Abstract
The spread of drug-resistant Plasmodium falciparum malaria has been a major impediment to malaria control and threatens prospects for elimination. We recently demonstrated the return of chloroquine-susceptible malaria in Malawi after chloroquine use was abandoned. In this study, we trace the origins of chloroquine-resistant and chloroquine-susceptible parasites in Malawi by sequencing the P. falciparum chloroquine resistance transporter gene (pfcrt) and by genotyping microsatellites flanking this gene in isolates from infections that occurred in Malawi from 1992 through 2005. Malaria parasites from 2005 harbored the expected wild-type pfcrt haplotype associated with chloroquine susceptibility and have maintained high levels of diversity without linkage disequilibrium, which suggests that the return of chloroquine susceptibility is not the result of a back mutation in a formerly resistant parasite or a new selective sweep. Chloroquine-susceptible parasites that predominate in Malawi likely represent a reexpansion of the susceptible parasites that survived in the population despite widespread drug pressure in the region.
The spread of drug-resistant Plasmodium falciparum malaria has been a major impediment to malaria control and contributed to the failure of the first global campaign to eradicate malaria. Malaria parasites have developed resistance to every drug that has been introduced on a large scale, including chloroquine and sulfadoxine-pyrimethamine, and now there is evidence that decreased sensitivity to the artemisinins is emerging [1, 2].
In 1993, Malawi became the first African country to stop the use of chloroquine as the first-line treatment for falciparum malaria, because of unacceptably high levels of treatment failure. We documented a decrease in the prevalence of the lysine to threonine substitution at position 76 in the gene encoding the P. falciparum chloroquine resistance transporter (pfcrt), which is associated with chloroquine resistance [3]. In 2005, we confirmed that chloroquine had 99% clinical efficacy [4], 12 years after it was removed from use due to a clinical efficacy of <50% [5]. This was the first conclusive evidence of the return of drug-susceptible malaria after the removal of drug pressure.
Previous studies of the evolution of antimalarial drug resistance have focused on indicators of selective sweeps in the P. falciparum genome as a result of the spread of resistance mutations through parasite populations [6–8]. During a selective sweep, alleles in proximity to advantageous polymorphisms increase in frequency along with the allele under selection, which results in reduced genetic variation in the regions surrounding the selected locus [9–12] and increased linkage disequilibrium between the selected mutation and flanking sites [13–15]. Alleles at flanking neutral sites can be examined to determine the origin of drug-resistant parasites. Such evaluation of haplotypes based on polymorphisms in the resistance-conferring genes and flanking microsatellites has provided evidence that chloroquine-resistant and high-level pyrimethamine-resistant parasites originated in South-east Asia and were imported into Africa. Resistant parasites from Africa carry haplotypes identical to those carried by resistant parasites from Southeast Asia at microsatellite loci flanking the genes responsible for chloroquine and pyrimethamine resistance, pfcrt and dihydrofolate reductase-thymidylate synthase, respectively [6, 8]. In contrast, chloroquine-susceptible and pyrimethamine-susceptible parasites from Africa are highly diverse in the regions flanking drug resistance genes, with linkage equilibrium between alleles. Recently, it was shown that highly resistant parasites can also emerge independently: dihydropteroate synthase polymorphisms that are associated with sulfadoxine resistance have multiple origins throughout Africa [16].
The purpose of this study was to investigate the population genetics underlying the return of chloroquine-susceptible P. falciparum in Malawi after cessation of chloroquine use. We aimed to test the hypothesis that the chloroquine-susceptible parasites currently found in Malawi represent an expansion of chloroquine-susceptible parasites that survived in the population despite widespread drug pressure in the region when chloroquine was being used as the first-line treatment for malaria. Other potential explanations for the return of chloroquine susceptibility include a back mutation of threonine to lysine at position 76 in the pfcrt gene of a previously resistant parasite that resulted in a chloroquine-susceptible phenotype, which appears to have happened at least once in a Sudanese isolate [17], or propagation of 1 or a few chloroquine-susceptible lineages with increased fitness. To test this hypothesis, we sequenced the polymorphic regions of the pfcrt gene and genotyped microsatellites surrounding this gene in P. falciparum parasites carrying chloroquine-resistant forms of pfcrt, as well as chloroquine-susceptible parasites that had been collected in Blantyre, Malawi, during the time period from 1992 through 2005, when chloroquine susceptibility returned to the area.
METHODS
Specimens
The specimens that were included in this study were collected from children with malaria. Specimens collected from 1992 through 1996 were obtained from thick malaria smears from children with severe malaria who were admitted to Queen Elizabeth Central Hospital, and specimens collected from 1997 through 2005 were obtained as blood spots on filter paper from children with uncomplicated malaria who were enrolled in drug efficacy studies conducted at the Ndirande Health Centre, on the outskirts of Blantyre, Malawi, as described elsewhere [3, 4]. The studies were reviewed and approved by the University of Malawi College of Medicine Research and Ethics Committee and the University of Maryland and/or Michigan State University Institutional Review Boards. Informed consent was obtained from the guardians of study participants prior to collection of study specimens. DNA suitable for polymerase chain reaction (PCR) amplification was extracted from thick smears or blood spots collected on filter paper by either the QIAamp DNA Blood Mini Kit protocol (Qiagen) or methanol fixation and heat extraction as described elsewhere [18]. Subsequent PCR amplification was performed using 1–5 μL of this material. Genomic DNA from 3D7, a chloroquine-susceptible P. falciparum parasite, and Dd2, a chloroquine-resistant parasite line of Asian origin, were used as controls (Malaria Research and Reference Reagent Resource Center; MRA-102G genomic DNA from 3D7, deposited by D. Carucci, and MRA-150G genomic DNA from Dd2, deposited by D. Walliker).
Sequencing pfcrt
Specimens collected from 1992 through 1998 and in 2005 underwent nested PCR followed by allele-specific endonuclease digestion to determine the allele at pfcrt position 76 [18]. As reported elsewhere [3, 8], chloroquine-resistant parasites collected from 1992 through 1998 included in the analyses reported here had a lysine to threonine amino acid change at position 76 that occurs in the context of a characteristic haplotype with amino acids at codons 72, 74, 75, 76, 220, 271, 326, and 371, (CIETSESI), whereas the susceptible-type lysine at codon 76 has the SMNKAQNR haplotype. To determine whether chloroquine-susceptible parasites that were found recently in Malawi were the result of a back mutation at pfcrt position 76 (as evidenced by the presence of a chloroquine-susceptible type K76 allele accompanied by resistant alleles at other polymorphic positions within pfcrt), a sub-group of the specimens were subjected to sequencing of the polymorphic regions of pfcrt. Of the specimens collected in 2005, 105 (50%) underwent analysis of polymorphic regions of pfcrt that are associated with chloroquine resistance, with the use of methods that are described elsewhere [4].
Microsatellite genotyping
We genotyped microsatellites on a subset of the chloroquine-susceptible specimens and all specimens from chloroquine-resistant infections that were identified based on their pfcrt genotype. Eleven microsatellites, located 2.8, 4.3, 10.8, 29.3, 55.1, and 79.7 kb upstream of pfcrt and 0.6, 10.4, 23.6, 78.9, and 103.0 kb downstream of pfcrt on chromosome 7, were genotyped by performing heminested PCR with fluorescently labeled primers followed by capillary electrophoresis, using primers described by Nash et al [7]. In the first PCR run, 3 primers were multiplexed, with final concentrations of 0.2 mmol/L dNTP, 3 mmol/L magnesium chloride, 0.1 μmol/L of each primer with 0.5 U of Taq polymerase, and 1 μL of DNA template in a final reaction volume of 10 μL. The cycling conditions for the first PCR run were as follows: 94°C for 2 min, followed by 25 cycles of 94°C for 30 s, 42°C for 30 s, 40°C for 30 s, and 65°C for 40 s, followed by 65°C for 2 min. In the heminested PCR run, each primer was used separately, with final concentrations of 0.2 mmol/L dNTP, 2.5 mmol/L magnesium chloride, 0.4 μmol/L of primer with 1.0 U of Taq polymerase, and 1 μL of DNA template in a final volume of 15 μL. The cycling conditions for the second PCR run were as follows: 94°C for 2 min, followed by 25 cycles of 94°C for 20 s, 45°C for 30 s, and 65°C for 30 s, followed by 65°C for 2 min. PCR was performed on a DNA Engine Tetrad 2 cycler (Bio-Rad Laboratories).
Fragment sizes were analyzed using an Applied Biosystems 3730XL high-throughput 96-capillary DNA sequencer and ABI software. The resulting electropherograms were analyzed using GeneMapper software (version 4.0; Applied Biosystems). Only data from infections with single or predominant parasite clones were included in the analysis, whereas multiple clone infections, which are common in this setting, were excluded because it was not possible to determine their haplotypes. An allele was considered predominant in the infection if the signal intensity of the majority allele was at least twice that of minority alleles. Haplotypes were included in analysis if genotyping was available for all of the 11 microsatellite loci.
Statistical analysis
Expected heterozygosity was calculated at each locus using the formula
where n is the number of infections and pi is the frequency of the ith allele. The variance of He was calculated as
He (±1 standard deviation) was calculated for 3 groups of parasites: parasites with chloroquine-resistant pfcrt genotypes that were collected during 1992–1998, those with chloroquine-susceptible pfcrt genotypes that were collected during 1992–1998, and chloroquine-susceptible parasites that were collected in 2005. Permutation was used to detect significant differences in diversity between the 3 groups of parasites by calculating diversity ratios, Hsensitive (1992–1998)/Hresistant, Hsensitive (2005)/Hresistant, and Hsensitive (2005)/Hsensitive (1992–1998), in the observed data and in 10,000 permuted data sets in which the alleles at each locus were reshuffled between parasites. P values were determined by calculating the proportion of permuted diversity ratios that were greater than the observed ratio [7, 19].
Extended haplotype homozygosity (EHH) [15] was calculated to examine patterns of linkage disequilibrium surrounding pfcrt using the EHH calculator [21]. The results for the 3 groups described above were compared.
RESULTS
Single-nucleotide polymorphism analysis of pfcrt codon 76
Specimens from a total of 308 infections, 98 from 1992 through 1998 and 210 from 2005, underwent PCR-restriction digest analysis to determine the allele at pfcrt codon 76. Among the 98 specimens from infections from 1992 through 1998, 83 (84.7%) were found to contain a chloroquine-susceptible allele at pfcrt codon 76. Specimens from 15 (15.3%) infections contained parasites with the resistance-conferring threonine amino acid substitution at pfcrt codon 76, and among those, 9 (9.2%) contained only the resistant allele at that locus. The predominance of chloroquine-susceptible genotypes reflects the larger number of specimens available later in the time period. All specimens from infections in 2005 had the chloroquine-susceptible allele at codon 76.
Haplotype determination of pfcrt
To determine whether chloroquine-susceptible parasites that have reemerged in Malawi are the result of a back mutation at pfcrt codon 76, specimens collected in 2005 underwent direct sequencing of pfcrt regions including codons 72–76, 97, 220, 271, 326, and 371. The chloroquine-susceptible type pfcrt haplotype at positions 72–76 that is typically observed in chloroquine-susceptible parasites, CMNVK, was found in all of these specimens. In addition, all isolates also had the expected chloroquine-susceptible amino acids at codons 97, 220, 271, 326, and 371, except for 1 specimen that was found to have an amino acid substitution at codon 220 (alanine to serine). The specimen with this haplotype was from an infection that had cleared following chloroquine treatment in vivo and was confirmed with repeated sequencing.
Microsatellite analyses
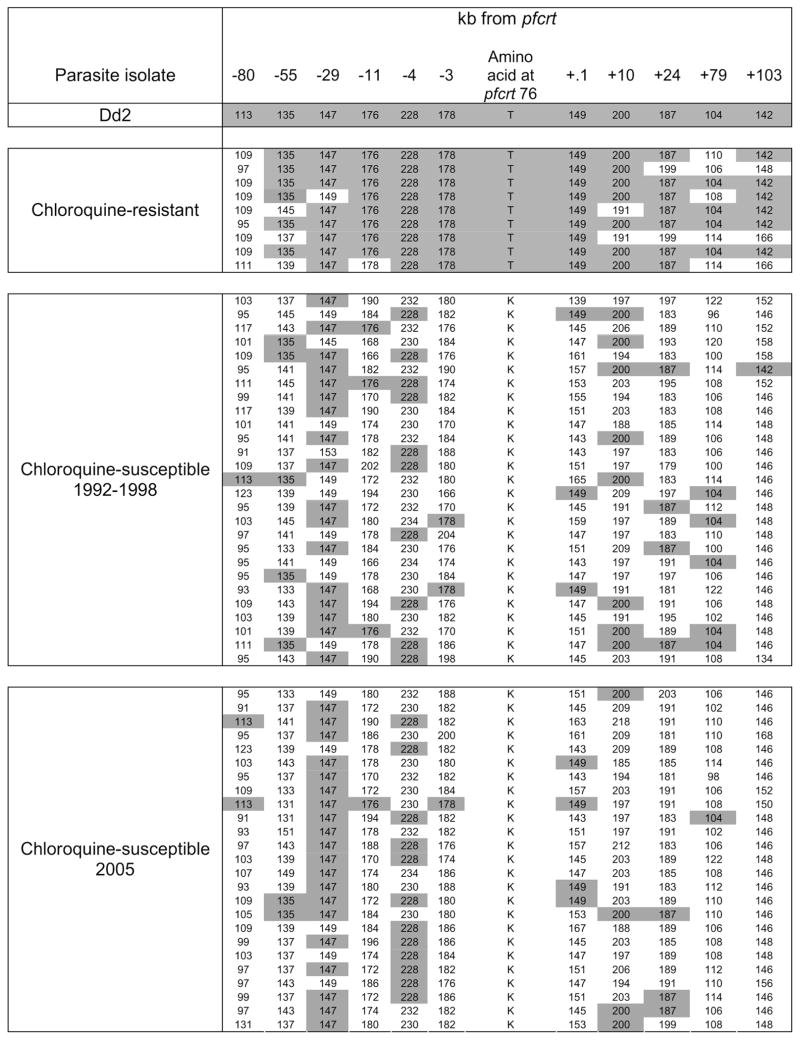
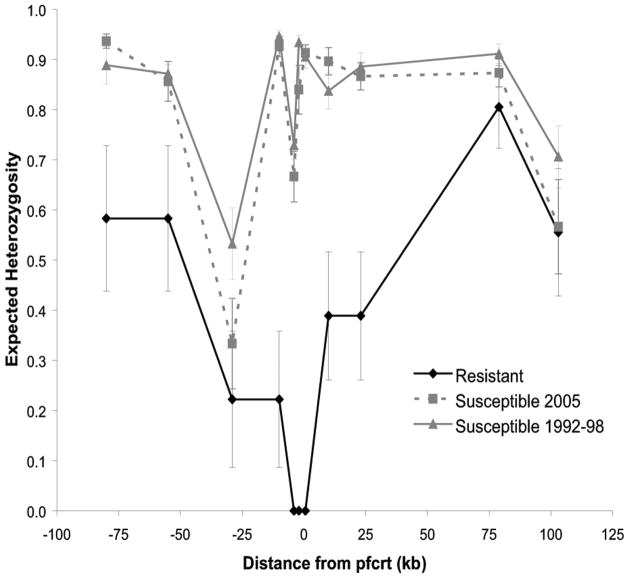
To determine whether the chloroquine-susceptible parasites that have reemerged in Malawi have resulted from a highly fit drug-susceptible parasite sweeping through the population, 152 specimens were subjected to microsatellite analysis, including all specimens collected from 1992 through 1998. Specimens from 61 infections had a single or predominant haplotype and were included in the analysis. The microsatellite haplotypes of the chloroquine-resistant parasites from Malawi and the chloroquine-resistant laboratory strain of Southeast Asian origin, Dd2, are shown in Figure 1. The expected heterozygosity (He) was calculated for each microsatellite locus in each of 3 groups: chloroquine-susceptible parasites collected from 1992 through 1998, chloroquine-resistant parasites collected from 1992 through 1998, and chloroquine-susceptible parasites collected in 2005. A decrease in heterozygosity suggestive of relatively recent selection was noted in the area surrounding pfcrt in the chloroquine-resistant parasites, which is consistent with previous studies [7, 8]. On the basis of permutation, there was a statistically significant reduction in diversity from 80 kb upstream to 23 kb downstream of pfcrt when comparing chloroquine-susceptible types collected during 1992–1998 and in 2005 to resistant parasites (Figure 2). A decrease in heterozygosity was observed at the microsatellite locus 29 kb upstream of pfcrt in all parasites, which is likely due to the lack of variation at this marker (Figure 1).
Figure 1.
Microsatellite haploptyes of chloroquine-resistant and chloroquine-susceptible Plasmodium falciparum parasites in the region of chromosome 7 surrounding the gene encoding the P. falciparum chloroquine resistance transporter (pfcrt). Alleles identical to the chloroquine-resistant Dd2 strain of P. falciparum of Asian origin are shown in gray. Microsatellite sizes are shown in nucleotide base pairs, as determined by capillary electrophoresis of polymerase chain reaction–amplified microsatellites.
Figure 2.
Expected heterozygosity in microsatellites around Plasmodium falciparum chloroquine resistance transporter (pfcrt) in chloroquine-resistant parasites and chloroquine-susceptible parasites from 1992 through 1995 and in 2005. Error bars indicate the 95% confidence intervals.
Chloroquine-susceptible parasites from all years were highly diverse. Figure 1 demonstrates the allelic variation among each group of isolates: chloroquine-resistant isolates, chloroquine-susceptible isolates collected from 1992 through 1998, and chloroquine-susceptible isolates collected in 2005. There was no evidence of a selective sweep of a single new sensitive genotype from the 1990s through 2005. Moreover, every haplotype among the susceptible parasites was unique. At each microsatellite locus, some of the chloroquine-susceptible parasites carried the same allele as the resistant parasites, but none carried an identical haplotype in the region from 29 kb upstream to 23 kb downstream of pfcrt. Both the older and the more recent chloroquine-susceptible parasites had similar levels of diversity, with no significant difference in the ratio Hsensitive (2005)/Hsensitive (1992–1998) at any of the genotyped loci (Figure 2).
EHH was calculated to examine patterns of linkage disequilibrium surrounding pfcrt in the 3 groups of parasites (Figure 3) [15, 20]. In chloroquine-susceptible parasites collected both from 1992 through 1998 and in 2005, EHH decreases to 0 outside of pfcrt, whereas EHH decays much more slowly within chloroquine-resistant parasites, indicating greater linkage disequilibrium surrounding pfcrt in resistant parasites, which is consistent with a recent selective sweep.
Figure 3.
Extended haplotype heterozygosity (EHH) around surrounding Plasmodium falciparum chloroquine resistance transporter (pfcrt) in chloroquine-resistant parasites and chloroquine-susceptible parasites from 1992 through 1995 and in 2005. Error bars indicate the 95% confidence intervals.
DISCUSSION
This study traces the origins of chloroquine-resistant and chloroquine-susceptible parasites in Malawi. We demonstrated that chloroquine-resistant parasites in Malawi had haplotypes that were nearly identical to that of a chloroquine-resistant isolate from Southeast Asia at microsatellite loci flanking pfcrt. The Asian origin of chloroquine resistance in Africa has been demonstrated elsewhere in other African countries [21], and our results extend the observation to include Malawi. The selective valley around chloroquine-resistant parasites in Malawi is ~80 kb upstream and 30 kb downstream of pfcrt, except for an area of decreased heterozygosity at the microsatellite locus 29 kb upstream of pfcrt in both chloroquine-resistant and chloroquine-susceptible isolates, which is likely due to the lack of variation at this marker. The selection valley was narrower than that observed by Wootton et al [8] in chloroquine-resistant parasites from Asia. This is likely due to more extensive parasite diversity in Africa than in Asia and more frequent recombination of nonidentical parasite lineages in Africa.
These results support the hypothesis that the return of chloroquine-susceptible malaria in Malawi following the removal of chloroquine drug pressure represents a reexpansion of a heterogeneous population of susceptible parasites that persisted in Malawi during the period when chloroquine was used. The predominant chloroquine-susceptible pfcrt genotype observed in Malawi contains the expected amino acid sequence at all polymorphic codons within the gene, suggesting that chloroquine susceptibility did not arise from a back mutation at codon 76. Diversity in the regions flanking pfcrt did not differ between chloroquine-susceptible parasites circulating in 2005 and chloroquine-susceptible parasites circulating at the time of chloroquine drug pressure, which indicates that chloroquine susceptibility did not result from propagation of a single chloroquine-susceptible parasite with increased fitness. An isolate from a single infection had a chloroquine-resistant allele at position 220 but otherwise had the expected chloroquine-susceptible haplotype, as has been reported for a small percentage of parasites in India [22]. The infection was successfully treated with chloroquine.
These findings have important implications for our understanding of the effect of the removal of drug pressure on the evolution and ecology of drug resistance. Drug-susceptible organisms may regain predominance as long as there is a population that survives despite prolonged drug pressure in the region that favors resistant parasites. This observation does not imply that parasites survive exposure to the drug; rather, it suggests that some parasites are not exposed to lethal drug concentrations, which supports the existence of a reservoir that is removed from drug pressure. In the case of countries where malaria is endemic, transmission is high, and acquired immunity is extensive, asymptomatic adults, who rarely become ill and who generally do not receive therapy, may provide such a reservoir for susceptible parasites to persist in the population. If nascent malaria elimination efforts in Africa result in lower transmission and less acquired immunity, then this reservoir may shrink and eventually disappear, allowing resistant alleles to become fully fixed in the population, as has been observed in both South America [23] and Southeast Asia [7]. It will be important to monitor the effects of malaria control and elimination interventions on parasite population diversity in order to track and predict the patterns of the rise and fall of drug resistance.
The renaissance of chloroquine-susceptible parasites was not expected. Earlier evidence had suggested that chloroquine-resistant parasites may have a survival advantage. In vitro, ex vivo, and ecological studies indicated that chloroquine-resistant parasites formed more gametocytes than did chloroquine-susceptible parasites, suggesting that resistance may increase infectiousness [24]. Buckling et al [25–27] found that the isolates of P. falciparum in vitro and Plasmodium chabaudi in mice that survived the nonlethal drug pressure with chloroquine were more likely to form gametocytes and more likely to be infectious to mosquitoes than untreated parasites. Our study did not examine the sexual stage of the parasite life cycle, but the rapid decline in the prevalence of chloroquine resistance in Malawi implies that any increase in infectiousness among resistant organisms must have been outweighed by other survival advantages of the susceptible genotypes.
The resurgence of susceptible parasites in the absence of drug pressure is best explained by a fitness cost associated with drug resistance that allows susceptible organisms to outcompete resistant organisms in the absence of drug pressure. Resistant organisms often evolve compensatory mutations that restore a variable degree of fitness to susceptible organisms [28]. In the case of chloroquine-resistant malaria, although the global spread of these parasites took decades, the return of susceptibility in Malawi was rapid—detectable within a year after drug pressure was removed [3]. P. falciparum with resistant forms of pfcrt are now undetectable by standard PCR methods at our study site and at many sites throughout the country (Dzinjalamala et al, unpublished data, 2007), although chloroquine-resistant parasites have been detected at a very low prevalence on the border with Tanzania [29] and highly sensitive techniques have suggested the persistence of minority variant resistant parasites at a similar study site [30]. The rapid disappearance of chloroquine resistance points to a high fitness cost of the resistance-conferring K76T mutation for which neither the constellation of other pfcrt mutations that do not cause resistance in the absence of K76T nor other mutations elsewhere in the genome adequately compensate.
The dramatic return of chloroquine-susceptible falciparum following the removal of chloroquine drug pressure in Malawi has not occurred in Southeast Asia or South America, where treatment policies were changed well before policies were changed in Africa. A modest increase in chloroquine-susceptible malaria had been documented in China following reduction of chloroquine use [31]. The reasons for these different patterns may lie in parasite, host, and/or environmental factors. Chloroquine resistance may have persisted in Asia and South America because of lower transmission (which is accompanied by smaller effective population sizes and less frequent polyclonal infections, and thus fewer opportunities for recombination, leading to fixation of the resistant genotype in the population), higher proportions of the parasite population being subjected to drug pressure in relatively nonimmune human populations, and/or persistent drug pressure with chloroquine or amodiaquine [32, 33]. It is also possible that the genetic backgrounds of parasites from Asia and South America have unique characteristics that favor the emergence or support the spread of resistance [33, 34]. There is some evidence of impaired DNA repair mechanisms in Asian parasites, compared with African parasites [35, 36]. New genomic epidemiological approaches and the exploration of rapid mutator or impaired DNA repair phenotypes may lead to the identification of a genetic or evolutionary basis for the consistent finding of emerging drug resistance in Asia that spreads across continents.
The results of this study have important implications for the future of antimalarial drug use in Africa. Almost all countries where malaria is endemic are now recommending the use of artemisinin-based combination therapy in place of the older drugs typically used as monotherapy—chloroquine and sulfadoxine-pyrimethamine. In a recent multinational survey, sub-Saharan African countries were found to have maintained parasite diversity with respect to pfcrt in that susceptible parasites are still present in the population, although they are in the minority [37]. We anticipate that as artemisinin-based combination therapies are successfully deployed throughout Africa, chloroquine-susceptible malaria will return to the region, a trend that has been documented in Kenya [38].
If chloroquine susceptibility does become widespread in Africa, the possibility of using chloroquine in the future promises to be a welcome addition to the limited armamentarium in the battle against malaria. However, any reintroduction of chloroquine for the treatment or prevention of malaria should be planned with attention to the evolution of drug resistance. Chloroquine could become an attractive option for use as a tool for malaria prevention in specific targeted populations, as it is safe and well tolerated, has a long duration of action to maximize the period of protection, and has a resistance profile that differs from that of most other drugs currently being used. The extent to which the use of chloroquine (alone or in combination) for intermittent preventive treatment in select vulnerable groups, such as pregnant women or infants, would contribute to the reemergence of chloroquine resistance is not known.
If chloroquine is used again for the treatment of malaria, it should be used in combination with a partner drug to protect against resistance. The combination of chloroquine plus an artemisinin derivative represents a gross mismatch of drug pharmacokinetic and pharmacodynamic profiles, with the artemisinins clearing from the blood in just a few hours and chloroquine being detectable for at least 4–6 weeks. In the presence of high rates ofmalaria transmission, chloroquine will effectively be present as monotherapy once the 3-d course of artemisinins is completed [39]. We are currently evaluating the efficacy of chloroquine combined with drugs with different pharmacokinetic and pharmacodynamic profiles for the treatment of falciparum malaria in Malawi, with the goal of identifying combinations that could deter the reemergence of resistance.
Acknowledgments
Financial support: National Institutes of Health (grants K23AI059316, U01AI044824, R29AI040539, R01AI44824M01, RR 16500, and K12RR023250); Doris Duke Charitable Foundation (grants to M.K.L. and C.V.P.); Howard Hughes Medical Institute (grant to C.V.P.).
We thank Dr Licheng Zhao for conducting the microsatellite genotyping and Dr Tim Anderson for sharing the polymerase chain reaction conditions for the microsatellite amplification. We are grateful to the children who participated in these studies and their families.
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 56th Annual Meeting of the American Society for Tropical Medicine and Hygiene, Philadelphia, PA, 7 November 2007 (abstract 969).
References
- 1.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359(24):2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361(5):455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187(12):1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 4.Laufer MK, Thesing PC, Eddington ND, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355(19):1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 5.Bloland PB, Lackritz EM, Kazembe PN, Were JB, Steketee R, Campbell CC. Beyond chloroquine: implications of drug resistance for evaluating malaria therapy efficacy and treatment policy in Africa. J Infect Dis. 1993;167(4):932–937. doi: 10.1093/infdis/167.4.932. [DOI] [PubMed] [Google Scholar]
- 6.Roper C, Pearce R, Bredenkamp B, et al. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361(9364):1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 7.Nash D, Nair S, Mayxay M, et al. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci. 2005;272(1568):1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418(6895):320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 9.Barton NH. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 2000;355(1403):1553–1562. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160(2):765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlotterer C. A microsatellite-based multilocus screen for the identification of local selective sweeps. Genetics. 2002;160(2):753–763. doi: 10.1093/genetics/160.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23(1):23–35. [PubMed] [Google Scholar]
- 13.Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167(3):1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McVean G. The structure of linkage disequilibrium around a selective sweep. Genetics. 2007;175(3):1395–1406. doi: 10.1534/genetics.106.062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 16.Pearce RJ, Pota H, Evehe MS, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6(4):e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djimde A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344(4):257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 19.Pearce R, Malisa A, Kachur SP, Barnes K, Sharp B, Roper C. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol Biol Evol. 2005;22(9):1834–1844. doi: 10.1093/molbev/msi177. [DOI] [PubMed] [Google Scholar]
- 20.Mueller JC, Andreoli C. Plotting haplotype-specific linkage disequilibrium patterns by extended haplotype homozygosity. Bioinformatics. 2004;20(5):786–787. doi: 10.1093/bioinformatics/btg481. [DOI] [PubMed] [Google Scholar]
- 21.Ariey F, Fandeur T, Durand R, et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittra P, Vinayak S, Chandawat H, et al. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J Infect Dis. 2006;193(9):1304–1312. doi: 10.1086/502979. [DOI] [PubMed] [Google Scholar]
- 23.Cortese JF, Caraballo A, Contreras CE, Plowe CV. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002;186(7):999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- 24.Koella JC. Costs and benefits of resistance against antimalarial drugs. Parasitol Today. 1998;14(9):360–364. doi: 10.1016/s0169-4758(98)01297-6. [DOI] [PubMed] [Google Scholar]
- 25.Buckling A, Ranford-Cartwright LC, Miles A, Read AF. Chloroquine increases Plasmodium falciparum gametocytogenesis in vitro. Parasitology. 1999;118(4):339–346. doi: 10.1017/s0031182099003960. [DOI] [PubMed] [Google Scholar]
- 26.Buckling AG, Read AF. The effect of chloroquine treatment on the infectivity of Plasmodium chabaudi gametocytes. Int J Parasitol. 1999;29(4):619–625. doi: 10.1016/s0020-7519(98)00230-6. [DOI] [PubMed] [Google Scholar]
- 27.Buckling AG, Taylor LH, Carlton JM, Read AF. Adaptive changes in Plasmodium transmission strategies following chloroquine chemotherapy. Proc Biol Sci. 1997;264(1381):553–559. doi: 10.1098/rspb.1997.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bridges DJ, Molyneux M, Nkhoma S. Low level genotypic chloroquine resistance near Malawi’s northern border with Tanzania. Trop Med Int Health. 2009;14(9):1093–1096. doi: 10.1111/j.1365-3156.2009.02340.x. [DOI] [PubMed] [Google Scholar]
- 30.Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13(6):872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Mu J, Li G, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg. 2005;72(4):410–414. [PubMed] [Google Scholar]
- 32.Sá JM, Twu O, Hayton K, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A. 2009;106(45):18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laufer MK, Plowe CV. Withdrawing antimalarial drugs: impact on parasite resistance and implications for malaria treatment policies. Drug Resist Updat. 2004;7(4–5):279–288. doi: 10.1016/j.drup.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94(17):9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trotta RF, Brown ML, Terrell JC, Geyer JA. Defective DNA repair as a potential mechanism for the rapid development of drug resistance in Plasmodium falciparum. Biochemistry. 2004;43(17):4885–4891. doi: 10.1021/bi0499258. [DOI] [PubMed] [Google Scholar]
- 36.Bethke LL, Zilversmit M, Nielsen K, et al. Duplication, gene conversion, and genetic diversity in the species-specific acyl-CoA synthetase gene family of Plasmodium falciparum. Mol Biochem Parasitol. 2006;150(1):10–24. doi: 10.1016/j.molbiopara.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 2009;8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mwai L, Ochong E, Abdirahman A, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8(1):106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyunt MM, Plowe CV. Pharmacologic advances in the global control and treatment of malaria: combination therapy and resistance. Clin Pharmacol Ther. 2007;82(5):601–605. doi: 10.1038/sj.clpt.6100361. [DOI] [PubMed] [Google Scholar]