Abstract
Background and Aims
The perianths of the Lardizabalaceae are diverse. The second-whorl floral organs of Sinofranchetia chinensis (Lardizabalaceae) are nectar leaves. The aim of this study was to explore the nature of this type of floral organ, and to determine its relationship to nectar leaves in other Ranunculales species, and to other floral organs in Sinofranchetia chinensis.
Methods
Approaches of evolutionary developmental biology were used, including 3′ RACE (rapid amplification of cDNA ends) for isolating floral MADS-box genes, phylogenetic analysis for reconstructing gene evolutionary history, in situ hybridization and tissue-specific RT-PCR for identifying gene expression patterns and SEM (scanning electron microscopy) for observing the epidermal cell morphology of floral organs.
Key Results
Fourteen new floral MADS-box genes were isolated from Sinofranchetia chinensis and from two other species of Lardizabalaceae, Holboellia grandiflora and Decaisnea insignis. The phylogenetic analysis of AP3-like genes in Ranunculales showed that three AP3 paralogues from Sinofranchetia chinensis belong to the AP3-I, -II and -III lineages. In situ hybridization results showed that SIchAP3-3 is significantly expressed only in nectar leaves at the late stages of floral development, and SIchAG, a C-class MADS-box gene, is expressed not only in stamens and carpels, but also in nectar leaves. SEM observation revealed that the adaxial surface of nectar leaves is covered with conical epidermal cells, a hallmark of petaloidy.
Conclusions
The gene expression data imply that the nectar leaves in S. chinensis might share a similar genetic regulatory code with other nectar leaves in Ranunculales species. Based on gene expression and morphological evidence, it is considered that the nectar leaves in S. chinensis could be referred to as petals. Furthermore, the study supports the hypothesis that the nectar leaves in some Ranunculales species might be derived from stamens.
Keywords: Nectar leaves, perianth, petals, Ranunculales, Lardizabalaceae, Sinofranchetia chinensis, MADS-box, expression pattern, evolutionary developmental biology
INTRODUCTION
The reproductive organs of most angiosperms are enclosed by a sterile outer structure, which is usually called the perianth. The perianth, as a remarkable novelty in angiosperms, generally contains two whorls and shows great morphological diversity. The perianth shape ranges in different lineages from undifferentiated to bipartite (i.e. differentiated into sepals and petals; Cronquist, 1988; Takhtajan, 1997; Zanis et al., 2003; Endress and Matthews, 2006; Ronse De Craene, 2008). The various morphologies of perianths in different angiosperm lineages have led to the suggestion that perianths could have different origins (Endress, 1994, 2006). Particularly, petals (the inner part of the perianth) have been thought to have evolved several times independently from sterile stamens (andropetals) or bracts (bracteopetals) during angiosperm evolution (e.g. Eames, 1961; Weberling, 1989; Friis and Endress, 1990; Takhtajan, 1991; Endress, 1994, 2001). However, the molecular evolutionary mechanisms responsible for the different origins of petals remain unclear.
Ranunculales, the earliest-diverging lineage in eudicots, displays extreme diversity in perianth morphology and has received increasing attention from evolutionary-developmental biologists (e.g. Albert et al., 1998; Kramer et al., 1998, 2003, 2007; Kramer and Irish, 1999, 2000; Theissen et al., 2002; Rasmussen et al., 2009; Kramer and Hodges, 2010; Sharma et al., 2011). This order is composed of seven families according to recent molecular phylogenetic studies: Ranunculaceae, Berberidaceae, Menispermaceae, Lardizabalaceae, Circaeasteraceae, Papaveraceae and Eupteleaceae (Angiosperm Phylogeny Group III, 2009; Wang et al., 2009). The majority of genera in the Ranunculaceae, Berberidaceae, Menispermaceae, Lardizabalaceae and Papaveraceae have bipartite perianths, whereas there is no clear differentiation of the perianth in Circaeasteraceae, and the flowers lack perianths in Eupteleaceae (Qin, 1997; Damerval and Nadot, 2007; Rasmussen et al., 2009; Takhtajan, 2009; Wang et al., 2009; Zhang and Ren, 2011). Diverse perianth architectures are present in Ranunculales, including petaloid sepals, floral nectariferous organs and spurs (Qin, 1997; Damerval and Nadot, 2007; Rasmussen et al., 2009; Takhtajan, 2009; Wang et al., 2009; Zhang et al., 2009; Zhang and Ren, 2011). The floral nectariferous organs are also called nectar leaves, which are nectar-bearing and formed between the perianth and androecium (Janchen, 1949). In Ranunculales, nectar leaves are found in some species of Ranunculaceae, Berberidaceae, Menispermaceae and Lardizabalaceae (Erbar et al., 1998; Ronse De Craene, 2010). Furthermore, nectar leaves in different species of Ranunculales display different morphologies: some are small and greenish, such as in Sinofranchetia (Lardizabalaceae); some are large and petaloid, such as in Ranunculus (Ranunculaceae); some are converted to long spurs, such as in Aquilegia (Ranunculaceae) (Leppik, 1988; Weberling, 1989; Zhang et al., 2009). The diversification of nectar leaves in different species of Ranunculales is also reflected in floral morphogenesis: some types of nectar leaves share common primordia with stamens, such as in Berberidaceae and Holboellia (Lardizabalaceae); some develop from individual primordia distinguished from that of stamens, such as in Sinofranchetia (Lardizabalaceae) (Zhang et al., 2009; Ronse De Craene, 2010; Zhang and Ren, 2011). In addition, the nectar leaves have ever been referred to as ‘petals’, especially in some species of Ranunculaceae and Berberidaceae, because these nectar leaves are colourful, sterile and positioned in the second whorl of the bipartite perianth, fitting the broader definition of petals (e.g. Cronquist, 1981; Kramer et al., 2007; Kramer and Hodges, 2010). However, some other studies considered that the term ‘petals’ for these organs is mainly related to the function of display rather than the morphological concept (Leppik, 1988; Weberling, 1989). Furthermore, some previous studies of the species of Ranunculaceae and Berberidaceae suggested that the nectar leaves might be derived from the sterilization of stamens (Endress, 1995; Ronse De Craene, 2010).
In the past two decades, great advances have been made in evolutionary developmental biology studies of angiosperm flowers. New models and theories have been proposed based on the evolution, expression and functional analyses of genes involved in floral development, besides floral morphology and morphogenesis (Albert et al., 1998; Baum and Whitlock, 1999; Kramer and Irish, 1999, 2000; Irish, 2003; Kramer et al., 2003; Soltis et al., 2004). These models and theories promote our understanding of the molecular mechanisms for the origin and evolution of angiosperm flowers. Among them, the best-known one is the floral ABCE model, which was proposed from genetic analyses of two core eudicot species, Arabidopsis thaliana and Antirrhinum majus. In this model, the identity of floral organs is determined by the combinations of four classes of genes: A + E class genes are responsible for the specification of sepals, A + B + E for petals, B + C + E for stamens and C + E for carpels (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Colombo et al., 1995; Ma and dePamphilis, 2000; Pelaz et al., 2000; Theissen, 2001b). Most of the A-, B-, C- and E-class genes are MIKCC-type MADS-box genes and they belong to the AP1/FUL (A-class), AP3/PI (B-class), AG (C-class) and SEP (E-class) MADS-box gene subfamilies, respectively (Litt and Irish, 2003; Kramer et al., 2004; Irish and Litt, 2005; Zahn et al., 2005a, b, 2006; Kramer and Zimmer, 2006; Shan et al., 2007, 2009). The ABCE model works relatively well for most core eudicot flowers with well-differentiated sepals and petals, but not for most species of basal eudicots and basal angiosperms with less derived perianth architecture (Soltis et al., 2007). Accordingly, some modified ABCE models have been put forward, such as the ‘sliding boundary’ model and ‘fading borders’ model (Soltis et al., 2007). The ‘sliding boundary’ model suggests that the boundary of B-class gene expression can slide across the developing flower from its pre-existing location to the outer perianth whorl (e.g. Kanno et al., 2003; Kramer et al., 2003; Ochiai et al., 2004; Kramer and Jaramillo, 2005; Hintz et al., 2006; Kramer and Zimmer, 2006; Ronse De Craene, 2007). In addition, the labile petal/stamen boundary in the flower has been suggested to correspond to the sliding of the A–C boundary (Goto et al., 2001; Theissen, 2001a; Kim et al., 2005; Chanderbali et al., 2006, 2009; Xu et al., 2006; Dubois et al., 2010). The ‘fading borders’ model suggests that the gradual transitions in floral organ morphology are due to the gradient in the expression levels of floral organ identity genes (Buzgo et al., 2004; Kim et al., 2005; Soltis et al., 2006; Soltis et al., 2007).
Both the ABCE model and its derived models emphasize the importance of B-class MADS-box genes for petal identity specification in core eudicots and for the development of petal-like structures in basal eudicots and basal angiosperms (Weigel and Meyerowitz, 1994; Albert et al., 1998; Kramer et al., 1998, 2003, 2007; Kramer and Irish, 2000; Theissen et al., 2002; Lamb and Irish, 2003; Aoki et al., 2004; Kim et al., 2004; Kramer and Jaramillo, 2005; Zahn et al., 2005b; Rasmussen et al., 2009; Sharma et al., 2011). Phylogenetic studies have indicated that two major gene duplication events occurred during the evolution of B-class genes, one before the origin of angiosperms giving rise to the AP3 and PI lineages, and the other before the divergence of core eudicots leading to the euAP3 and TM6 lineages (Kramer et al., 1998, 2006; Kramer and Irish, 2000; Aoki et al., 2004; Kim et al., 2004; Stellari et al., 2004). These gene duplication events and subsequent functional diversification have resulted in modifications of floral organ identity programmes in different plant groups (Kramer et al., 1998, 2003; Rasmussen et al., 2009; Specht and Bartlett, 2009). It was also found that two recent gene duplication events had occurred during the evolution of AP3-like genes in Ranunculales, giving rise to three AP3 lineages (AP3-I, -II and -III), which in turn enable gene subfunctionalization (Kramer et al., 2003; Rasmussen et al., 2009; Sharma et al., 2011). The recent studies in Ranunculales have suggested that the diversification of AP3-like genes is responsible for petaloidy diversity (e.g. Rasmussen et al., 2009; Specht and Bartlett, 2009). In particular, the genes from the AP3-III lineage are found to be petal-specific in the Ranunculaceae and Berberidaceae (Kramer et al., 2003; Rasmussen et al., 2009; Sharma et al., 2011). Therefore, AP3-like genes are good candidates for studying the molecular mechanisms regulating petal or petal-like structure specification across different species.
In this study, as an initial step towards understanding the development of nectar leaves in Lardizabalaceae at the molecular level, we identified the floral MADS-box genes in Sinofranchetia chinensis and especially studied the evolution of the B-class MADS-box genes in Ranunculales. Furthermore, we investigated the expression patterns of the floral MADS-box genes and observed the epidermal cell morphology of floral organs in S. chinensis. By integrating molecular, developmental and morphological data, we hope to explore the nature of the nectar leaves in S. chinensis, and reveal its relationship to nectar leaves in other Ranunculales species, and to other floral organs in S. chinensis.
MATERIALS AND METHODS
Plant materials
Sinofranchetia chinensis is a liana with unisexual flowers. The functionally unisexual flowers are bisexual in organization at the floral bud stages, and the unisexuality is found in mature flowers (Zhang et al., 2009). A Sinofranchetia flower has petaloid sepals with a purple margin in the first whorl, greenish nectar leaves in the second whorl, stamens or sterile staminodes in the third whorl, and rudimentary carpels or carpels in the forth whorl (Figs 1 and 6A). Floral buds and mature flowers of S. chinensis at different developmental stages were collected from Taibai Mountain (1200–1500 m a.s.l.), Meixian County, Shaanxi Province, China. They were treated in one of three ways: fixed in FAA (formalin to acetic acid to 50% alcohol in the ratio of 5 : 6 : 89) for morphological observation; stored in liquid nitrogen for RNA isolation; or fixed in PFA (4 % paraformaldehyde) and embedded in Paraplast (Sigma, St Louis, MO, USA) for in situ hybridization. Plant materials of Holboellia grandiflora and Decaisnea insignis were also collected for RNA extraction.
Fig. 1.
Morphology of Sinofranchetia chinensis flowers: (A) plant with inflorescences; (B) female inflorescence; (C) male inflorescence; (D) female flower; (E) male flower. Red asterisks indicate sepals; red arrows indicate nectar leaves. Scale bars = 2 mm.
Fig. 6.
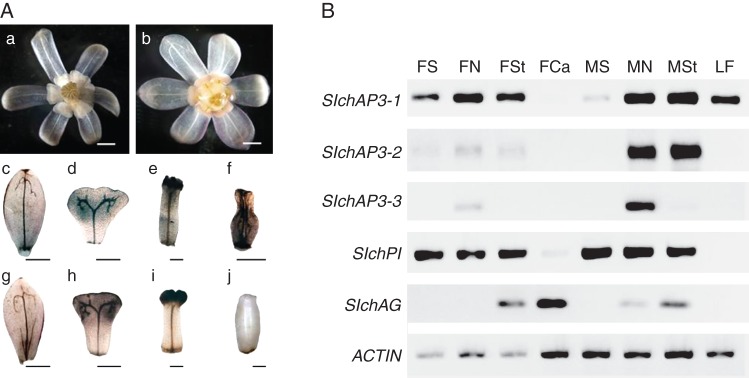
Flowers and floral organs with corresponding expression analyses in Sinofranchetia chinensis: (A) FAA-fixed flowers and floral organs of S. chinensis: (a) male flower; (b) female flower; (c–f) floral organs in male flower; (g–j) floral organs in female flower; (c) and (g) sepal; (d) and (h) nectar leaf; (e) stamen; (f) rudimentary carpel; (i) staminode; (j) carpel. Scale bars = 2 mm. (B) Tissue-specific RT-PCR results for SIchAP3-1, SIchAP3-2, SIchAP3-3, SIchPI, SIchAG and ACTIN (control gene). Abbreviations: FS, female flower sepal; FN, female flower nectar leaf; FSt, female flower staminode; FCa, female flower carpel; MS, male flower sepal; MN, male flower nectar leaf; MSt, male flower stamen; LF, leaf; male flower rudimentary carpel is not included in RT-PCR.
Gene cloning
Total RNA of floral buds and young flowers was extracted using Plant RNA Reagent (Invitrogen, Carlsbad, CA, USA). Poly(A) mRNA was purified from total RNA using the Oligotex mRNA Mini Kit (Qiagen, Hilden, Germany). First-strand cDNA was synthesized using SuperScriptTM III Reverse Transcriptase (Invitrogen). We performed 3′ RACE (rapid amplification of cDNA ends) PCR with the degenerate primers and the adapter primer AP (5′-CCGGATCCTCTACAGCGGCCGC-3′). To clone B-class MADS-box genes, hemi-nested PCR assay was carried out with the B-class gene-specific degenerate primer B1 and the adapter primer AP. The PCR reaction was heated at 94 °C for 4 min, followed by ten cycles of 94 °C for 30 s, 48 °C for 30 s and 72 °C for 1 min, and then 25 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min, and finally extended at 72 °C for 10 min. A second B-class gene-specific degenerate primer B2 and AP were then used to amplify the PCR products obtained in the first step. The PCR was performed with 35 cycles, and the annealing temperature was set at 52 °C. The amplified fragments over 800 bp were purified and cloned into the pGEM-T easy vector (Promega, Madison, WI, USA). The plasmid DNA was isolated by alkaline lysis precipitation (Sambrook et al., 1989). The positive clones were identified by restriction enzyme analysis of the plasmids, and at least three independent clones were sequenced for each identified locus using forward primer T7 (5′-TAATACGACTCACTATAGGG-3′) and reverse primer SP6 (5′-ATTTAGGTGACACTATAG-3′). In a similar way, other floral MADS-box genes were cloned. Only nucleotide sequences with Phred quality scores >20 were used for further analysis. The degenerate primers for B-, C- and E-class gene amplification and the number of sequenced clones for each gene are listed in Supplementary Data Table S1.
Sequence retrieval and alignment
Aside from all the floral MADS-box genes cloned from S. chinensis, H. grandiflora and D. insignis, we also obtained homologous sequences from other species using BLAST against the publicly available databases, including NCBI (http://www.ncbi.nlm.nih.gov) and the TIGR plant transcript assembly database (http://plantta.jcvi.org/). The full-length amino acid sequences for phylogenetic analyses of A-, B-, C- and E-class MADS-box genes (referred to as global analysis hereafter), AP3-like genes in Ranunculales (referred to as AP3 analysis hereafter) and PI-like genes in Ranunculales (referred to as PI analysis hereafter) were aligned with ClustalX 1.83 using the default parameters (Thompson et al., 1997). Alignments were adjusted manually using GeneDoc (Nicholas and Nicholas, 1997). The corresponding DNA matrices were generated by aa2dna (https://homes.bio.psu.edu/people/faculty/nei/software.htm) using the well-aligned amino acid matrices. In addition, we used ClustalX 1.83 to estimate the column scores of the amino acid matrices for the global analysis, AP3 analysis and PI analysis, respectively, and the residues with higher-than-12 quality scores were kept in the alignment (Thompson et al., 1997; Zahn et al., 2005a; Shan et al., 2007). Based on these residues, we generated the corresponding nucleotide matrices for further phylogenetic analyses, and the length of the dataset for the global analysis was 588 bp, 606 bp for the AP3 analysis and 612 bp for the PI analysis.
Phylogenetic analysis
Phylogenetic analyses were performed for each DNA matrix using the maximum likelihood (ML) method in PhyML version 2.4.4 (Guindon and Gascuel, 2003). The best fit model of nucleotide evolution for the DNA matrices was GTR + I + Γ, which was chosen by running MODELTEST version 3.06 (Posada and Crandall, 1998). The proportion of invariable sites and gamma distribution parameter values for the GTR + I + Γ model were optimized by using MODELTEST. Each ML analysis used a BIONJ tree as a starting point (Gascuel, 1997). The support value for each tree branch was based on the best ML tree filtered through 1000 bootstrap replicates in PhyML (Felsenstein, 1985).
In situ hybridization
The floral buds of S. chinensis at various developmental stages were used for in situ hybridization. The gene-specific primers were designed for amplifying the partial CDS (coding sequences) and 3′ UTR (untranslated regions) of Sinofranchetia floral MADS-box genes to make RNA probes (Supplementary Data Table S2). The sense and antisense digoxigenin-labelled RNA probes for in situ hybridization were synthesized using a DIG northern starter kit (Roche Diagnostics, Mannheim, Germany). Embedding of plant materials, pretreatment, hybridization and washing of the sections were performed as previously described (Li et al., 2005; Shan et al., 2006) with slight modifications.
Tissue-specific RT-PCR
The expression patterns of AP3, PI and AG homologues in the mature flowers of S. chinensis were investigated by using tissue-specific RT-PCR. The RNA used in RT-PCR was extracted from sepals, petals, staminodes and carpels of mature female flowers, and sepals, petals and stamens of mature male flowers and from young leaves. The rudimentary carpels in the male flowers are too small to be collected, so they were not included in the analysis. The extraction of total RNA, purification of poly (A) mRNA, and synthesis of the first-strand cDNA were performed according to the methods described above. The amounts of templates were normalized using the control gene ACTIN. Gene-specific forward and reverse primers were used to detect gene expression (Supplementary Data Table S2). The PCR thermocycling conditions used were: initial denaturation at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 55–60 °C (depending on the melting temperature of primer pairs) for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were fractionated in 1 % agarose gels and digitally photographed. We repeated the RT-PCR experiments three times independently.
Scanning electron microscopy
The characteristics of the epidermal cells are an important criterion for identifying morphological equivalents or homologues among different floral organs (Endress, 1994; Krizek et al., 2000; Pelaz et al., 2000; Jaramillo and Kramer, 2004; Geuten et al., 2006). We therefore performed SEM (scanning electron microscopy) analysis of epidermal cell shapes for the sepal, nectar leaf, sterile staminode/stamen and carpel/rudimentary carpel in the female and male flowers of S. chinensis. Young flowers were collected at 7-d intervals and immediately fixed with FAA. Subsequently, the materials were dehydrated in an alcohol–isoamyl acetate series, treated by critical point drying in CO2, mounted, and then coated with gold. Observations of the epidermal cell morphology of different floral organs were performed using a Hitachi S-800 scanning electron microscope.
RESULTS
A-, B-, C- and E-class MADS-box genes in S. chinensis
Six floral MADS-box genes were isolated from S. chinensis, four from H. grandiflora and four from D. insignis. Through BLAST against NCBI databases, these genes were preliminarily grouped into B-, C- and E-class MADS-box genes, and these classifications were further supported by ML analysis (Fig. 2). The studied species, gene name and accession number for 14 newly isolated genes in this study, two previously published genes from S. chinensis (Shan et al., 2007) and 92 representative floral MADS-box genes from other species downloaded from databases are listed in Supplementary Data Table S3. All floral MADS-box genes analysed here formed four distinct well-supported clades in the ML tree, corresponding to the AP1/FUL (A-class), AP3/PI (B-class), AG (C-class) and SEP (E-class) subfamilies of MADS-box genes (Fig. 2). Furthermore, the relationships among these genes are largely consistent with the species phylogeny (Fig. 2). Accordingly, the eight floral MADS-box genes from S. chinensis were classified as two FUL-like genes (SIchFL1 and SIchFL2), three AP3-like genes (SIchAP3-1, SIchAP3-2 and SIchAP3-3), one PI-like gene (SIchPI), one AG-like gene (SIchAG) and one SEP3-like gene (SIchSEP3); the four floral MADS-box genes from H. grandiflora were one AP3-like gene (HOgrAP3), one PI-like gene (HOgrPI) and two AG-like genes (HOgrAG1 and HOgrAG2); and the four floral MADS-box genes from D. insignis were two AP3-like genes (DEinAP3-1 and DEinAP3-2), one PI-like gene (DEinPI) and one AG-like gene (DEinAG) (Fig. 2). The phylogenetic analysis indicated that the following paralogous gene pairs might be the result of gene duplication events that took place before the divergence of the Lardizabalaceae (Fig. 2): SIchFL1 and SIchFL2; SIchAP3-1, SIchAP3-2 and SIchAP3-3; HOgrAG1 and HOgrAG2; and DEinAP3-1 and DEinAP3-2.
Fig. 2.
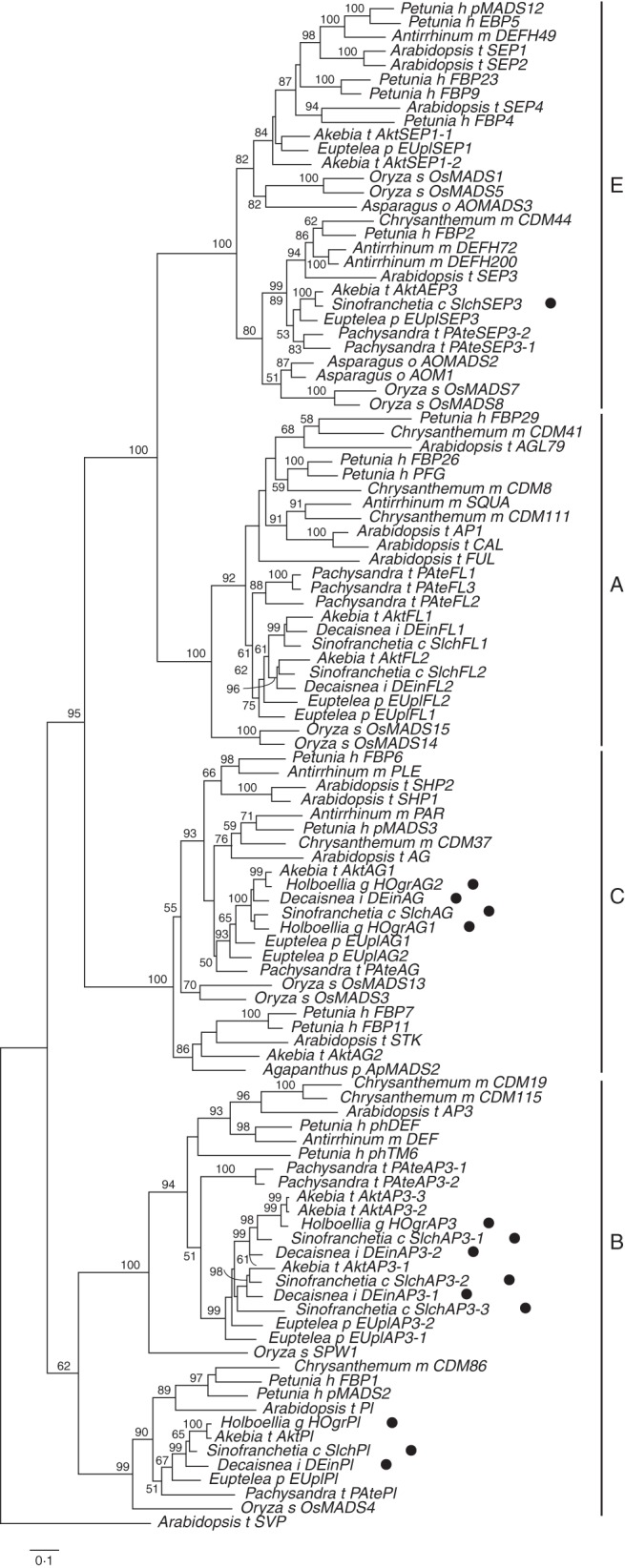
Phylogenetic relationships of 108 representative floral MADS-box genes. Bootstrap values (>50 %) are shown above the branches. Genes obtained in this study are indicated with dots.
Sequence and phylogenetic analysis of B-class MADS-box genes in Ranunculales
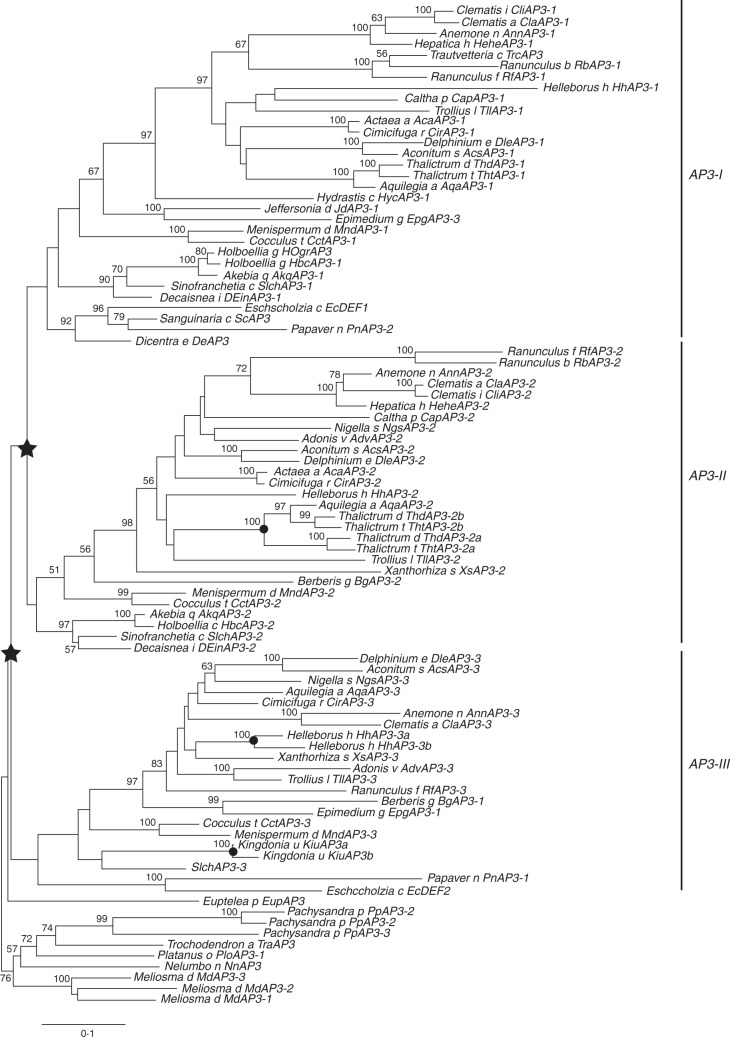
Our phylogenetic analysis of AP3-like genes in Ranunculales showed that the three AP3-like genes from S. chinensis were grouped into three paralogous AP3 lineages of Ranunculales (Fig. 3 and Supplementary Data Table S4). SIchAP3-1 belongs to the AP3-I lineage, SIchAP3-2 to the AP3-II lineage, and SIchAP3-3 to the AP3-III lineage. Furthermore, SIchAP3-1 and SIchAP3-2 protein sequences show 67 % identity; SIchAP3-2 and SIchAP3-3 share 62 % identity; and SIchAP3-1 and SIchAP3-3 share 56 % identity. The highly conserved MADS domain and K domain, as well as PI-derived motif and paleoAP3 motif, were found in the three SIchAP3 proteins from multiple sequence alignment with other AP3-like proteins of Ranunculales; however, the SIchAP3-2 protein lacks 29 amino acids in the K domain (Supplementary Data Fig. S1). At least four families (Ranunculaceae, Berberidaceae, Menispermaceae and Lardizabalaceae) of Ranunculales have all three paralogues of AP3-like genes, but the relationships among these three AP3 lineages in Ranunculales are still uncertain (Fig. 3).
Fig. 3.
Maximum-likelihood tree of AP3-like genes in Ranunculales. Bootstrap values (>50 %) are shown above the branches. In Ranunculales, the inferred two major, successive gene duplication events are highlighted by stars, and the inferred small-scale gene duplication events are indicated with dots.
In this study, we have added three new PI homologues from Lardizabalaceae to the phylogenetic analysis of PI-like genes of Ranunculales (Supplementary Data Table S5 and Fig. S2). These were from three different species of Lardizabalaceae, only one copy in each species (Supplementary Data Fig. S2).
Spatiotemporal expression patterns of floral MADS-box genes in S. chinensis
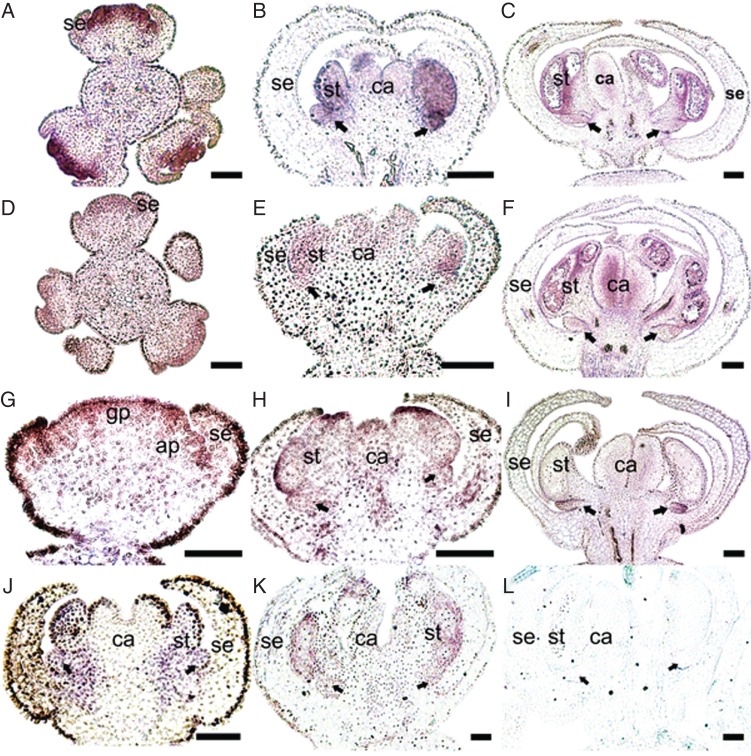
To detect the spatiotemporal expression patterns of floral MADS-box genes in S. chinensis at the floral bud stages, we performed in situ hybridization experiments. The results for B-class MADS-box genes are presented in Fig. 4. At the early stages, SIchAP3-1 is highly expressed in the primordia of the nectar leaves, as well as in the androecial and gynoecial primordia (Fig. 4A). Later on, the expression signals of SIchAP3-1 were mainly detected in the nectar leaves and developing stamens, but the expression level in the carpels reduces gradually during floral development (Fig. 4B). At the late stages, SIchAP3-1 expression is mainly restricted to the nectar leaves and stamens (Fig. 4C). The expression pattern of SIchAP3-2 is mostly like that of SIchAP3-1, but weak expression of this gene was also detected in the sepal primordia, and its expression level in the carpels is constant during the late stages (Fig. 4D–F). SIchAP3-3 expression is ubiquitously in the whole flower (Fig. 4G) and gradually narrows to the inner three whorls (Fig. 4H). At the late stages, SIchAP3-3 is expressed strongly in the nectar leaves, but very weakly in the stamens and carpels (Fig. 4I). SIchPI is expressed in the nectar leaves and stamens but not in the carpels (Fig. 4J, K).
Fig. 4.
Expression patterns of B-class MADS-box genes of Sinofranchetia chinensis as revealed by in situ hybridization analyses: (A–C) SIchAP3-1; (D–F) SIchAP3-2; (G–I) SIchAP3-3; (J, K) SIchPI; (L) negative control with sense probe for SIchPI. (A) and (D) show a young inflorescence with multiple flowers, whereas all other panels show only one flower. Abbreviations: gp, gynoecial primordium; ap, androecial primordium; se, sepal; st, stamen; ca, carpel. Arrows indicate nectar leaves. Scale bars = 100 µm.
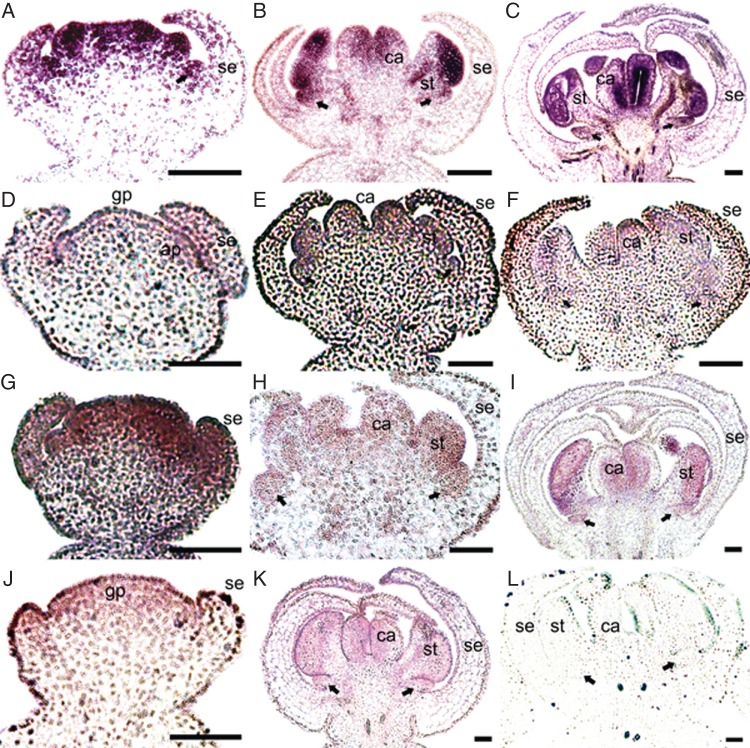
The expression patterns of A-, C- and E-class MADS-box genes in S. chinensis are shown in Fig. 5. SIchFL1 has very strong expression signals throughout the floral organ primordia at the early stages (Fig. 5A), and high expression levels are maintained in the nectar leaves, stamens and carpels at the late stages (Fig. 5B, C). The expression level of SIchFL2 is lower than that of SIchFL1, and its transcripts were found in the floral meristems and all the floral organs (Fig. 5D–F). Initially, SIchAG is highly expressed throughout all the floral organ primordia (Fig. 5G). During the later stages of development, SIchAG is expressed in the nectar leaves, stamens and carpels (Fig. 5H, I). SIchSEP3 is ubiquitously expressed in the whole flower, at the early and late stages (Fig. 5J, K).
Fig. 5.
Expression patterns of A-, C- and E-class MADS-box genes of Sinofranchetia chinensis as revealed by in situ hybridization analyses; (A–C) SIchFL1; (D–F) SIchFL2; (G–I) SIchAG; (J, K) SIchSEP3; (L) negative control with sense probe for SIchAG. Abbreviations: gp, gynoecial primordium; ap, androecial primordium; se, sepal; st, stamen; ca, carpel. Arrowheads indicate nectar leaves. Scale bars = 100 µm.
Furthermore, we investigated the expression patterns of B- and C-class MADS-box genes of S. chinensis in mature flowers by tissue-specific RT-PCR (Fig. 6). The mature male and female flowers can be distinguished from each other, and B- and C-class MADS-box genes are differentially expressed in the male and female flowers (Fig. 6B). SIchAP3-1 is highly expressed in the nectar leaves and stamens/staminodes in both male and female flowers, whereas there is very low expression of SIchAP3-1 in the carpels of the female flowers. Its expression was also found in the sepals, and the expression level is higher in the female flowers than in the male flowers. In addition, a high expression signal of SIchAP3-1 was also observed in the leaves. SIchAP3-2 shows higher expression in the nectar leaves and stamens in the male flowers, and lower expression in the sepals, nectar leaves and staminodes in the female flowers. No expression signal of SIchAP3-2 was found in the carpels or leaves. In comparison, the expression of SIchAP3-3 is mainly restricted in the nectar leaves in both male and female flowers, and the expression level is higher in the male flowers. Very low expression of SIchAP3-3 was also detected in the stamens of the male flowers, but not at a significant level and not in the staminodes of the female flowers. SIchPI is expressed at high level in the sepals, nectar leaves and stamens/staminodes in both male and female flowers and at low levels in the carpels of the female flowers. In the female flowers, the transcripts of SIchAG were found in the staminodes and carpels. In comparison, SIchAG is expressed at relatively low levels in the nectar leaves and stamens in the male flowers (Fig. 6B).
Epidermal cell morphology of different floral organs in S. chinensis
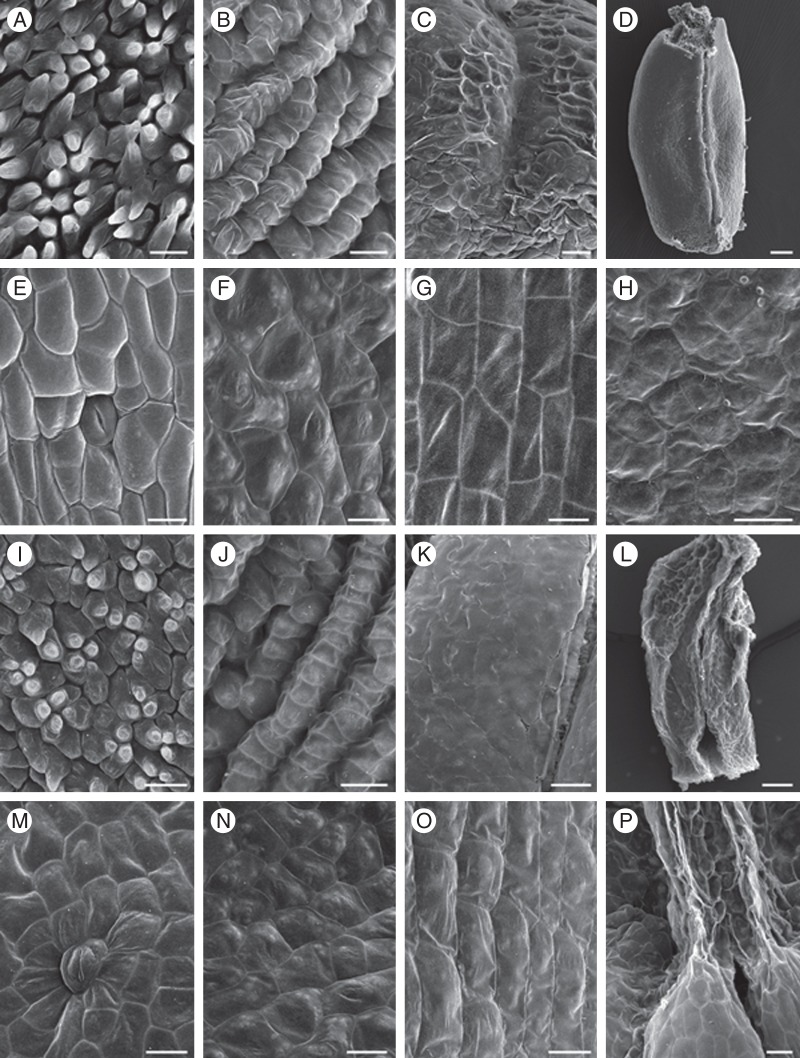
There is no significant difference in morphology of the epidermal cells of different floral organs between female flowers and male flowers (Fig. 7). The adaxial epidermal sepal cells are asymmetrically conical-papillate (Fig. 7A, I); the abaxial ones are flat, irregular and with slightly sunken stomata (Fig. 7E, M). The conical epidermal cells were found on the adaxial surface of the nectar leaf (Fig. 7B, J). The abaxial surface of the nectar leaf consists of flat and irregular cells (Fig. 7F, N). The epidermal cells of the anther are irregular in shape (Fig. 7C, K), and those of the filament are flat and rectangular (Fig. 7G, O). The epidermis of the carpel/rudimentary carpel is covered with irregularly shaped cells (Fig. 7D, H, L, P).
Fig. 7.
Epidermal cell morphology of floral organs of female and male flowers in Sinofranchetia chinensis under SEM: (A–H) female floral organs; (I–P) male floral organs. (A) adaxial and (E) abaxial surface of the sepal; (B) adaxial and (F) abaxial surface of the nectar leaf; (C) anther and (G) filament of the staminode; (D) carpel of female flower; (H) surface of the carpel; (I) adaxial and (M) abaxial surface of the sepal; (J) adaxial and (N) abaxial surface of the nectar leaf; (K) anther and (O) filament of the stamen; (L) rudimentary carpel of male flower; (P) surface of the carpel. Scale bars: (A–C, E–K, M–O) = 20 µm; (P) = 50 µm; (L) = 100 µm; (D) = 250 µm.
DISCUSSION
In this study, the A-, B-, C- and E-class MADS-box genes were found to be expressed in the nectar leaves of S. chinensis at different levels during different developmental stages (Figs 4–6). It implies that these floral MADS-box genes might contribute to the developmental regulation of the nectar leaves in S. chinensis. In addition, the A- and E-class genes display relatively broad expression patterns in most of the floral organs in S. chinensis, while the B- and C-class genes have major expression regions, and some even show relatively specific expression in the nectar leaves at mature stages. Therefore, the B- and C-class genes might be preferential candidates to explore the nature of the nectar leaves in S. chinensis at the molecular level.
Three AP3-like genes were identified in S. chinensis, and, importantly, SIchAP3-3 is the first representative gene of the AP3-III lineage from Lardizabalaceae, based on the updated phylogenetic tree of AP3-like genes in Ranunculales (Fig. 3). Meanwhile, our phylogenetic analysis suggested that the two major gene duplication events involved in the evolution of AP3-like genes in Ranunculales occurred at least before the divergence of the Ranunculaceae, Berberidaceae, Menispermaceae and Lardizabalaceae (Fig. 3; Rasmussen et al., 2009; Sharma et al., 2011). Furthermore, the three AP3-like genes of S. chinensis show distinct and complex expression patterns (Figs 4A–I and 6B). They were all detected in the floral meristems as the sepals initiated, and their expression domains diverged gradually during floral development, especially at mature stages (Figs 4A–I and 6B). These results suggest that the three copies of AP3-like genes in S. chinensis might have undergone subfunctionalization after gene duplication; similar cases were also reported in other Ranunculales species, such as Aquilegia vulgaris (Ranunculaceae) and Papaver somniferum (Papaveraceae) (Force et al., 1999; Lynch and Force, 2000; Kramer et al., 2003, 2007; Moore et al., 2005; Drea et al., 2007).
In addition, we compared the expression pattern of AP3-like genes in S. chinensis with that in A. vulgaris, a well-studied Ranunculaceae species. Both of these two species have three copies of AP3-like genes and nectar leaves, except that the nectar leaves in A. vulgaris have become spurs and are larger (Kramer et al., 2007). At early stages, the expression of SIchAP3-2 and SIchAP3-3 is broader than that of corresponding Aquilegia genes, AqvAP3-2 and AqvAP3-3 (Fig. 4D, G; Kramer et al., 2007), and SIchAP3-1 shares the same expression pattern with AqvAP3-1 (Fig. 4A; Kramer et al., 2007). At mature stages, the expression domains of SIchAP3-1, SIchAP3-2 and SIchAP3-3 generally resemble those of AqvAP3-1, AqvAP3-2 and AqvAP3-3, respectively, except for the expression of SIchAP3-1 in leaves (Fig. 6B; Kramer et al., 2007). SIchAP3-3 and AqvAP3-3 are all mainly expressed in nectar leaves and weakly expressed in stamens but not in staminodes (Fig. 6B; Kramer et al., 2007). And another B-class gene, SIchPI, which is a PI-like gene, is strongly expressed in the nectar leaves and stamens/staminodes at late stages, like AqvPI (Figs 4K and 6B; Kramer et al., 2007). In general, the expression pattern of B-class genes in S. chinensis and A. vulgaris is very similar, especially at mature stages. More interestingly, the expression pattern of B-class genes mentioned above is also found in other Ranunculales species with nectar leaves, such as Trollius laxus and Xanthorhiza simplicissima (Ranunculaceae), Berberis gilgiana and Epimedium grandiflora (Berberidaceae) (Rasmussen et al., 2009). These gene-expression data imply that the development of the nectar leaves in S. chinensis might be under a similar gene regulatory programme as other nectar leaves in Ranunculales species, although the shapes of these nectar leaves are variable in different species. Taking account of the expression pattern for B-class genes in other eudicots, the expression in nectar leaves and stamens in some Ranunculales species seems more or less corresponding to the expression in petals and stamens in core eudicots (e.g. Goto and Meyerowitz, 1994; Kramer et al., 1998; Kramer and Irish, 1999, 2000; Lamb and Irish, 2003). It might support the reference to ‘nectar leaves’ as ‘petals’ (e.g. Cronquist, 1981; Kramer et al., 2007; Kramer and Hodges, 2010).
What is more, the expression for the C-class gene SIchAG of S. chinensis was observed not only in the stamens/staminodes and carpels, but also in the nectar leaves (Figs 5G–I and 6B). Compared with the relatively conserved and concentrated expression in stamens and carpels of AG-like genes in most angiosperms, it seems that the SIchAG gene shifts the expression domain outwards, which just fits the ‘shifting boundary’ model (Kramer et al., 2003, 2004). Furthermore, it has been suggested that the shifts in the expression domain of genes could lead to the genetic shifts in floral architecture and result in changes in floral structure, or even homeotic transformations (Bowman, 1997; Albert et al., 1998; Kramer et al., 2003). For example, the ectopic expression of C-class genes in the second whorl of the flower has resulted in staminoid petals or even true stamens instead of petals in Antirrhinum (Bradley et al., 1993). Therefore, the morphological similarity between nectar leaves and stamens/staminodes in S. chinensis (Figs 1 and 6A) might have some relationships with the expression pattern of SIchAG, which needs to be investigated by gene-function analysis in the future. More importantly, the nectar leaves share more common expressed floral MADS-box genes with the stamens/staminodes than other floral organs in S. chinensis, which might reflect the close genetic relationship between nectar leaves and stamens (Figs 4–6). To some degree, it suggests that the nectar leaves in S. chinensis might be derived from stamens, which is consistent with the hypothesis reported for species of Ranunculaceae and Berberidaceae (Endress, 1995; Ronse De Craene, 2010).
Based on the morphological data, there are sepals, nectar leaves, stamens/staminodes and carpels from outer whorl to inner whorl in S. chinensis. Since a colourful perianth could enhance the attractiveness to potential pollinators, the showy petaloid sepals of S. chinensis might take more responsibility for attracting potential pollinators, compared with the small greenish nectar leaves (Glover and Martin, 1998). Moreover, the nectar leaves could secrete nectar, which could be a ‘reward’ to the pollinators, and there may be a trade-off between size/attractiveness and nectar production for the nectar leaves (Nepi et al., 2009). In addition, the nectar leaves and sepals are all covered with cone-shaped cells on the adaxial surface, which can be easily distinguished from the morphology of the stamen/staminode epidermis (Fig. 7). The conical epidermal cell has been suggested to be a hallmark for petaloidy, possibly aiding pollinator orientation on the flowers in previous studies (Glover and Martin, 1998; Geuten et al., 2006; Kramer et al., 2007; Kramer and Hodges, 2010; Whitney et al., 2011). It seems that the nectar leaves more or less play the role, just like petals. Our analysis also showed that the major cells on the abaxial surface of the sepals, nectar leaves and stamens in S. chinensis are generally similar, except that the sepals have stomata in their epidermis (Fig. 7).
In this study, SIchAP3-3, the first representative gene of the AP3-III lineage from Lardizabalaceae, was observed to be specifically expressed in the nectar leaves at mature stages, which is very similar to the expression pattern of AP3-III members in other Ranunculales species with nectar leaves (Fig. 6B; Kramer et al., 2007; Rasmussen et al., 2009). It suggests that the development of nectar leaves in S. chinensis might share a similar genetic regulatory code with other nectar leaves in Ranunculales species. Our study suggests that the nectar leaves in S. chinensis could be referred to as petals, and they might preserve the genetic footprint of the stamen ancestor. However, all of these need to be explored in further and more comprehensive studies.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Prof. Yi Ren and Dr Xiaohui Zhang for helping with the collection of plant materials, Dr Liang Zhao for providing the pictures of plant materials in the field, Prof. Hongzhi Kong, Dr Wei Wang, Dr Wenheng Zhang and Prof. Pam Soltis for reading the early draft of the manuscript. We also thank two anonymous referees for helpful comments on the manuscript. This work was supported by the National Nature Science Foundation of China (grant numbers 40830209, 30530090, 31100171 and 31061160184), and CAS Visiting Professorship for Senior International Scientists (2011T1S24).
LITERATURE CITED
- Albert VA, Gustaffson MGH, Di Laurenzio L. Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II. New York, NY: Kluwer Academic; 1998. pp. 349–374. [Google Scholar]
- Angiosperm Phylogeny Group III. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Aoki S, Uehara K, Imafuku M, Hasebe M, Ito M. Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. Journal of Plant Research. 2004;117:229–244. doi: 10.1007/s10265-004-0153-7. [DOI] [PubMed] [Google Scholar]
- Baum DA, Whitlock BA. Plant development: genetic clues to petal evolution. Current Biology. 1999;9:525–527. doi: 10.1016/s0960-9822(99)80327-3. [DOI] [PubMed] [Google Scholar]
- Bowman JL. Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. Journal of Biosciences. 1997;22:515–527. [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Buzgo M, Soltis DE, Soltis PS, Ma H. Towards a comprehensive integration of morphological and genetic studies of floral development. Trends in Plant Science. 2004;9:164–173. doi: 10.1016/j.tplants.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Chanderbali AS, Kim S, Buzgo M, et al. Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae) International Journal of Plant Sciences. 2006;167:1075–1089. [Google Scholar]
- Chanderbali AS, Albert VA, Leebens-Mack J, Altman NS, Soltis DE, Soltis PS. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae) Proceedings of the National Academy of Sciences of the USA. 2009;106:8929–8934. doi: 10.1073/pnas.0811476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, et al. The petunia MADS box gene FBP11 determines ovule identity. The Plant Cell. 1995;7:1859–1868. doi: 10.1105/tpc.7.11.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronquist A. An integrated system of classification of flowering plants. New York, NY: Columbia University Press; 1981. [Google Scholar]
- Cronquist A. The evolution and classification of flowering plants, 2nd edn. New York, NY: The New York Botanical Garden; 1988. [Google Scholar]
- Damerval C, Nadot S. Evolution of perianth and stamen characteristics with respect to floral symmetry in Ranunculales. Annals of Botany. 2007;100:631–640. doi: 10.1093/aob/mcm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Hileman LC, De Martino G, Irish VF. Functional analyses of genetic pathways controlling petal specification in poppy. Development. 2007;134:4157–4166. doi: 10.1242/dev.013136. [DOI] [PubMed] [Google Scholar]
- Dubois A, Raymond O, Maene M, et al. Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS ONE. 2010;5:e9288. doi: 10.1371/journal.pone.0009288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames AJ. Morphology of the angiosperms. New York, NY: McGraw-Hill; 1961. [Google Scholar]
- Endress PK. Floral structure and evolution of primitive angiosperms: recent advances. Plant Systematics and Evolution. 1994;192:79–97. [Google Scholar]
- Endress PK. Floral structure and evolution in Ranunculaceae. Plant Systematics and Evolution. 1995;9(Suppl.):47–61. [Google Scholar]
- Endress PK. Origins of flower morphology. Journal of Experimental Zoology. 2001;291:105–115. doi: 10.1002/jez.1063. [DOI] [PubMed] [Google Scholar]
- Endress PK. Angiosperm floral evolution: morphological developmental framework. Advances in Botanical Research. 2006;44:1–61. [Google Scholar]
- Endress PK, Matthews ML. Elaborate petals and staminodes in eudicots: diversity, function, and evolution. Organisms, Diversity, and Evolution. 2006;6:257–293. [Google Scholar]
- Erbar C, Kusma S, Leins P. Development and interpretation of nectary organs in Ranunculaceae. Flora. 1998;194:317–332. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis EM, Endress PK. Origin and evolution of angiosperm flowers. Advances in Botanical Research. 1990;17:99–162. [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Geuten K, Becker A, Kaufmann K, et al. Petaloidy and petal identity MADS-box genes in the balsaminoid genera Impatiens and Marcgravia. The Plant Journal. 2006;47:501–518. doi: 10.1111/j.1365-313X.2006.02800.x. [DOI] [PubMed] [Google Scholar]
- Glover BJ, Martin C. The role of petal cell shape and pigmentation in pollination success in Antirrhinum majus. Heredity. 1998;80:778–784. [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Development. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Goto K, Kyozuka J, Bowman JL. Turning floral organs into leaves, leaves into floral organs. Current Opinion in Genetics and Development. 2001;11:449–456. doi: 10.1016/s0959-437x(00)00216-1. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systems Biology. 2003;52:596–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hintz M, Bartholmes C, Nutt P, et al. Catching a ‘hopeful monster’: shepherd's purse (Capsella bursa-pastoris) as a model system to study the evolution of flower development. Journal of Experimental Botany. 2006;57:3531–3542. doi: 10.1093/jxb/erl158. [DOI] [PubMed] [Google Scholar]
- Irish VF. The evolution of floral homeotic gene function. BioEssays. 2003;25:637–646. doi: 10.1002/bies.10292. [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Current Opinion in Genetics and Development. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Janchen E. Die systematische Gliederung der Ranunculaceen und Berberidaceen. Denkschriften (Österreichische Akademie der Wissenschaften. Mathematisch-Naturwissenschaftliche Klasse) 1949;108:1–82. [Google Scholar]
- Jaramillo MA, Kramer EM. APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae) Evolution and Development. 2004;6:449–458. doi: 10.1111/j.1525-142X.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- Kanno A, Saeki H, Kameya T, Saedler H, Theissen G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana) Plant Molecular Biology. 2003;52:831–841. doi: 10.1023/a:1025070827979. [DOI] [PubMed] [Google Scholar]
- Kim S, Yoo MJ, Albert VA, Farris JS, Soltis PS, Soltis DE. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. American Journal of Botany. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
- Kim S, Koh J, Yoo MJ, et al. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. The Plant Journal. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Hodges SA. Aquilegia as a model system for the evolution and ecology of petals. Philosophical Transactions of the Royal Society – Biological Sciences. 2010;365:477–490. doi: 10.1098/rstb.2009.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. Evolution of genetic mechanisms controlling petal development. Nature. 1999;399:144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. International Journal of Plant Sciences. 2000;161:S29–S40. [Google Scholar]
- Kramer EM, Jaramillo MA. Genetic basis for innovations in floral organ identity. Journal of Experimental Zoology. 2005;304B:526–535. doi: 10.1002/jez.b.21046. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Zimmer EA. Gene duplication and floral developmental genetics of basal eudicots. Advances in Botanical Research. 2006;44:353–384. [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Di Stilio VS, Schluter PM. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. International Journal of Plant Sciences. 2003;164:1–11. [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Su HJ, Wu CC, Hu JM. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evolutionary Biology. 2006;6(30) doi: 10.1186/1471-2148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. The Plant Cell. 2007;19:750–766. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. The Plant Cell. 2000;12:1357–1366. doi: 10.1105/tpc.12.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RS, Irish VF. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proceedings of the National Academy of Sciences of the USA. 2003;100:6558–6563. doi: 10.1073/pnas.0631708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppik EE. A study on floral evolution in relation to pollination ecology. New Delhi: International Books & Periodicals Supply Service; 1988. [Google Scholar]
- Li GS, Meng Z, Kong HZ, Chen ZD, Theissen G, Lu AM. Characterization of candidate class A, B and E floral homeotic genes from the perianthless basal angiosperm Chloranthus spicatus (Chloranthaceae) Development Genes and Evolution. 2005;215:437–449. doi: 10.1007/s00427-005-0002-2. [DOI] [PubMed] [Google Scholar]
- Litt A, Irish VF. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Zhang J, Zhang N, et al. Interactions among proteins of floral MADS-Box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Molecular Biology and Evolution. 2010;27:1598–1611. doi: 10.1093/molbev/msq044. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, dePamphilis C. The ABCs of floral evolution. Cell. 2000;101:5–8. doi: 10.1016/S0092-8674(00)80618-2. [DOI] [PubMed] [Google Scholar]
- Moore RC, Grant SR, Purugganan MD. Molecular population genetics of redundant floral-regulatory genes in Arabidopsis thaliana. Molecular Biology and Evolution. 2005;22:91–103. doi: 10.1093/molbev/msh261. [DOI] [PubMed] [Google Scholar]
- Nepi M, von Aderkas P, Wagner R, Mugnaini S, Coulter A, Pacini E. Nectar and pollination drops: how different are they? Annals of Botany. 2009;104:205–219. doi: 10.1093/aob/mcp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB. Genedoc: a tool for editing and annotating multiple sequence alignments. 1997 Distributed by the author. [Google Scholar]
- Ochiai T, Nakamura T, Mashiko Y, et al. The differentiation of sepal and petal morphologies in Commelinaceae. Gene. 2004;343:253–262. doi: 10.1016/j.gene.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qin HN. A taxonomic revision of the Lardizabalaceae. Cathaya. 1997;8–9:1–214. [Google Scholar]
- Rasmussen DA, Kramer EM, Zimmer EA. One size fits all? Molecular evidence for a commonly inherited petal identity program in Ranunculales. American Journal of Botany. 2009;96:96–109. doi: 10.3732/ajb.0800038. [DOI] [PubMed] [Google Scholar]
- Ronse De Craene LP. Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Annals of Botany. 2007;100:621–630. doi: 10.1093/aob/mcm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronse De Craene LP. Homology and evolution of petals in the core eudicots. Systematic Botany. 2008;33:301–325. [Google Scholar]
- Ronse De Craene LP. Floral diagrams: an aid to understanding flower morphology and evolution. New York, NY: Cambridge University Press; 2010. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. New York, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shan HY, Su KM, Lu WL, Kong HZ, Chen ZD, Meng Z. Conservation and divergence of candidate class B genes in Akebia trifoliata (Lardizabalaceae) Development Genes and Evolution. 2006;216:785–795. doi: 10.1007/s00427-006-0107-2. [DOI] [PubMed] [Google Scholar]
- Shan HY, Zhang N, Liu CJ, et al. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Molecular Phylogenetics and Evolution. 2007;44:26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Shan HY, Zahn L, Guindon S, et al. Evolution of plant MADS box transcription factors: evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Molecular Biology and Evolution. 2009;26:2229–2244. doi: 10.1093/molbev/msp129. [DOI] [PubMed] [Google Scholar]
- Sharma B, Guo C, Kong H, Kramer EM. Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytologist. 2011;191:870–883. doi: 10.1111/j.1469-8137.2011.03744.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Chanderbali A, Kim S, Buzgo M, Soltis PS. The ABC model and its applicability to basal angiosperms. Annals of Botany. 2007;100:155–163. doi: 10.1093/aob/mcm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW, Endress PK, Crane PR. The diversification of flowering plants. In: Cracraft J, Donoghue MJ, editors. Assembling the tree of life. Oxford: Oxford University Press; 2004. [Google Scholar]
- Soltis PS, Soltis DE, Kim S, Chanderbali A, Buzgo M. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC-function. Advances in Botanical Research. 2006;44:483–506. [Google Scholar]
- Specht CD, Bartlett ME. Flower evolution: the origin and subsequent diversification of the angiosperm flower. Annual Review of Ecology Evolution and Systematics. 2009;40:217–243. [Google Scholar]
- Stellari GM, Jaramillo MA, Kramer EM. Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Molecular Biology and Evolution. 2004;21:506–519. doi: 10.1093/molbev/msh044. [DOI] [PubMed] [Google Scholar]
- Takhtajan A. Evolutionary trends in flowering plants. New York, NY: Columbia University History Press; 1991. [Google Scholar]
- Takhtajan A. Diversity and classification of flowering plants. New York, NY: Columbia University Press; 1997. [Google Scholar]
- Takhtajan A. Flowering plants. 2009 Springer Verlag. [Google Scholar]
- Theissen G. Development of floral organ identity: stories from the MADS house. Current Opinion in Plant Biology. 2001a;4:75–85. doi: 10.1016/s1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- Theissen G. Genetics of identity. Nature. 2001b;414:491–491. doi: 10.1038/35107163. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Winter KU, Muenster T, Kirchner C, Saedler H. How the land plants learned their floral ABCs: the role of MADS box genes in the evolutionary origin of flowers. In: Cronk QC, Bateman RM, Hawkins JM, editors. Developmental genetics and plant evolution. London: Taylor & Francis; 2002. pp. 173–206. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu AM, Ren Y, Endress ME, Chen ZD. Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspectives in Plant Ecology Evolution and Systematics. 2009;11:81–110. [Google Scholar]
- Weberling F. Morphology of flowers and inflorescences. New York, NY: Cambridge University Press; 1989. [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Whitney HM, Bennett KMW, Dorling M, et al. Why do so many petals have conical epidermal cells? Annals of Botany. 2011;108:609–616. doi: 10.1093/aob/mcr065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsdell WC. The origin of the perianth of flowers, with special reference to the Ranunculaceae. New Phytologist. 1903;2:42–48. [Google Scholar]
- Xu YF, Teo LL, Zhou J, Kumar PP, Yu H. Floral organ identity genes in the orchid Dendrobium crumenatum. The Plant Journal. 2006;46:54–68. doi: 10.1111/j.1365-313X.2006.02669.x. [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack J, et al. The evolution of the SEPALLATA subfamily of MADS-box genes: a pre-angiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005a;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, dePamphilis CW, Ma H, Theissen G. To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. Journal of Heredity. 2005b;96:225–240. doi: 10.1093/jhered/esi033. [DOI] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack JH, Arrington JM, et al. Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evolution and Development. 2006;8:30–45. doi: 10.1111/j.1525-142X.2006.05073.x. [DOI] [PubMed] [Google Scholar]
- Zanis MJ, Soltis PS, Qiu YL, Zimmer E, Soltis DE. Phylogenetic analyses and perianth evolution in basal angiosperms. Annals of the Missouri Botanical Garden. 2003;90:129–150. [Google Scholar]
- Zhang XH, Ren Y. Comparative floral development in Lardizabalaceae (Ranunculales) Botanical Journal of the Linnean Society. 2011;166:171–184. [Google Scholar]
- Zhang XH, Ren Y, Tian XH. Floral morphogenesis in Sinofranchetia (Lardizabalaceae) and its systematic significance. Botanical Journal of the Linnean Society. 2009;160:82–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.