vector was constructed using the pXcmkn12 (AB107950·1) vector obtained from Dr S. Yasuda (National Instutute of Genetics, Shizuoka, Japan). The Zea mays ubiquitin gene signal peptide-coding sequence was synthesized from two 102 bp single-stranded oligonucleotides (ZmSP-1 and ZmSP-2) annealed to each other by slowly decreasing the temperature from 94 °C to 50 °C over 44 min. The obtained double-stranded oligonucleotide was used as a template for PCR with primers ZmSP_F and ZmSP_R containing EcoRI and KpnI restriction sites, respectively (see Supplementary Data Table S1 for sequences) using Encyclo DNA polymerase (Evrogen, PK001), and the product was cloned into the pGEM-T Easy vector (Promega, A1360). After digestion of recombinant pGEM-T_Easy_ZmSP vector, the EcoRI–KpnI fragment was moved into the pXcmkn12 plasmid treated with the same enzymes to obtain the pXcmkn12-ZmSP vector. The green fluorescent protein (GFP) coding sequence, together with the transcription terminator, was amplified with Encyclo DNA polymerase from the psmGFP plasmid (www.arabidopsis.org, CD3-326) using GFP_F and GFP_R primers containing XbaI and KpnI restriction sites, respectively. The product was cloned into a pGEM-T Easy vector. After digestion of recombinant pGEM-T_Easy_GFP vector, the XbaI–KpnI fragment was moved into the pXcmkn12-ZmSP plasmid cleaved with the same enzymes. To obtain the final pMono_T-vector, a PCR fragment amplified with ZmSP_F and GFP_R primers from the pXcmkn12-ZmSP-GFP plasmid was cloned into pGEM-T Easy vector containing two NotI restriction sites flanking the cloning site.
pUbiP2 T-DNA-containing vector was constructed using a pMLBart binary vector backbone (Gleave, 1992). The Z. mays ubiquitin promoter and the downstream intron required for efficient transcription were amplified from the pIPKb002 binary vector (Himmelbach et al., 2007) using two variants: first using UbiP_F1 and UbiP_R1 primers, and secondly using UbiP_F2 and UbiP_R2 primers. The first set of forward and reverse primers contained NotI and SpeI restriction sites, respectively, and the second set contained SacI and NotI sites, respectively. Both amplification products were cloned into the pGEM-T Easy vector to obtain pGEM-T Easy_UbiP1 and pGEM-T Easy_UbiP2 plasmids. The NOS promoter from the pMLBart vector was replaced with the ubiquitin promoter from pGEM-T Easy_UbiP1 treated with NotI and SpeI restriction enzymes to obtain pUbiP1 vector. The SacI–NotI fragment from pGEM-T Easy_UbiP2 was moved into the pUbiP1 vector with the same restriction enzymes to obtain pUbiP2 binary vector.
Aspergillus nidulans genes were amplified by Encyclo DNA polymerase (Evrogen) from plasmids extracted from Pichia pastoris strains (Bauer et al., 2006) obtained from The Fungal Genetics Stock Center (www.fgsc.net) using appropriate forward and reverse primers (Supplementary Data Table S1). All forward primers started from the second nucleotide of the ATG start codon to fit into the same open reading frame with the signal peptide sequence. Obtained binary vectors were transformed into Agrobacterium tumefaciens EHA101 cells by electroporation.
Genomic DNA genotyping, reverse transcription–PCR (RT–PCR) and flanking sequence tag isolation
Total DNA from leaf tissue was extracted using cetyl trimethylammonium bromide (CTAB) isolation buffer according to the protocol of Stewart and Via (1993). PCR genotyping of transformed plants was performed using appropriate gene-specific primers (see Supplementary Data Table S1) using the following conditions: one repeat: 94 °C 3 min; 31 repeats: 94 °C 10 s, 56 °C 30 s, 72 °C 90 s; and one repeat: 72 °C 4 min, using GoTaq Green Master Mix (Promega, M7122). Total RNA was extracted using the SV Total RNA Isolation System (Promega, Z3101), and first-strand cDNA synthesis was carried out using the SuperScript® III First-Strand Synthesis System (Invitrogen, 18080051) following the manufacturer's recommendations. Flanking sequence tags were amplified by miPCR as previously described (Pogorelko and Fursova, 2008) using EcoRV restriction enzyme for digestion of genomic DNA, and pUbiP_Forw1 + pUbiP_Rev1 primers for the first step PCR and pUbiP_Forw2 + pUbiP_Rev2, pUbiP_Forw2 + pUbiP_Rev3 for the second and third PCR steps, respectively (see Supplementary Data Table S2 for sequences).
Apoplastic fluid preparation
Harvested leaves (approx. 0·5 g) from individual plants were cut into 3–6 mm segments using a sharp razor blade and placed vertically into a 10 mL syringe sealed with parafilm. A 5 mL aliquot of pre-cooled extraction buffer (25 mm Tris–HCl, 50 mm EDTA, 150 mm MgCl2, pH 7·4) was added and the syringe was placed under vacuum twice for 30 min at 4 min intervals. After vacuum infiltration, the buffer was carefully drained and the syringe placed into a centrifuge tube and centrifuged at 1000 g for 10 min. Apoplastic fluid, which accumulated at the bottom of the tube, was collected into a new tube and kept on ice until it was assayed for activity. All the above steps were performed at 4 °C. Protein concentration was determined using the Bradford assay (Bradford, 1976). For western analysis, total protein was precipitated from apoplastic extract by addition of trichloroacetic acid (TCA) to a final concentration of 10 % (v/v). Proteins were pelleted by centrifugation at 8000 g for 20 min, washed with acetone, and air-dried.
Western blot analysis of apoplastic proteins
Western blot hybridization was performed according to Magi and Liberatori (2005). TCA-precipitated apoplastic proteins were re-suspended in 1× Laemmli's buffer [125 mm Tris–HCl (pH 6·8); 1 % SDS; 37·7 % glycerol; 1 % β-mercaptoethanol; 0·01 % bromophenol blue], boiled for 5 min, and used immediately for SDS–PAGE. A 20 µg aliquot of total protein was loaded per well on a 10 % SDS–PAGE Bio-Rad Mini Gel. Following separation, proteins were electrophoretically transferred to a Trans-Blot Transfer Medium membrane (Bio-Rad, 162-0112) using the Trans-Blot Cell (Bio-Rad) with a standard transfer buffer. Primary anti-GFP monoclonal antibodies (Covance, CA, USA, MMS-118P) diluted 1:5000 and secondary anti-mouse IgG (whole molecule)–peroxidase antibody (Sigma, A9044) diluted 1:40 000 were used for detection of GFP fusion proteins. Membranes were treated with the HyGlo Quick Spray Reagent A + B for peroxidase activity (Denville, E2400) and visualized using ChemiDoc XRS System (BioRad, 170-8265).
Assays for enzymatic activity
α-Galactosidase (AnGAL), rhamnogalacturonan acetylesterase (AnRAE) and α-l-arabinofuranosidasae (AnAF) activity were all assayed by measuring the concentration of 4-nitrophenol (pNP) released from a specific substrate; pNP-galactopyranoside (Sigma, N3016) was used for AnGAL, pNP-acetyl (Sigma, N3641) for AnRAE, and pNP-arabinofuranoside (Sigma, N8130) for AnAF. For each reaction, apoplastic extract from transgenic plants or wild-type plants was incubated in 0·1 m sodium phosphate buffer (0·042 m NaH2PO4; 0·058 m Na2HPO4; pH 7·0), and 2 mm of the specific pNP-substrate, in a total volume of 100 µL. In each reaction, apoplastic fluids were added to obtain: 3·24 × 10−2 mg of total protein for AnGAL assay, 1·28 × 10−1 mg of total protein for AnRAE assay, and 4·41 × 10−2 mg of total protein for AnAF assay. A reaction mixture containing boiled apoplastic extract was used as a negative control for each assay. The reactions were carried out at 37 °C and absorbance measured at 420 nm at various time points using a microplate reader. Activity was described as the amount of pNP nmoles released by 1 mg of protein in 1 min.
Analysis of cell wall composition
Cell walls were purified from plant material remaining after apoplastic fluid extractions as described in Zabotina et al. (2008). To determine monosaccharide composition, 1 mg of dry cell wall was hydrolysed in 2 n trifluoroacetic acid at 120 °C for 2 h. After acid evaporation, hydrolysate was re-dissolved in water and analysed by HPAEC-PAD using a CarboPac PA-20 column (Dionex, CA, USA) as described previously (Zabotina et al., 2008). Monosaccharide standards included l-fucose, l-rhamnose, l-arabinose, d-galactose, d-glucose, d-xylose, d-mannose, d-galacturonic acid and d-glucuronic acid (all from Sigma). To determine response factors, standard curves were created using mixtures of all standard monosaccharides at different concentrations.
The acetyl content of cell walls was estimated using an assay adapted from McComb and McCready (1957). Briefly, 2 mg of dry cell wall material was mixed with 250 µL of 0·5 m hydroxylamine hydrochloride, followed by the gradual addition of 250 µL of 2 n sodium hydroxide. After 30 min incubation at 21 °C, 100 µL of the resulting mixture was mixed with 40 µL of water and 100 µL of acid methanol solution (35·2 µL of 70 % perchloric acid in 464·8 µL of absolute methanol). A 260 µL aliquot of ferric perchlorate solution (32·8 µL of 2·36 % ferric perchlorate (non-yellow) in 3·8 % perchloric acid diluted with 227·2 µL of absolute methanol) was added in small increments, mixing after each addition. After 30 min, the absorbance of formed ferric acetohydroxamic complex was determined at 510 nm using a microplate reader. The acetyl content was calculated using a standard curve created with glucose penta-acetate as a standard.
Fluorescence microscopy
Brachypodium plants expressing hydrolase–GFP fusion proteins (4–6 weeks old) were examined with a Leica DMIRE2 fluorescence microscope equipped with a Chroma 41028 Yellow GFP BP (10c/topaz) filter, using 488 nm excitation and 520–550 nm emission wavelengths for GFP. Images were captured with a Nikon DS-L1 camera.
Saccharification assay
Dry cell wall material (10 mg) was suspended in 0·3 mL of sodium citrate buffer (pH 4·9) containing 4 U of cellulase (from Trichoderma reesei, Sigma, C2730) and 1 U of cellobiase (from Aspergillus niger, Sigma, C6105). Sample mixture was incubated on a shaker at 25 °C. At each time point, the reaction was terminated by heating at 100 °C for 15 min and supernatants were collected by centrifugation at 10 000 g.
Reducing sugar determination was performed using the PAHBAH assay (Lever, 1972) with minor modifications. Briefly, 15 µL of supernatant was mixed with 135 µL of freshly prepared PAHBAH reagent (1 vol. of 5 % p-hydroxy benzoic acid hydrazide in 5 % HCl), 9 vols of solution (1·25 % trisodium citrate, 0·11 % calcium chloride and 2 % sodium hydroxide), and heated at 100 °C for 6 min. Absorbance was measured at 410 nm.
RESULTS
Vector system for plant cell wall-directed protein expression
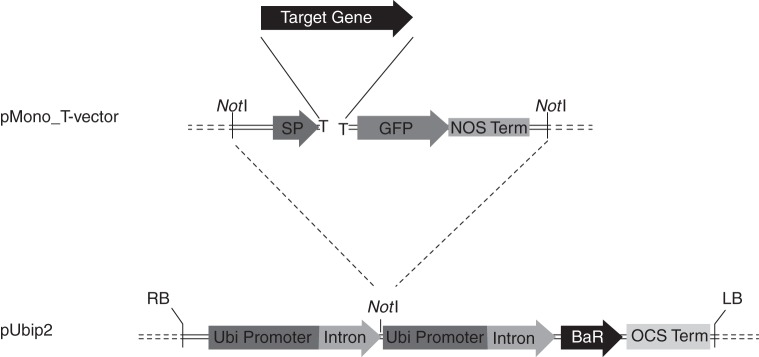
To express a wide range of different proteins in cell walls of monocotyledonous plants, we have created a new system of two vectors for quick and efficient gene cloning. The pMono_T-vector (GenBank ID: JN681269) is designed for direct A-tailed PCR fragments and contains the maize expansin-β signal peptide and GFP gene sequences. Amplicons with a truncated start codon missing the first nucleotide were cloned in-frame with both the upstream signal peptide and the downstream GFP nucleotide sequences (Fig. 1). A fusion cassette excised from the recombinant pMono_T-vector by NotI restriction digestion was moved into the linearized pUbiP2 (GenBank ID: JN681270), also cut with NotI, and placed under the control of the maize ubiquitin gene promoter. The Basta herbicide resistance gene was included as a selectable marker.
Fig. 1.
The expression cassette of the vector developed for Brachypodium transformation. Abbreviations: LB, left border; OCS Term, octopine synthase terminator; BaR, gene for herbicide resistance (Basta, dl-phosphinothricin); Ubi Promoter, maize ubiquitin promoter; nos term, terminator from nopaline synthase; GFP, green fluorescent protein; Target Gene, gene of interest; SP, maize expansin-B signal peptide; RB, right border.
Transformation of mature seeds
To transform seeds, we first prepared the Agrobacterium cultures (50 mL) containing each construct of interest. The culture was grown in LB medium containing 100 mg L−1 spectinomycin and 50 mg L−1 kanamycin at 28 °C to OD600 = 1, sedimented by centrifugation at 5000 g for 10 min, washed with an equal volume of infiltration buffer (10 mm MES, 10 mm MgCl2, pH 5·6) and re-suspended in 50 mL of infiltration buffer containing 50 µm acetosyringone, 0·01 % Silwet-L77 and extracts from Nicotiana tabacum leaves. The tobacco extracts were added to induce the agrobacteria to initiate gene transfer in monocots. To make the extracts, N. tabacum plants were grown in aseptic conditions in half-strength Murashige and Skoog (MS) medium containing 0·5 % gelrite and 1 % sucrose for 4–6 weeks. Then 30–50 g of fresh leaves were cut into 1–3 cm2 squares and pre-incubated in infiltration buffer prior to co-cultivation for 2 h. The liquid phase after addition of acetosyringone and Silwett-L77 was used to re-suspend washed, pelleted, Agrobacterium culture.
Next, we prepared the Brachypodium seeds for transformation. Fifty Brachypodium seeds were soaked in deionized water for 2 h to remove the lemma and awn. They were then rinsed with 70 % ethanol, sterilized for 8 min in 1·3 % (v/v) hypochlorite solution, washed three times in sterile water and immediately used for trimming (Fig. 2). A quarter of the upper part of each Brachypodium seed was removed with scissors in aseptic conditions, avoiding drying. Trimmed seeds, the bottom portion, were co-cultivated with the prepared A. tumefaciens culture (EHA101 strain) carrying the construct of interest. During co-cultivation, the seeds and culture were incubated for 30 h at 21 °C and shaken at 200 rpm.
Fig. 2.

Illustration of Brachypodium seed with the cut area and the part used for the co-cultivation with Agrobacterium cells. (A) Whole initial spikelet; (B) whole seed with removed lemma and awn; (C) trimmed seed prepared for co-cultivation. Scale bar = 3 mm.
We chose 30 h as the duration of co-cultivation to allow the agrobacteria to penetrate the intracellular spaces of the seed tissues and transform embryo cells. After co-cultivation, seeds were rinsed in infiltration buffer and placed on MS medium containing 0·5 % gelrite and 225 mg L−1 timentin for 3 weeks to kill any remaining agrobacteria. At the end of the second week, the surviving seeds germinated and then 1-week-old seedlings were transferred to soil and grown for 2–3 weeks to produce plant mass for analysis. On average, 10 % of co-cultivated seeds were able to germinate. We did not use Basta selection at this stage in order to avoid additional stress on the seedlings.
Fluorescence, genotyping and analysis of hydrolase expression in infiltrated seedlings
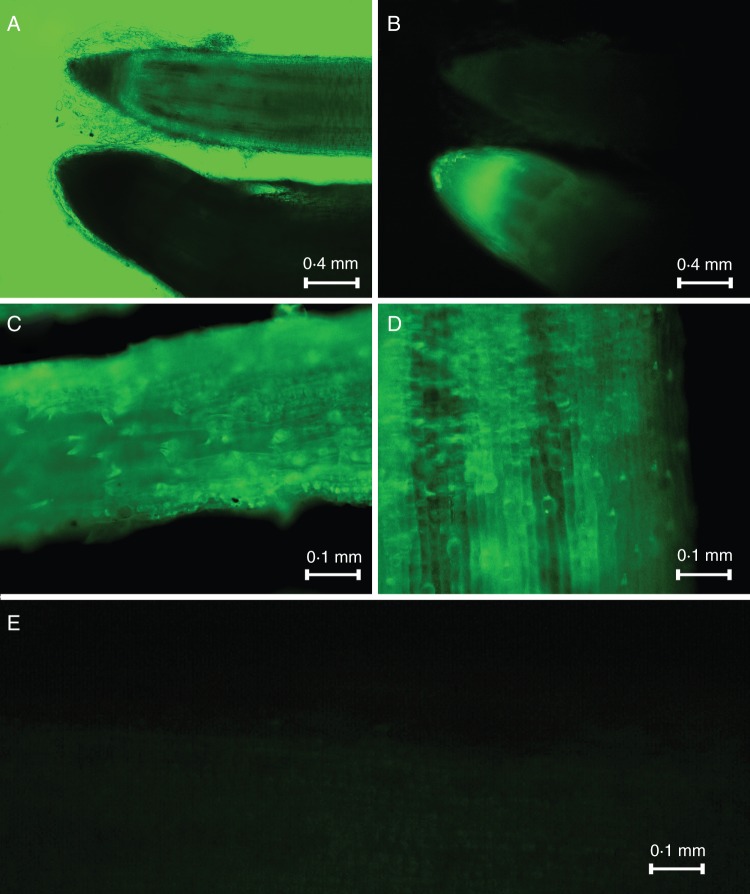
Two-week-old Brachypodium seedlings obtained from transformed seeds were observed by fluorescence microscopy (Fig. 3). Visualization of the roots and leaves confirmed that GFP protein is expressed in roughly 50 % of the plants that survived after infiltration.
Fig. 3.
Fluorescence images of root and leaves of Brachypodium plants expressing AnRAE (A, B), AnAF (C) and AnGAL (D) proteins fused with GFP and wild-type leaf (E). (A) Image without fluorescence excitation; (B) fluorescent image; (A) and (B) show AnRAE (bottom) and wild-type plant roots (top).
To confirm that the fluorescent plants carried the hydrolase expression cassette, genomic DNA of three independent leaves from two independent plants for each construct were used for PCR analysis (Fig. 4A). PCR analysis shows a T-DNA insertion with the hydrolase expression cassette present in the corresponding transgenic lines. Additionally, the flanking sequence tags were isolated for each type of transiently expressed gene, cloned and sequenced (see Supplementary Data Table S2). The results confirmed that the amplified signals originated from plant genomic DNA and not from Agrobacterium cells that survived in plants grown on media containing timentin.
Fig. 4.
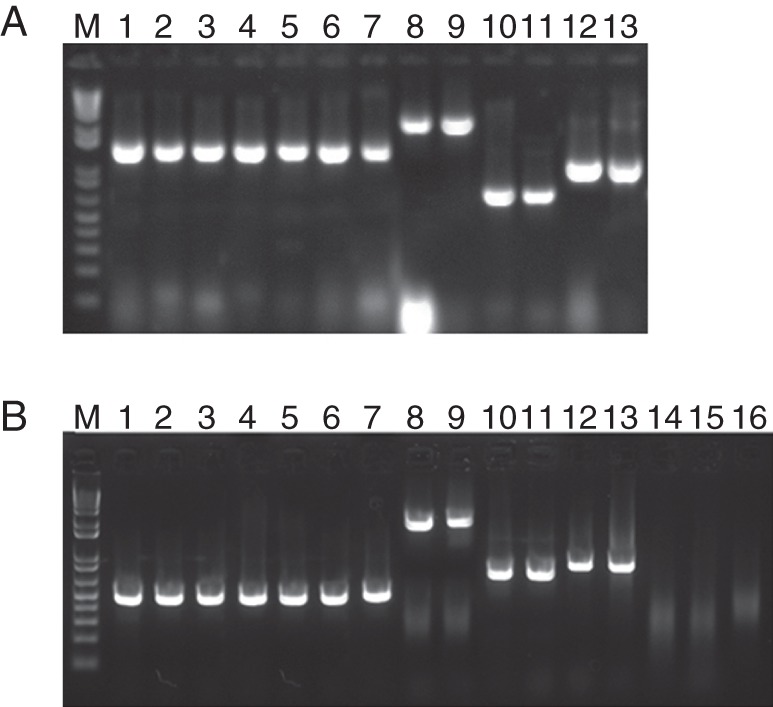
(A) PCR analysis using genomic DNA and (B) RT–PCR using total RNA extracted from the wild-type Brachypodium plants and the plants expressing microbial hydrolases (see Supplementary Data Table S1 for sequences; M = marker). (A) Lanes 1–7: amplification of a glyceraldehyde phosphate dehydrogenase (GADPH) housekeeping gene fragment from genomic DNA of Brachypodium plants carrying an AnGAL expression cassette (lanes 1 and 2), an AnRAE expression cassette (lanes 3 and 4), an AnAF expression cassette (lanes 5 and 6) and a wild-type plant (lane 7). Lanes 8–13: amplification of a hydrolase gene from genomic DNA of Brachypodium plants carrying an AnGAL expression cassette (lanes 8 and 9), an AnRAE expression cassette (lanes 10 and 11) and an AnAF expression cassette (lanes 12 and 13). (B) Lanes 1–7: amplification of a GADPH housekeeping gene fragment from cDNA of Brachypodium plants carrying an AnGAL expression cassette (lanes 1 and 2), an AnRAE expression cassette (lanes 3 and 4), an AnAF expression cassette (lanes 5 and 6), and a wild type plant (lane 7). Lanes 8–13: amplification of hydrolase genes from cDNA of Brachypodium plants carrying an AnGAL expression cassette (lanes 8 and 9), an AnRAE expression cassette (lanes 10 and 11) and an AnAF expression cassette (lanes 12 and 13). Lanes 14–16: amplification of hydrolase genes from cDNA of wild-type Brachypodium plants using gene-specific primers for AnGAL (lane 14), AnRAE (lane 15) and AnAF (lane 16).
To confirm gene expression, total RNA was isolated from transfected plants and used for RT–PCR. The obtained cDNA products showed the expected sizes corresponding to expressed hydrolase genes, whereas the wild-type sample did not show any positive signals (Fig. 4B).
Transiently expressed hydrolases maintain their hydrolytic activity in apoplast of Brachypodium plants
To confirm that transiently expressed proteins accumulated in the cell wall, apoplastic fluids were prepared from transfected plants and analysed by western blot with anti-GFP monoclonal antibodies. Apoplastic fluids were prepared from the fluorescent tissues of plants expressing transformed constructs, and western blotting showed a band corresponding to the size of the expressed protein linked with GFP. The predicted size of α-l-arabinofuranosidase is 27 kDa, rhamnogalacturonan acetylesterase is 36 kDa, α-galactosidase is 83 kDa and GFP is 26 kDa. Thus, the GFP fusion proteins have sizes of 62, 53 and 109 kDa, respectively (Fig. 5).
Fig. 5.
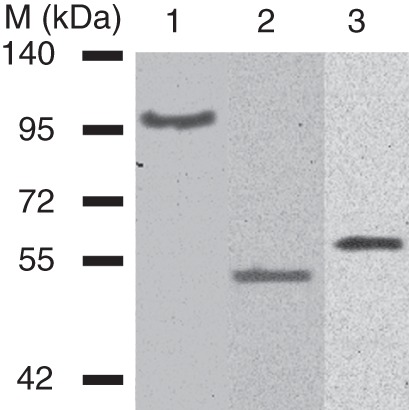
Western blot analysis of total proteins from apoplast of Brachypodium plants expressing microbial hydrolases. On the left are shown protein size markers (M). Corresponding microbial proteins fused with GFP were found in all Brachypodium plants selected by fluorescence: lane 1, a plant expressing an AnGAL–GFP fusion protein (62 kDa); lane 2, a plant expressing an AnRAE–GFP fusion protein (53 kDa); lane 3, a plant expressing an AnAF–GFP fusion protein (109 kDa). Proteins were detected using anti-GFP monoclonal antibodies (at a dilution of 1 : 5000).
To confirm that the expressed hydrolases maintained their activity, apoplastic fluids extracted from three different leaves of two independent plants for each hydrolase expression cassette were used for enzymatic activity assays. All conditions for the assays were first established using commercially available enzymes with a similar hydrolytic activity.
The activities of A. nidulans rhamnogalacturonan acetylesterase, α-l-arabinofuranosidase and α-glucosidase were determined using the substrates pNP-acetyl, pNP-α-furanoside and pNP-α-galactopyranoside, respectively. Enzymatic activity was estimated by measuring the amount of pNP released as a function of time. Transgenic plants expressing each of these enzymes displayed significantly higher activity in the apoplastic fluids compared with wild-type plants (Fig. 6). When apoplastic fluids were boiled for 10 min, hydrolytic activities of expressed proteins were depleted (see Supplementary Data Figure S1).
Fig. 6.
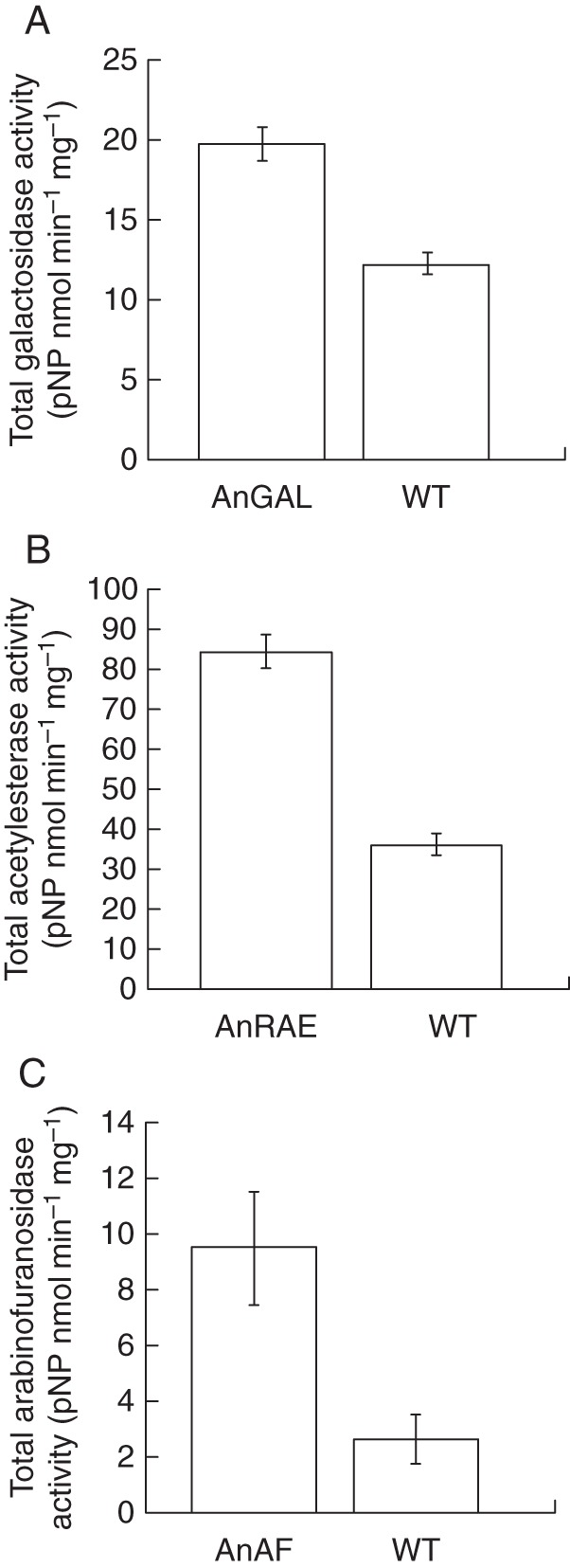
Hydrolytic activities in the apoplast of Brachypodium wild-type plants and plants expressing microbial hydrolases. (A) α-Galactosidase activity (pNP-α-galactopyranoside was used as a substrate) in the plants expressing AnGAL and in wild-type plants. (B) Rhamnogalacturonan acetylesterase activity (pNP-acetyl was used as a substrate) in the plants expressing AnRAE and in wild-type plants. (C) α-Arabinofuranosidase activity (pNP-arabinoside was used as a substrate) in the plants expressing AnAF and in wild-type plants. In all assays, apoplast fluids were prepared from three independent batches of leaves collected from two independent regenerated plants, and three independent batches of leaves collected from two independent wild-type plants; all plants were the same age. Mean ± s.d. (t-test, n = 6, P < 0·05). Result for negative control reactions are presented in Supplementary Data Figure S1.
Transient expression of A. nidulans hydrolases affects cell wall composition
To examine the effect of expressed hydrolases on the plant cell wall, we analysed the composition of cell wall material isolated from wild-type and transfected plants. Cell walls were extracted from the plant tissues remaining after apoplastic fluid preparations. Both α-galactosidase and α-l-arabinosidase (AnGAL and AnAF) caused significant cell wall alterations in comparison with the wild type (Table 1). Transfected plants expressing AnGAL showed a 29 % reduction of galactose content in comparison with wild-type plants (Table 1). Cell walls isolated from AnAF-expressing plants had a 12 % reduction of arabinose content in comparison with wild-type plants (Table 1).
Table 1.
Monosaccharide composition (mol %) of cell wall from infiltrated Brachypodium plants expressing AnGAL (12 weeks old), AnAF (8 weeks old) and wild-type plants
| Fuc | Ara | Rha | Gal | Glc | Xyl | Man | GalA | |
|---|---|---|---|---|---|---|---|---|
| WT-1 | 0·68 ± 0·06 | 19·26 ± 1·17 | 0·96 ± 0·13 | 6·61 ± 0·40* | 12·28 ± 0·48 | 52·07 ± 1·35 | 4·79 ± 0·55 | 3·33 ± 0·05 |
| AnGAL | 0·58 ± 0·13 | 16·65 ± 2·54 | 0·71 ± 0·22 | 4·70 ± 0·11* | 14·60 ± 2·38 | 53·38 ± 0·76 | 5·94 ± 0·53 | 3·28 ± 0·03 |
| WT-2 | 0·20 ± 0·03 | 10·78 ± 0·29* | 2·90 ± 0·45 | 3·42 ± 0·31 | 11·75 ± 0·78 | 65·16 ± 0·44 | 2·01 ± 0·17 | 3·59 ± 0·31 |
| AnAF | 0·16 ± 0·04 | 9·46 ± 0·13* | 2·58 ± 0·02 | 2·78 ± 0·58 | 10·13 ± 1·46 | 68·97 ± 1·91 | 2·22 ± 0·10 | 3·79 ± 0·04 |
* Significantly different (t-test, P < 0·05, n = 6).
Plants expressing AnRAE did not show statistically different acetyl content in their cell walls compared with wild-type plants (Fig. 7A). This is probably due to the insignificant amount of pectins in Brachypodium cell walls.
Fig. 7.
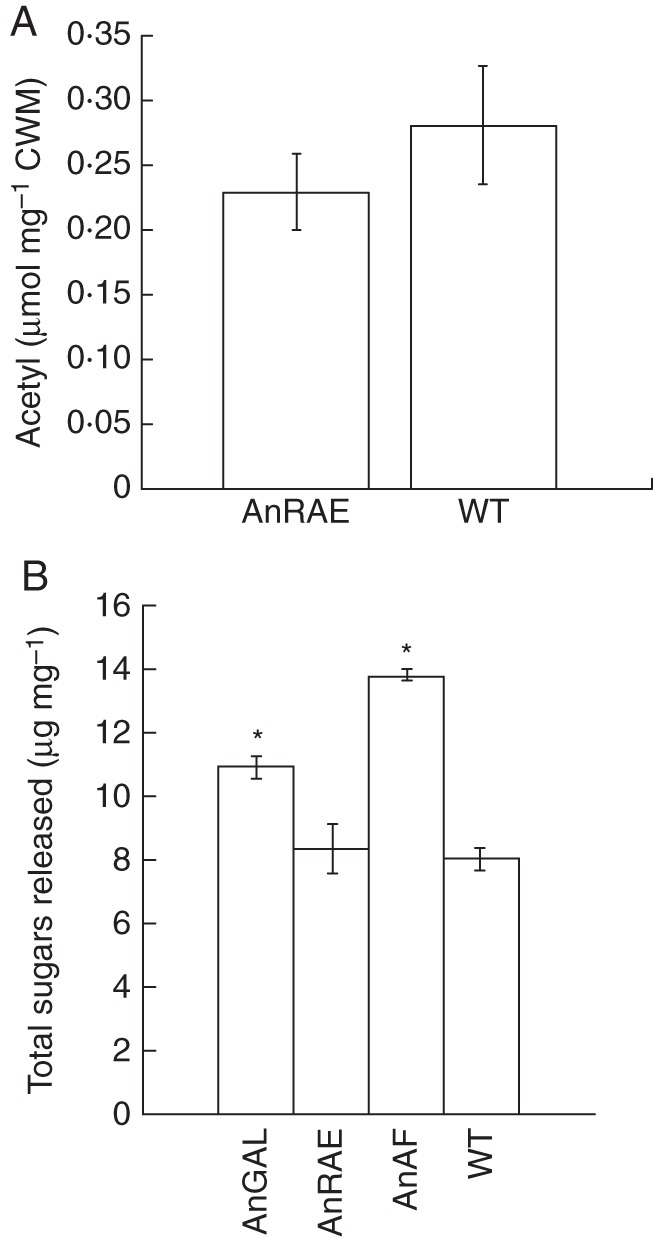
(A) Acetyl content (μmol mg−1 of dry cell wall material) in the cell wall of Brachypodium plants expressing AnRAE and of wild-type plants. Analyses were done using three independent leaves collected from two independent regenerated plants or wild-type plants grown simultaneously. Bars show the mean ± standard deviation. The difference between infiltrated and wild-type plants is not significant (t-test, n = 6, P > 0·05). (B) Amount of reducing sugars released after 8 h incubation of 10 mg of dry cell wall material with cellulase mixture (4 U of cellulase from T. reesei and 1 U of cellobiase from A. niger). *Difference is significant (t-test, P < 0·05; n = 6).
Modification of the cell wall by transiently expressed active hydrolases reduces cell wall recalcitrance
To determine whether the modification of cell wall composition by expression of microbial hydrolases led to changes in plant biomass digestibility, a saccharification assay was carried out on cell wall samples. Dry cell wall material was treated with a cellulase mixture for 8 h, and then the amount of released reducing sugars was measured as described in the Materials and Methods (Fig. 7B). Brachypodium plants expressing AnGAL and AnAF showed significantly higher amounts of released sugars in comparison with wild-type plants by 36 and 73 %, respectively. There was no significant difference between plants expressing AnRAE and wild-type plants.
DISCUSSION
We have developed a technique for Agrobacterium-mediated transient gene expression and applied it to B. distachyon plants. This technique allowed us to generate transfected chimeric plants carrying the genes of three A. nidulans hydrolases. Expression of GFP–hydrolase fusion proteins allowed a quick and easy selection of transformed tissues using a fluorescence microscope. We confirmed that using the maize β-expansin signal peptide ensured the localization of expressed proteins to the cell wall. Agrobacterium penetrates into the plant through sites of wounding and multiplies in the apoplast, then initiates gene transfer into the plant cells.
The main goal in developing our approach was to achieve gene transfer from Agrobacterium cells to Brachypodium cells. This gene transfer is activated by the TRA gene cluster, which is controlled by promoters sensitive to plant metabolites. Normally, Agrobacterium does not recognize monocotyledonous plants as a host organism. In dicot plants, specific phenolic metabolites from extracellular spaces are essential for the activation of gene transfection from Agrobacterium to plant cells (Tinland, 1996), but metabolites from monocots are unable to initiate T-DNA transfer. Accordingly, we added N. tabacum leaf extract and acetosyringone to Agrobacterium culture prior to co-cultivation with trimmed seeds to initiate T-DNA transfection.
This transfection has to be done at the earliest, embryonic stage of plant development, when the embryo has a small number of cells that can be transformed and carry the insertion through subsequent divisions. Generative organs developed from the transformed area can produce transgenic seeds and thus stable transformants in the next generation. By trimming the seeds and leaving the intact embryo, as well as a sufficient portion of the cotyledon to support seed germination, we facilitated Agrobacterium penetration into the embryonic cells.
After co-cultivation of the Brachypodium seeds with Agrobacterium culture, approx. 10 % of the seeds germinated. Analysis of the T0 plants by fluorescence microscopy allowed for a quick selection of the plants with transformed leaf tissues and demonstrated 50 % transformation efficiency. Fluorescent tissues were confirmed to carry the corresponding gene insertion into the genomic DNA and to express active proteins localized in cell walls.
Currently, the widely used technique to introduce genetic constructs into the genome of monocotyledonous plants, including Brachypodium, is Agrobacterium-mediated transformation of embryonic callus. This approach applied to Brachypodium can have a transformation efficiency of up to 36·5 % (Vogel and Hill, 2008), and transformation of wheat has significantly lower efficiency, with a maximum of 6 % (Jones et al., 2005). Transformation via particle bombardment usually has efficiencies in the range of 5·3–14 % depending on the species (Christiansen et al., 2005). The main downside of this approach is the requirement for initiation and handling of callus cultures in sterile conditions and maintaining them for a long period. In addition, up to 8 months are required to obtain transformed seeds (Bevan et al., 2010). The attempt to adopt the floral dip protocol developed for dicots to be useful for wheat transformation showed dramatically lower transformation efficiency at 0·44 % (number of transformants/number of florets dipped) (Zale et al., 2009). However, the advantage of the latter is the ability to obtain next-generation transformed plants much faster in comparison with callus transformation methods. Another described method for wheat transformation avoiding callus requires piercing of soaked seeds with a needle to inject Agrobacterium into a seed (Supartana et al., 2006). The reported efficiency of transformation was approx. 50 %, but was not statistically analysed due to the limited number of plants. Delivery of Agrobacterium culture directly to the embryonic cells via piercing facilitates transformation in comparison with transformation of callus followed by regeneration of transformed plants. However, this method requires substantial experience in piercing of the seeds to avoid embryo damage, and is a comparatively tedious and slow procedure. Recently, a high-throughput Agrobacterium-mediated transient gene expression system (Chen et al., 2010) using inoculation of young switchgrass seedlings with Agrobacterium culture has been reported. Transient expression of the β-glucuronidase (GUS)-encoding gene was increased from 6 % in untreated seedlings to 54 % after a combination of wounding treatments and the addition of 100 µm acetosyringone. However, GUS activity was observed only within 5–6 d after co-incubation of the seedlings with Agrobacterium, and then disappeared. In our method, co-incubation of mature Brachypodium seeds with Agrobacterium produced transgenic plant tissues that maintained expression of active proteins for a long period, up to 12 weeks, which allows analyses at different stages of plant development.
Our method has some other advantages in comparison with the previously described approaches for Agrobacterium-mediated transient gene expression in monocotyledonous plants, and possibly for stable transformation in the future. Our method of transient gene expression allows, for example, determination or confirmation of gene function in a short period of time (2–3 weeks) after co-cultivation of seeds with Agrobacterium culture. This time period is significantly shorter than the 2–3 months required for transformation via callus and regeneration of plants. Using seed trimming instead of needle injection makes the procedure substantially simpler and does not require extensive time or experience. Although our method has a lower number of germinating seeds after co-cultivation with Agrobacterium cells, the greater number of transformed plants among the survivors provides a higher transformation efficiency compared with other non-callus transformation methods. Due to the fact that after co-cultivation with Agrobacterium the embryonic cells in the seeds are transformed randomly, giving a different transformed/non-transformed cell ratio, we did not use selection for herbicide tolerance during seed germination, in order not to lose those transformants with a low number of transformed embryonic cells. Instead, we used GFP fused with the expressed proteins to screen for transformed plants. The seeds obtained from regenerated chimeric plants will be subjected to selection in the future to recover potential stable transformants.
Applying the transformation technique, we expressed three A. nidulans hydrolases, rhamnogalacturonan acetyl esterase (AnRAE), α-galactosidase (AnGAL) and α-arabinofuranosidase (AnAF) in Brachypodium plant tissues (leaves and roots), and demonstrated that they localize in the cell wall, maintain hydrolytic activities and cause cell wall modifications. We observed that tissues with detectable GFP fluorescence always showed a band of the corresponding size on western blots and showed a higher level of enzymatic activity in their apoplastic fluid in comparison with apoplastic fluid prepared from wild-type plants. The correlation between the detected fluorescence and the presence of active proteins of the correct size confirms the feasibility of using GFP fusion proteins for a quick selection of transformants, as was demonstrated earlier for expression of microbial hydrolases in Arabidopsis (Pogorelko et al., 2011). Two of the selected Aspergillus genes, AnAF and AnGAL, caused a statistically significant reduction of arabinose and galactose, respectively, in transgenic Brachypodium tissues, but expression of AnRAE did not decrease the acetyl content of the cell wall despite the high activity of the expressed enzyme in transformed tissues. Cell walls of monocotyledonous plants contain a very low amount of pectic polysaccharides; therefore, acetyl groups from rhamnogalacturonan represent a minor portion of the total acetyl groups present in hemicellulosic polysaccharides. Most probaby, AnRAE specifically recognizes rhamnogalacturonan, and de-esterification of a minor component does not produce significant changes in the total acetyl content of the Brachypodium cell wall.
Expression of specific microbial hydrolases in the plant apoplast is a promising approach to modifying cell wall recalcitrance to biodegradability. Saccharification assays performed with cell walls prepared from transgenic Brachypodium tissues demonstrated a significant effect of two expressed hydrolases, AnAF and AnGAL, on digestibility. There are a few examples when expression of microbial hydrolases, particularly in plant cell walls, increased biomass digestibility in maize, tobacco, Festuca arundinacea and arabidopsis (Sticklen, 2007; Jung et al., 2010; Buanafina et al., 2010; Pogorelko et al., 2011). An almost 70 % increase in cell wall digestibility in AnAF-expressing Brachypodium plants can have great potential; however, currently we do not know how it will affect plant morphology in the stable transformants.
Conclusions
Here we describe a new technique for transient gene expression in the monocotyledonous plant B. distachyon; in addition, this technique may produce stable transformants in the next generation. This method is a rapid and easy way to express a gene of interest in plant tissues, and we demonstrated its applicability for expression of specific enzymes. Transiently expressed hydrolases from A. nidulans were expressed in all tissues and directed to the apoplast by fusion with an apoplast-specific signal peptide. Fusion of the protein with GFP allows for rapid selection of transformed plants or tissues and does not disrupt the protein's activity. Modification of Brachypodium cell walls by expressed hydrolases increased plant biomass degradability. These results confirmed the applicability and efficiency of our approach for rapid screening of different genes, for example hydrolases, which can facilitate their functional characterization for biotechnological applications such as biomass modification or pre-treatment for biofuel production. We believe that this approach can be applied to other monocotyledonous species and in the future will lead to development of efficient methods for stable transformation of monocots.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr S. Yasuda (National Instutute of Genetics, Shizuoka, Japan) for the pXcmkn12 (AB107950·1) vector and Rachel Ann Morris for help in manuscript preparation. This work was supported by the grant #09-3384 obtained from Roy J. Carver Charitable Trust (2009–2011), and we express our gratitude for their valuable support.
LITERATURE CITED
- An G, Lee S, Kim SH, Kim SR. Molecular genetics using T-DNA in rice. Plant and Cell Physiology. 2005;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- Azhaguvel P, Lia W, Rudda JC, Gill BS, Michels GJ, Weng Y. Aphid feeding response and microsatellite-based genetic diversity among diploid Brachypodium distachyon accessions. Plant Genetic Resources. 2009;7:72–79. [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proceedings of the National Academy of Sciences, USA. 2006;103:11417–11422. doi: 10.1073/pnas.0604632103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW, Garvin DF, Vogel JP. Brachypodium distachyon genomics for sustainable food and fuel production. Current Opinion in Biotechnology. 2010;21:211–217. doi: 10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buanafina M, Langdon T, Hauck B, Dalton S, Timms-Taravella E, Morris P. Targeting expression of a fungal ferulic acid esterase to the apoplast, endoplasmic reticulum or Golgi can disrupt feruloylation of the growing cell wall and increase the biodegradability of tall fescue (Festuca arundinacea) Plant Biotechnology Journal. 2010;8:316–331. doi: 10.1111/j.1467-7652.2009.00485.x. [DOI] [PubMed] [Google Scholar]
- Charles M, Tang H, Belcram H, Paterson A, Gornicki P, Chalhoub B. Sixty million years in evolution of soft grain trait in grasses: emergence of the softness locus in the common ancestor of Pooideae and Ehrhartoideae. Molecular Biology and Evolution. 2009;26:1651–1661. doi: 10.1093/molbev/msp076. [DOI] [PubMed] [Google Scholar]
- Chen X, Equi R, Baxter H, Berk K, Han J, Agarwal S, Zale J. A high-throughput transient gene expression system for switchgrass (Panicum virgatum L.) seedlings. Biotechnology for Biofuels. 2010;21:963–977. doi: 10.1186/1754-6834-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen P, Andersen CH, Didion T, Folling M, Nielsen KK. A rapid and efficient transformation protocol for the grass Brachypodium distachyon. Plant Cell Reports. 2005;23:751–758. doi: 10.1007/s00299-004-0889-5. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology. 2001;127:1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Faricelli ME, Valarik M, Dubcovsky J. Control of flowering time and spike development in cereals: the earliness per se Eps-1 region in wheat, rice, and Brachypodium. Functional and Integrated Genomics. 2009;10:293–306. doi: 10.1007/s10142-009-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin DF, Gu Y-Q, Hasterok R, et al. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Plant Genome. 2008;48:69–84. [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology. 1992;6:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wanjugi H, Coleman-Derr D, Kong X, Anderson OD. Conserved globulin gene across eight grass genomes identify fundamental units of the loci encoding seed storage proteins. Functional and Integrated Genomics. 2010;10:111–122. doi: 10.1007/s10142-009-0135-x. [DOI] [PubMed] [Google Scholar]
- Hess D, Dressler K, Nimmrichter R. Transformation experiments by pipetting Agrobacterium into the spikelets of wheat (Triticum aestivum L.) Plant Science. 1990;72:233–244. [Google Scholar]
- Himmelbach A, Zierold U, Hensel G, et al. A set of modular binary vectors for transformation of cereals. Plant Physiology. 2007;145:1192–1200. doi: 10.1104/pp.107.111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Doherty A, Wu H. Review of methodologies and a protocol for the Agrobacterium-mediated genetic transformation of wheat. Plant Methods. 2005;1:5. doi: 10.1186/1746-4811-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Kim S, Bae H, Lim HS, Bae HJ. Expression of thermostable bacterial beta-glucosidase (BglB) in transgenic tobacco plants. Bioresource Technology. 2010;101:7155–7161. doi: 10.1016/j.biortech.2010.03.140. [DOI] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco DL, Vensel WH. Globulins are the main seed storage proteins in Brachypodium distachyon. Theoretical and Applied Genetics. 2008;117:555–563. doi: 10.1007/s00122-008-0799-y. [DOI] [PubMed] [Google Scholar]
- Lever M. New reaction for colorimetric determination of carbohydrates. Analytical Biochemistry. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- McComb EA, McCready RM. Determination of acetyl in pectin and in acetylated carbohydrate polymers. Analytical Chemistry. 1957;29:819–821. [Google Scholar]
- Mur LAJ, Xu R, Casson SA, Stoddart WM, Routledge APM, Draper J. Characterisation of a proteinase inhibitor from Brachypodium distachyon suggests the conservation of defence signalling pathways between grasses and dicots. Molecular Plant Pathology. 2004;5:267–280. doi: 10.1111/j.1364-3703.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- Olsen P, Lenk I, Jensen CS, et al. Analysis of two heterologous flowering genes in Brachypodium distachyon demonstrates its potential as a grass model plant. Plant Science. 2006;170:1020–1025. [Google Scholar]
- Parker D, Beckmann M, Enot DP, et al. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nature Protocols. 2008;3:435–445. doi: 10.1038/nprot.2007.499. [DOI] [PubMed] [Google Scholar]
- Pogorelko GV, Fursova OV. A highly efficient miPCR method for isolating FSTs from transgenic Arabidopsis thaliana plants. Journal of Genetics. 2008;87:133–140. doi: 10.1007/s12041-008-0020-8. [DOI] [PubMed] [Google Scholar]
- Pogorelko G, Fursova O, Lin M, Pyle E, Jass J, Zabotina OA. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Molecular Biology. 2011;77:433–445. doi: 10.1007/s11103-011-9822-9. [DOI] [PubMed] [Google Scholar]
- Schwartz CJ, Doyle MR, Manzaneda AJ, Rey PJ, Mitchell-Olds T, Amasino RM. Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioenergy Research. 2010;3:38–46. [Google Scholar]
- Stewart CN, Jr, Via L. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques. 1993;14:748–749. [PubMed] [Google Scholar]
- Sticklen M. Feedstock crop genetic engineering for alcohol fuels. Crop Science. 2007;47:2238–2248. [Google Scholar]
- Supartana P, Shimizu T, Nogawa M, et al. Development of simple and efficient in planta transformation method for wheat using Agrobacterium tumefaciens. Journal of Bioscience and Bioengineering Japan. 2006;102:162–170. doi: 10.1263/jbb.102.162. [DOI] [PubMed] [Google Scholar]
- Taylor NJ, Fauquet CM. Microparticle bombardment as a tool in plant science and agricultural biotechnology. DNA Cell Biology. 2002;21:963–977. doi: 10.1089/104454902762053891. [DOI] [PubMed] [Google Scholar]
- Thole V, Alves SC, Worland B, Bevan MW, Vain P. A protocol for efficiently retrieving and characterizing flanking sequence tags (FSTs) in Brachypodium distachyon T-DNA insertional mutants. Nature Protocols. 2009;4:650–661. doi: 10.1038/nprot.2009.32. [DOI] [PubMed] [Google Scholar]
- Tinland B. The integration of T-DNA into plant genomes. Trends in Plant Science. 1996;1:178–184. [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Reports. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Xu JH, Messing J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theoretical and Applied Genetics. 2009;119:1397–1412. doi: 10.1007/s00122-009-1143-x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ohlrogge JB. Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis β-oxidation mutants. Plant Physiology. 2009;150:1981–1989. doi: 10.1104/pp.109.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina O, Malm E, Drakakaki G, Bulone V, Raikhel N. Identification and preliminary characterization of a new chemical affecting glucosyltransferase activities involved in plant cell wall biosynthesis. Molecular Plant. 2008;1:977–989. doi: 10.1093/mp/ssn055. [DOI] [PubMed] [Google Scholar]
- Zale JM, Agarwal S, Loar S, Steber CM. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Reports. 2009;28:903–913. doi: 10.1007/s00299-009-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.