Abstract
There is intense interest in developing methods to regulate proliferation and differentiation of stem cells into neuronal fates for the purposes of regenerative medicine. One way to do this is through in vivo pharmacological engineering using small molecules. However, a key challenge is identification of relevant signaling pathways and therein druggable targets to manipulate stem cell behavior efficiently in vivo. Here, we use the planarian flatworm as a simple chemical-genetic screening model for nervous system regeneration to show that the isoquinoline drug praziquantel (PZQ) acts as a small molecule neurogenic to produce two-headed animals with integrated CNSs following regeneration. Characterization of the entire family of planarian voltage-operated Ca2+ channel α subunits (Cavα), followed by in vivo RNAi of specific Cav subunits, revealed that PZQ subverted regeneration by activation of a specific voltage-gated Ca2+ channel isoform (Cav1A). PZQ-evoked Ca2+ entry via Cav1A served to inhibit neuronally derived Hedgehog signals, as evidenced by data showing that RNAi of Cav1A prevented PZQ-evoked bipolarity, Ca2+ entry, and decreases in wnt1 and wnt11-5 levels. Surprisingly, the action of PZQ was opposed by Ca2+ influx through a closely related neuronal Cav isoform (Cav1B), establishing a novel interplay between specific Cav1 channel isoforms, Ca2+ entry, and neuronal Hedgehog signaling. These data map PZQ efficacy to specific neuronal Cav complexes in vivo and underscore that both activators (Cav1A) and inhibitors (Cav1B) of Ca2+ influx can act as small molecule neurogenics in vivo on account of the unique coupling of Ca2+ channels to neuronally derived polarity cues.
Introduction
Methods that generate large numbers of specific cell types as immunologically matched replacements for diseased tissue have clear therapeutic potential, especially for neurodegenerative conditions. Three broad strategies that achieve this encompass protein-based approaches (growth factor “cocktails”), genetic reprogramming (via specific transcription factors), and pharmacological engineering (small molecules that bias differentiation). While each method has advantages, the inherent appeal of small molecule-based approaches translates to their potential for use in vivo with lesser risks than exogenous genetic reprogramming. Key challenges are identifying “druggable” signaling pathways that regulate stem cell expansion and differentiation, and understanding the functional interplay of such pathways in vivo.
Ca2+ signaling exemplifies a well dissected pathway to nuclear reprogramming in differentiated neurons (Greer and Greenberg, 2008). In neural stem/progenitor cells also, in vitro screens have uncovered Ca2+ signaling modulators that regulate proliferation and adoption of neuronal cell fates (Diamandis et al., 2007; Schneider et al., 2008). While these insights derive from studying multipotent cells in vitro, it is important to discern whether similar principles hold in vivo. This is an important distinction as stem cell fate in vivo is controlled by cues inherent to the local microenvironment such that the efficacy of pharmacological agents identified in vitro will be modified by signals unique to the stem cell niche.
An attractive screening model for small molecule neurogenics is the planarian flatworm. Planarians exhibit impressive regenerative abilities owing to the maintained plasticity of their pluripotent stem cells (“neoblasts”), which differentiate into ∼30 cell types during homeostasis and enforced tissue regeneration. These worms afford the opportunity to study regeneration of an entire nervous system by simple amputation assays, rather than simply the regrowth/repair of a single neuron (Newmark and Sánchez Alvarado, 2002; Cebrià, 2007). Planarians hold great fascination for neuroscientists: they express a diverse array of neurotransmitters (Collins et al., 2010), occupy a unique evolutionary niche in terms of emergence of a centralized nervous system, and have behavioral screening potential. Further, most planarian genes (∼80%) show greater similarity to vertebrate orthologs relative to invertebrate sequences (Sánchez Alvarado et al., 2002; Friedländer et al., 2009).
Previously, while investigating the undefined mechanism of action of praziquantel (PZQ)—a drug used to treat schistosomiasis—we found that PZQ subverted regeneration to produce viable, two-headed worms with integrated CNSs (Nogi et al., 2009). Initial data suggested that PZQ miscued regeneration by modulating voltage-operated Ca2+ entry (Nogi et al., 2009). However, the lack of molecular information about voltage-operated Ca2+ channels (Cavs) in this system precluded functional genetic testing of this hypothesis. Here, we define the planarian family of Cavα subunits and employ in vivo RNAi to show that PZQ subverts regeneration by selective activation of a Cavα isoform (Cav1A) to dysregulate neuronal Hedgehog signaling. This effect was opposed by another neuronal Cav1 isoform (Cav1B). These data support a unique interplay between specific Cav1 channels and neuronal Hedgehog signaling and justify analysis of Cav1 channels in vivo as targets for small molecule neurogenics and for PZQ, the mainstay therapeutic for treating a disease that infects 200 million people worldwide.
Materials and Methods
Worm husbandry.
An asexual clonal GI strain (Gifu, Iruma river) of Dugesia japonica was maintained (∼5000 worms in 5 L of water) at room temperature (20–23°C) and fed strained chicken liver puree (∼10 ml) once a week. Regenerative assays were performed using 5 d-starved worms in pH-buffered artificial water at 22°C (1× Montjuïch salts: 1.6 mm NaCl, 1.0 mm CaCl2, 1.0 mm MgSO4, 0.1 mm MgCl2, 0.1 mm KCl, 1.2 mm NaHCO3, pH 7.4 buffered with 1.5 mm HEPES). PZQ, sourced from Sigma (P4668), was used as a racemic mixture. The basic planarian methods used in these experiments have been described by Chan and Marchant (2011).
In situ hybridization.
Whole-mount in situ hybridization was performed at 55°C in hybridization solution (50% formamide, 5× SSC, 100 μg/ml yeast tRNA, 100 μg/ml heparin sodium salt, 0.1% Tween 20, 10 mm DTT, 5% dextran sulfate sodium salt) incorporating digoxigenin (DIG)-labeled antisense riboprobe (40 ng/ml) denatured at 72°C for 15 min before use (Nogi et al., 2009). A standard mixture of BCIP/NBT in chromogenic reaction solution was used for color development, followed by paraformaldehyde fixation. DIG-labeled antisense riboprobe was synthesized by RNA polymerase (Roche) from linearized pGEM-T Easy plasmid as the template. Probe regions were as follows: PC2 (1–2285 bp); Hox9 (1–1491 bp); ndk (122–1692 bp); nlg (1–1204 bp); Inx7 (1–1528 bp); MHC (4879–5905 bp); wnt1 (1–1162 bp); wnt11-5 (1–1050 bp); Hh (59–1370 bp from AB504739.1); Cav1A (1027–1865 bp; 2229–4133); Cav1B (2722–4010 bp; 4380–6059); and Cav2A (120–962 bp). Staining was resolved and archived using a Leica MZ16F stereomicroscope and a QiCAM 12 bit cooled color CCD camera.
Cloning strategies.
Total RNA was isolated from 20 intact planarians using TRIzol and cDNA subsequently synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen). Novel Cavα cDNAs were identified by PCR amplification (LA Taq polymerase) using degenerative primers designed from regions with high sequence identity based on alignment with a published Schistosoma mansoni Cav1 sequence (Kohn et al., 2001b) and putative annotations in the S. mansoni (Berriman et al., 2009) and Schmidtea mediterranea genomes (Robb et al., 2008). Products were cloned into the pGEM-T Easy vector (Promega) for sequencing. On the basis of initial sequence data, further sequence was predicted from genomic annotations and verified by screening either a cDNA library prepared from regenerating D. japonica fragments or freshly synthesized cDNA if not represented in the existing library. Final sequences of the Cav clones were assembled in pGEM-T Easy vector and resequenced three times. For RACE analysis, mRNA was separated and purified using Oligotex mRNA mini Kits (Qiagen) and used to synthesize cDNA with gene-specific primers according to the manufacturer's instructions (5′/3′ RACE Kit, Roche). Oligo-dT primers were used to synthesize 3′RACE cDNA, and nested PCR to amplify 5′ RACE cDNA. Gene-specific primers for RACE were as follows: 5′RACE: Cav1A (5′CATTTTCTTCATCGCTGAGTTCGTCA-3, 5′-TTCTCCGCTAAGAACACCAAGAATTA-3′, 5′-TTGTTTCCTTGTGCATCATTTATCCA-3), Cav1B (5′-TCTGCATTGTTACCTTCTTCTTCTTC-3′, 5′-CGTTCAATAAGATGTAGTTTCCGCAA-3′, 5′-GACTTCATTCCAATCTTCACCAGTTAG-3′, 5′-GAGCA-ATGACTGCCAAAAACTATCAAA-3′); 3′RACE: Cav1A (5′-GGATTGGGAGCATTAGTTTCCTTGTA-3′, 5′-GGCTGCTGAAGACCCAATAAGAAC-3′, 5′-TATCTTCGTTGGTTTTGTCATCGTT-3′), Cav1B (5′-GCCTGGCTTATTATACAAATCAATCG-3′, 5′-TTACAGTGGCACATATAATAATCGACC-3′, 5′-GTTTTGTCATCGTTACGTTTCAGCA-3′). Products were gel purified (High Pure PCR Product Purification Kit, Roche) and fragments ligated into pGEM-T Easy vector for sequencing. A similar approach was used to isolate D. japonica βcatenin-1, adenomatous polyposis coli (APC), hedgehog (Hh), patched (ptc), wnt1, and wnt11-5. Wnt nomenclature is from Gurley et al. (2010). For in vivo RNAi, sequences were amplified using gene-specific primers incorporating Kozak sequence and cloned into the IPTG-inducible vector pDONRdT7 using Gateway BP Clonase (Invitrogen).
RNAi methods.
In vivo RNAi was performed following a feeding protocol described previously (Nogi et al., 2009). Over several feeding/regeneration cycles, worms were fed a chicken liver and bovine red blood cell mixture containing transformed HT115 bacteria induced to express individual dsRNA constructs. Comparison of phenotypic scoring and drug effects were examined using paired t tests, with differences considered significant at p < 0.05 (*) and p < 0.01 (**). All data are presented as mean ± SEM for the indicated number of experiments. As a control for nonspecific RNAi effects, a Schmidtea mediterranea six-1 (Smed-six-1) construct was used. This construct did not produce a phenotype in D. japonica regenerants, owing to nucleotide divergence between the six-1 genes in the two planarian species. Targeted sequences were as follows: Cav1A (1042–1831 bp; 2229–4133 bp), Cav1B (2722–4010 bp; 4380–6059 bp; 6194–7219 bp), Cav2A (120–962 bp), βcatenin-1 (1–1351 bp), APC (1–2413 bp), ptc (95–2572 bp), Smed-six-1 (1–506 bp). For assessment of knockdown following RNAi, cDNA from experimental cohorts of 10 worms, or from 40 posterior blastemas for wnt analysis, was used. Quantitative real-time PCR (qPCR) was performed using a ABI 7500 real-time PCR system (Applied Biosystems) and SYBR GreenER qPCR SuperMix Universal (Invitrogen). Primers for qPCR were as follows: Cav1A: 5′-ACTCGACCAAAGATTATCAATCCGAT-3′, 5′-CCACCAAACATTTGCATACCAAGAAG-3′; Cav1B: 5′-CTTTCAAAGAAGATTACAGTGGCACA-3′, 5′-ACCAAACTCGGTATCTGAAACTCTGTT-3′; Cav2A: 5′-TACGATGGAAGGGTGGACAGATGTT-3′, 5′-AAGCTCGTCTTTTCTCTACTCTTTCTC-3′; β-actin: 5′-GGTAATGAACGATTTAGATGTCCAGAAG-3′, 5′-TCTGCATACGATCAGCAATACCTGGAT-3′; wnt-1: 5′-ATCGCACAGGATTGGTTGTTGCT-3′, 5′-GTTCCATAATTGTTTTCGATCTCGT-3′; wnt11-5:5′-TTGGTGTCAGACATCAAGGATTTCA-3′, 5′-GCCTTGACAGTTCCAAACGTGGTT-3′. In all cases, at least one qPCR primer was localized outside the sequence of the RNAi construct. For absolute qPCR analyses, cDNA (not containing the RNAi targeted sequence) for each construct was cloned into pGEM-T Easy vector (Promega) and used as a template to create gene-specific standard curves for assessing mRNA levels in samples isolated at equivalent regenerative time points from different worms. The mRNA levels of specific genes was compared with controls using D. japonica β-actin to normalize RNA input. As a further calibration of absolute qPCR results, data were compared to those from a semiquantitative RT-PCR analysis of the same sample (data not shown).
Confocal Ca2+ imaging and 45Ca2+ assays.
Dissociated planarian cells were obtained by cutting worms into ∼10 fragments in Ca2+-free Holtfreter's solution (5/8 dilution) supplemented with 1 mm EDTA, 1% BSA (w/v), and 1% FBS (v/v). Fragments were washed in Ca2+-free Holtfreter's (5/8) and dissociated to single cells by incubation in 0.25% (w/v) trypsin for 15 min at room temperature, pipetting periodically with a Pasteur pipette. Enzymatic digestion was arrested with the addition of 10% (v/v) FBS and the dissociated cells were filtered though a 40 μm nylon mesh, followed by centrifugation (7 min, 300 × g). Cells were resuspended to a density of ∼106 cells/ml. For Ca2+ imaging, dissociated cells were plated onto poly-d-lysine-coated 35 mm Petri dishes (MatTek) and stored at room temperature for 18–24 h. Adhered cells were loaded with fluo4-AM (90 min, 4 μm) in Ca2+-free Holtfreter's (5/8) containing 2% BSA (w/v) and 0.025% pluronic, and then washed in Holtfreter's (5/8) supplemented with Ca2+ (1 mm) for 30 min before imaging. Dishes were imaged using an Andor Revolution spinning disc confocal microscope and changes in fluorescence monitored (λex = 488 nm, λem = 525 nm) following addition of either PZQ or vehicle controls. Fractionation of the crude dissociation sample was performed by serial centrifugation (10 min at 100 × g, 200 × g, 1000 × g, 3000 × g, 14,000 × g sequentially), such that the supernatant of each centrifugation step was removed and spun at increasing speeds to separate fractions by size. Fractions were fixed (8% paraformaldehyde in PBS), stained with a NeuroTrace green fluorescent Nissl stain (Invitrogen, 1:100 dilution in PBS, 40 min), and counterstained in DAPI (1 μg/ml, 10 min). For 45Ca2+ experiments, intact cells were incubated in Ca2+-containing Holtfreter's (5/8) solution [1 mm Ca2+ supplemented with 45Ca2+ (9 μCi/ml)] in the absence or presence of various concentrations of PZQ (100 nm-100 μm). After 30 min, cells were harvested by filtration in ice-cold sucrose-citrate solution (GF/B, Whatman) using a Brandel Harvester (Marchant et al., 1997) and cellular 45Ca2+ uptake assessed by liquid scintillation counting. Protein was quantified by Bradford assays for normalization of data between different fractions.
Results
We serendipitously discovered that trunk fragments of the planarian Dugesia japonica exposed to the drug PZQ immediately after amputation regenerated as viable, two-headed animals (Fig. 1A) with dual integrated CNSs (Nogi et al., 2009). The effect was highly penetrant (94 ± 4% bipolar, 70 μm PZQ for 48 h) and the duplication of the CNS by external drug application was clearly shown by in situ hybridization of a CNS marker (Fig. 1A). Mechanistic explanation of this effect is lacking: notably, the in vivo target(s) of PZQ remain unresolved despite its usage as a clinical drug for >30 years (Day et al., 1992; Cioli and Pica-Mattoccia, 2003; Caffrey, 2007). Prior in vitro evidence has implicated several possible molecular targets (Wiest et al., 1992; McTigue et al., 1995; Angelucci et al., 2007; Tallima and El Ridi, 2007; Gnanasekar et al., 2009), including activation of Ca2+ influx in muscle (Pax et al., 1978; Kohn et al., 2001a), however the in vivo relevance of such pathways has not been determined owing to a lack of functional genetic data. Our initial data—PZQ increased 45Ca2+ uptake, and bipolarity was phenocopied by depolarization and attenuated by either the L-type Ca2+ channel antagonist nicardipine or RNAi of accessory Cavβ subunits (Nogi et al., 2009)—was supportive of the Ca2+ hypothesis and justified analysis of a requirement for specific Cav channels. However, the lack of any molecular data about the Ca2+ channels themselves precluded such analysis, despite the fact that the striking duplication of the CNS achieved by PZQ exposure provided a simple visual screen for RNAi analysis of molecules needed for PZQ efficacy in vivo.
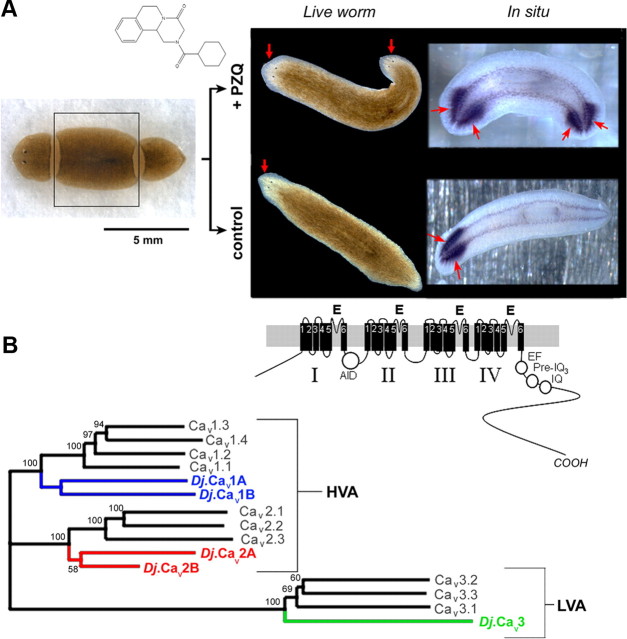
Figure 1.
PZQ-evoked bipolarity and characterization of planarian Cavα subunits. A, Left, Overview of regenerative assay. Trunk fragment (boxed) was isolated and incubated with PZQ (structure, top) in samples to be compared with control worms (bottom). Right, Images of bipolar worms (head, arrowed) produced after PZQ exposure (top) in live worms (left) and samples stained for the CNS marker PC2 (right). Control worms are shown for comparison (bottom). B, Phylogenetic analysis of sequence homology between planarian and human Cavα sequences aligned using MUSCLE and displayed as an unrooted tree assembled by a neighbor joining algorithm (Geneious 5.0). Bootstrap values are indicated at nodes. The lower value for D. japonica Cav2 subunits relates to the use of partial sequences. Accession numbers are referenced in Table 1, with addition of Cav2.1 (O00555), Cav2.2 (Q00975), Cav3.2 (O95180), and Cav3.3 (Q9P0X4). The D. japonica subunit previously referred to as Cav1.1 (Nogi et al., 2009) is renamed Cav2A to standardize nomenclature with the Schistosome literature (Kohn et al., 2001b). Inset, Schematic of Cav1 architecture (domains I–IV) and motifs (α-interaction domain, AID; EF-hand motif, EF; pre-IQ3 and IQ motif).
Planarian Cav channels
To enable a candidate RNAi approach, we characterized the entire family of planarian voltage-operated Ca2+ channel α subunits. Five discrete Cavα subunits (GenBank accession numbers: Dugesia japonica Cav1A, HQ724315; Cav1B, HQ724316; Cav2A, HQ724317; Cav2B, HQ724318; Cav3, HQ724319) were identified using degenerate PCR (Fig. 1B). Four subunits displayed high similarity to high-voltage-activated Cavα subunits (HVA, Fig. 1B), two of which clustered with vertebrate L-type Cavα channels (christened Cav1A and Cav1B) and two with non-L type sequences (Cav2A and Cav2B). The remaining Cavα subunit most closely resembled a T-type subunit (Cav3), representing the first low-voltage-activated (LVA) Cavα subunit reported in flatworms. Compared with established invertebrate models, which express only single representatives from the three Cavα gene families [EGL-19, UNC-2, CCA-1 in Caenorhabditis elegans (Yeh et al., 2008); Dmca1D, Dmca1A, and Ca-α1T in Drosophila (King, 2007)], the molecular repertoire of Cavα subunits in planarians was clearly more expansive.
As a first step toward investigating physiological roles for individual Cavα subunits, we focused on the two Cav1 subunits (Cav1A and Cav1B). Full-length sequences for Cav1A and Cav1B encoded proteins of 1812 (Cav1A) and 2652 (Cav1B) amino acids such that Cav1B potentially represents the largest HVA Ca2+ channel identified to date in any species (Zheng et al., 1995). Each Cav1 subunit possessed an architecture characteristic of the voltage-gated ion channel superfamily, comprising four repeated domains (I–IV) of six transmembrane spanning helices (S1–S6) with overall high sequence homology (Figs. 1B, 2). Diagnostic Cav features included the following: (1) a reentrant P-loop, located between S5 and S6, harboring the conserved glutamic acid residue that contributes to the EEEE selectivity gate in all HVA Cavα subunits; (2) essential residues within the α-interacting domain between domain I and II that are crucial for Cavβ interaction; and (3) the isoleucine-glutamine (IQ) domain, preIQ3 domain, and EF-hand consensus motifs within the cytoplasmic COOH terminus that mediate Ca2+-regulation of L-type Cav channels. Despite this architectural similarity, the functional properties of planarian Cav channels are likely unique from their vertebrate counterparts. For example, a definitive pharmacology of vertebrate Cav1 channels is modulation by dihydropyridines. A critical methionine residue for dihydropyridine binding in IIIS6 that is found in Cav1 channels from rat (M1161 in α1c) as well as C. elegans (M1056 in EGL-19) was represented by isoleucine in both the D. japonica Cav1 subunits (Fig. 2). Having been isolated as a resistant polymorphism (M1056I) toward nemadipine-evoked growth retardation in C. elegans (Kwok et al., 2008), this isoleucine substitution likely confers dihydropyridine insensitivity to the D. japonica Cav1 subunits. Therefore, extrapolation of pharmacological and regulatory properties from vertebrate Cav subtypes to their planarian orthologues is unsupported from overall sequence homology, which never surpassed 50% identity (Table 1).
Figure 2.

Sequence alignment of flatworm Cav1 subunits. Sequence alignment of Dugesia japonica Cav1A (Dj-Cav1A) and Cav1B (Dj-Cav1B) with Schistosoma mansoni Cav1A (Sm.Cav1A, AF361884) and Cav1B (Sm.Cav1B, CAZ34413.1). Sequences were aligned in Jalview using MUSCLE. Features were assigned with reference to a rat brain Cav1.2 subunit (GenBank #AAA18905) and indexed in the right hand column. These include highlighted residues that illustrate conservation of key residues within the α-interaction domain (AID, magenta), the EEEE ion selectivity filter motif (red), cytoplasmic COOH-terminal EF (blue), preIQ3 (purple), and IQ (green) regulatory domains, as well as 20 residues required for dihydropyridine interaction that are identical (yellow) or divergent (orange) in flatworm Cav sequences from rat Cav1.2 sequence. The isoleucine residue present in flatworm Cav1 subunits that may be relevant to DHP sensitivity is found as residue I1106 in Cav1A and I1317 in Cav1B.
Table 1.
Comparison of flatworm and vertebrate Cavα identity
|
Dj |
Sm |
Hs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cav1A | Cav1B | Cav1A | Cav1B | Cav2A | Cav1.1 | Cav1.2 | Cav1.3 | Cav1.4 | Cav2.3 | Cav3.1 | |
| Cav1 Dj | |||||||||||
| Cav1A | 52% | 57% | 54% | 39% | 49% | 48% | 49% | 48% | 38% | 23% | |
| Cav1B | 50% | 61% | 34% | 49% | 46% | 47% | 48% | 37% | 22% | ||
Sequences were aligned using the BLOSUM62 scoring matrix (ClustalW MSA), and amino acid identities were computed. Sm.Cav1 was renamed Sm.Cav1A, owing to the in silico predication of a second Cav1 subunit in Schistosoma mansoni (named Sm.Cav1B, GenBank #CAZ34413.1). Subunits for which complete coding sequences have been biologically verified are shown in bold. Groupings with the highest identity are underlined. The following accession numbers were used: Schistosoma mansoni (Sm): Sm.Cav1A (AF361884), Sm.Cav1B (CAZ34413.1), Sm.Cav2A (AF361883); Homo sapiens (Hs): Hs.Cav1.1 (Q13698), Hs.Cav1.2 (Q13936), Hs.Cav1.3 (Q1668), Hs.Cav1.4 (O60840), Hs.Cav2.3 (Q15878), Hs.Cav3.1 (O43497).
Unique roles of Cav1 subunits
In vivo RNAi was used to investigate whether Cav1 subunit function impacted PZQ-evoked bipolarity. RNAi constructs targeting multiple regions of Cav1A and Cav1B were designed and worms were fed bacteria expressing dsRNA against individual Cavα subunits (Sánchez Alvarado and Newmark, 1999). Constructs serving as phenotypically positive [Dj-six-1, a transcription factor required for eye regeneration (Mannini et al., 2004)] and negative (Smed-six-1, the same gene from Schmidtea mediterranea but with no phenotypic outcome owing to sequence divergence) RNAi controls were included in each assay. The RNAi protocol comprised two dual feeding and regenerative cycles, lasting ∼1 month in total duration (Fig. 3A). Before the final regenerative cycle, a cohort of worms was removed for real time PCR analysis to assess gene knockdown at the point of assaying the bipolarizing effect of PZQ. This protocol permitted screening individual RNAi constructs for optimal effectiveness and selectivity: Cav1A mRNA were decreased by ∼60% with no significant change in Cav1B mRNA; Cav1B proved more resistant to knockdown—levels of mRNA were decreased by ∼30% but with only a 4% change in Cav1A levels in the same samples (Fig. 3B).
Figure 3.
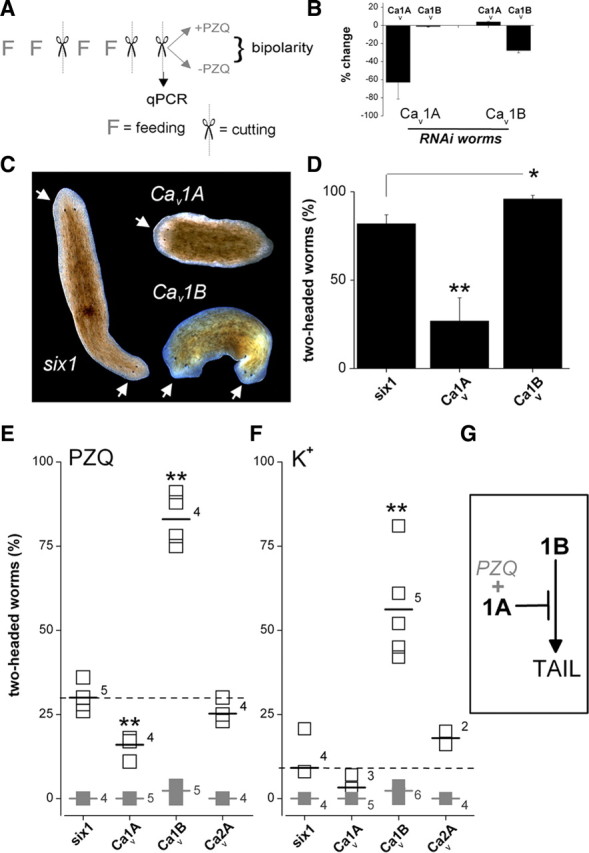
RNAi of Cav1 subunits modulates bipolar regeneration. A, Protocol for in vivo RNAi. Two cycles of dual feeding (F) and regeneration (scissors) were followed by a regenerative assay in the presence or absence of PZQ. Bipolarity was scored 7 d after the final regeneration. A cohort of trunk fragments was removed before drug exposure to assess the effectiveness and specificity of ablation of individual mRNAs at the time of drug addition using qPCR. All RNAi assays followed this protocol (FFxFFxx) with exceptions of APC (FFFFx), Ptc (FFFFxFFFx), and Hh (FFFxFFxx). B, Assessment of changes in mRNA abundance for Cav1 subunits in worms fed either Cav1A(RNAi) or Cav1B(RNAi) constructs, respectively. C, Images showing bipolar worms (head, arrowed) regenerating in Smed-six-1 and Cav1B, but not Cav1A cohorts. D, Percentage of two-headed worms in Smed-six-1 (control), Cav1A or Cav1B cohorts following exposure to PZQ (90 μm for 24 h). For clarity only the Smed-six-1 control cohort is shown—similar results were obtained in naive and Dj-six-1 worms (see text). Asterisks indicate probability of similarity at p < 0.05 (*) and p < 0.01 (**). E, Two-headed regenerants in the absence (gray, solid) or presence (black, open) of a lower dose of PZQ (50 μm for 24 h) in six-1 (control), Cav1A, or Cav1B worms. Numbers report the number of independent trials, and short horizontal lines indicate the arithmetic mean of these experiments. The dashed line indicates the mean value of control (six-1) dataset. F, Effect of depolarizing conditions. Trunk fragment regeneration in media with elevated [K+] (supplemented by 30 mm, 24 h) is shown. PZQ was not present in these assays. G, Working model for Cav1 channels in regenerative polarity.
Following Cav1 subunit RNAi, the effectiveness of PZQ in evoking bipolarity was evaluated. PZQ induced two-headed worms with high efficacy in the positive phenotype (eyeless) control cohort (Dj-six-1, 80 ± 8%), the negative (RNAi) control cohort (Smed-six-1, 82 ± 5%), and naive worms (82 ± 3% with 90 μm PZQ for 24 h). In contrast, Cav1A knockdown markedly antagonized the bipolarizing ability of PZQ (Fig. 3C), with approximately threefold fewer bipolar regenerants in Cav1A(RNAi) worms compared with controls (Fig. 3D). Bipolar worms occurred in high numbers with Cav1B(RNAi) worms, and moreover the penetrance of this effect (96 ± 2%, n = 6 trials) appeared greater than controls (p < 0.05, Fig. 3D). Given the difficulty of confirming potentiation to such a high dose of PZQ, we repeated these experiments using a lower PZQ concentration (50 μm, 24 h) such that both decreases and increases in bipolar worm numbers could be easily assayed.
In this suppressor-enhancer screen, all trunk fragments from RNAi control and naive worms regenerated with normal head–tail polarity in the absence of PZQ (Fig. 3E), whereas the lower dose of PZQ produced a smaller proportion of two-headed worms (30 ± 2%, n = 5 trials) as expected. Surprisingly, knockdown of the two discrete Cav1 isoforms produced different effects on PZQ-evoked bipolar regeneration. Knockdown of Cav1A again attenuated the anteriorization effect of PZQ (16 ± 2%, n = 4 trials; Fig. 3E), whereas RNAi of Cav1B increased the number of bipolar regenerants (83 ± 4%, n = 4 trials), an ∼2.8-fold potentiation over the control cohort (Fig. 3E). The opposing effects of Cav1A and Cav1B on PZQ-evoked bipolarity were not mimicked by knockdown of at least one other Cav isoform. RNAi targeting Cav2A (Nogi et al., 2009), which resulted in a ∼60% decrease in Cav2A mRNA, did not substantially change the proportion of PZQ-evoked two-headed worms (25 ± 2%, n = 4 trials) relative to controls (29 ± 2%, n = 5 trials). Therefore, RNAi targeting of three different Cavα subunits yielded three different outcomes (Cav1A, attenuation; Cav1B, potentiation; Cav2A, no effect) on PZQ-evoked bipolarity.
If PZQ acts to activate voltage-operated Ca2+ entry, then other depolarizing stimuli should also effect anteriorization. Therefore, we analyzed the effects of elevated [K+] (by 30 mm, a sufficient depolarizing stimulus in flatworms (Novozhilova et al., 2010)) in both control and Cav(RNAi) worms (Fig. 3F). Elevated [K+] yielded only a few two-headed worms in control and Cav1A(RNAi) worms (9 ± 4% and 3 ± 2%, respectively). However, a majority of Cav1B(RNAi) worms displayed bipolar regeneration (56 ± 7%) with the same treatment. First, these data show that Cav1 RNAi differentially miscued regenerative polarity in response to PZQ (Fig. 3D,E) or depolarization (Fig. 3F). Second, the contrast between the effectiveness of PZQ and K+ exposure in control worms was noteworthy: PZQ produced two-headed worms with high effectiveness in control worms (Fig. 3D,E), whereas depolarization alone was a far less effective stimulus (Fig. 3F). These data suggest selectivity in PZQ action on a subset of Cavα subunits (Cav1A), compared to elevated K+ acting as a nonselective depolarizing stimulus on a population of Ca2+ channels with opposing functions (Cav1A vs Cav1B).
A simple model based on RNAi data is shown in Figure 3G. The key feature is the opposing roles of Cav1A and Cav1B. Consistent with Cav1B knockdown potentiating the number of two-headed worms produced by PZQ and K+ treatments (Fig. 3), as well as the formation of a smaller number of bipolar regenerants during normal regeneration (2.4 ± 0.7%, Fig. 3E,F), Cav1B is assigned to function in a posteriorization pathway. Consistent with Cav1A RNAi blocking PZQ efficacy (Fig. 3), PZQ likely activates Cav1A, and this effect is likely selective for Cav1A over Cav1B on account of the different penetrance of PZQ versus K+ as depolarizing cues. The assignment of Cav1A in a pathway antagonistic to Cav1B function is also consistent with the observation of a cohort of Cav1A(RNAi) worms with inhibited head regeneration (2.2 ± 0.7%, n = 5 trials). This simple scheme provides a conceptual framework to define how voltage-gated Ca2+ entry modulates regenerative outcomes.
PZQ activates Ca2+ influx via Cav1A in a neuronally enriched cell fraction
In what cell type(s) are the Cav1 channels active in planaria? There exists surprisingly little knowledge about the cellular physiology of different planarian cell types, likely due to the lack of cell culturing methods since differentiated cells do not divide (neoblasts are the only mitotically active cells). Therefore, to address this question, we performed confocal Ca2+ imaging experiments in acutely dissociated samples prepared from entire worms. Planaria were dissociated into a heterogenous cell mixture that was plated on glass-bottomed dishes for confocal imaging. In samples loaded with the high-affinity Ca2+ indicator fluo-4 (Kd for Ca2+ ∼345 nm), addition of PZQ resulted in an increase in fluorescence in a subset of cells (Fig. 4A). Analysis of time-resolved fluorescence ratios from responding cells revealed that PZQ (100 μm), but not vehicle controls, evoked a rapid Ca2+ transient (F/F0 = 2.75 ± 0.5, n = 18 cells).
Figure 4.
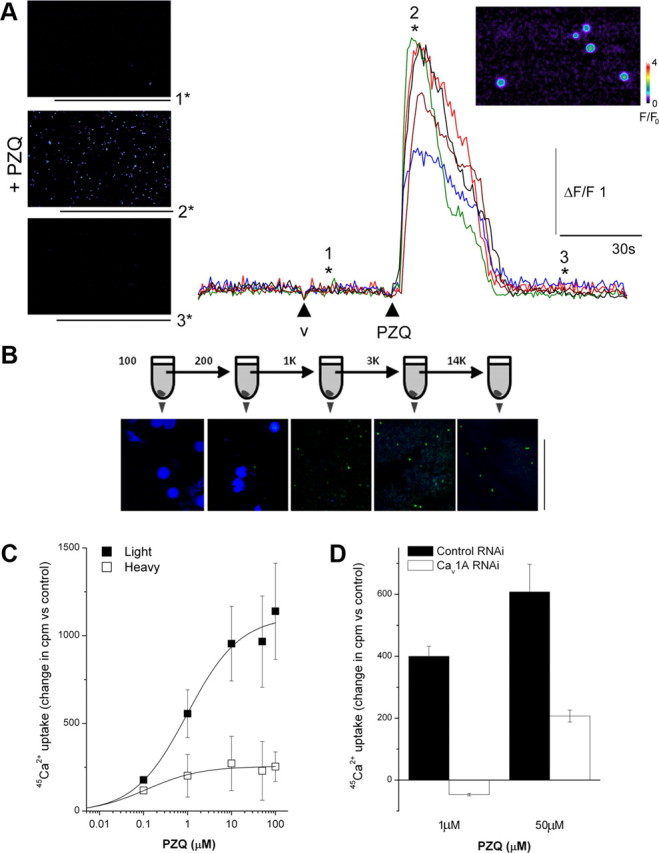
PZQ activates Ca2+ entry via Cav1A channels. A, Left, Confocal Ca2+ imaging of dissociated planarian cells depicting fluo-4 fluorescence on application of control vehicle (image “1”), as well as during (“2”) and after (“3”) application of PZQ (100 μm). Right, Traces show fluorescence profiles from five discrete cells, with the time points of images as indicated (v = vehicle). Inset, Higher-magnification image of responsive cells from which fluorescence profiles were collected (position “2*”). Fluo-4 fluorescence is represented on a pseudocolor scale where increasing warm coloration represents greater fluorescence emission. Fluorescence (F) and change in fluorescence (ΔF) are calibrated relative to fluorescence at time = 0 (F0). B, Schematic of serial centrifugation protocol. The dissociated planarian suspension was first spun at 100 × g to yield a pellet, which was retained for staining, while the supernatant was spun at the next higher speed. This procedure was repeated up to a final step at 14,000 × g. The constituents of the pellet was visualized at each stage using DAPI (blue) and NeuroTrace (green). Scale bar, 50 μm. C, Comparison of 45Ca2+ uptake in response to increasing concentrations of PZQ in equivalent “light” (solid) and “heavy” fraction samples (open) in the same preparation. D, Cav1A RNAi inhibited absolute 45Ca2+ uptake in response to submaximal (1 μm) and maximal (50 μm) concentrations of PZQ. Data are generated from the “light” fraction and expressed relative to untreated samples from the same preparative fractionation.
To obtain large quantities of this subpopulation, we performed fractionation experiments to isolate the responsive cells. Serial centrifugation experiments determined that this fraction was well separated from larger cells simply by centrifugation at higher speeds (Fig. 4B). Centrifugation yielded discrete “heavy” (centrifugation, <300 × g for 5 min) and “light” (centrifugation >300 × g for 5 min) fractions for analysis, with the latter containing the responsive population identified in the single cell Ca2+ imaging assays. Notably, the “light” fractions stained positively with a NeuroTrace (Nissl) stain, with little staining in the “heavy” fraction (Fig. 4B). Therefore both the size and staining profile suggested that the responsive fraction was neuronally derived (Morita and Best, 1966; Best and Noel, 1969). 45Ca2+ uptake experiments were then performed on both the “heavy” and “light” fractions to compare the efficacy of PZQ in both fractions. Increasing concentrations of PZQ evoked a progressive uptake of 45Ca2+ in cells present within the “light” fraction (EC50 = 0.98 μm). The extent of 45Ca2+ entry was significantly smaller (approximately fourfold) in the “heavy” fraction sample, underscoring the initial observation made by Ca2+ imaging (Fig. 4A) that PZQ activates Ca2+ entry into a discrete subset of planarian cells.
Having optimized the population-level 45Ca2+ assay, it was possible to compare results in different RNAi backgrounds. On the basis of the RNAi data implying that PZQ activates Cav1A (Fig. 3F), we compared 45Ca2+ uptake in control RNAi (Smed-six-1) and Cav1A RNAi backgrounds in response to PZQ. Submaximal PZQ (1 μm) failed to enhance 45Ca2+ uptake over control levels in Cav1A RNAi worms, in contrast to the increase observed in control RNAi worms. Similarly, 45Ca2+ uptake evoked by a maximal concentration of PZQ (50 μm) was inhibited by Cav1A RNAi. The inhibition of PZQ-evoked Ca2+ entry by Cav1A knockdown in response to PZQ supports the interpretation of RNAi data (Fig. 3F) to suggest that PZQ activates Ca2+ entry via Cav1A.
Cav1 channels modulate early patterning decisions
Where does Ca2+ act to miscue regeneration? Recently, immense progress has been made in elucidating signaling events regulating planarian regeneration. The crucial breakthrough was identification of a βcatenin isoform in Schmidtea mediterranea (Smed-βcatenin-1) essential for posterior (tail) specification during regeneration, likely by controlling transcriptional activation of a posterior fate circuit. Knockdown of Smed-βcatenin-1 yielded animals with head structures that regenerated from each wound (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008).
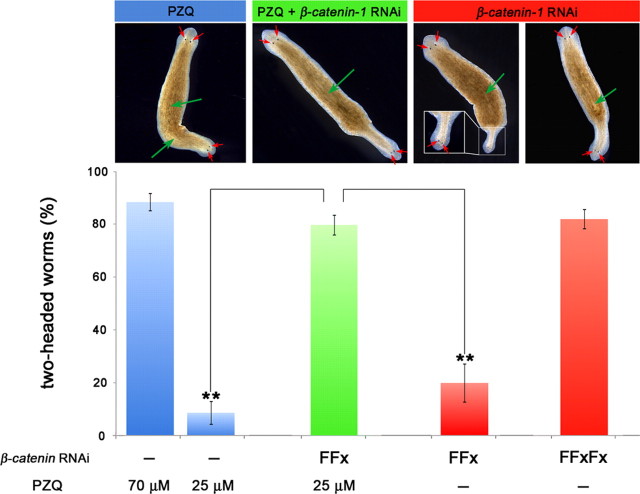
To probe the locus of action of PZQ, we compared two-headed phenotypes resulting from either pharmacological or genetic treatments. Both PZQ exposure and βcatenin-1 RNAi yielded two-headed animals with high penetrance (∼90%) from regenerating trunk fragments (Fig. 5A). For PZQ, this occurred over a single regeneration cycle and reflected a rapid and complete remodeling of the entire anterior–posterior (AP) axis. This entailed duplication of the pharynx and anteriorization of gut structures within the regenerating trunk, in addition to regeneration of the dual, integrated CNS and head structures from the blastema (Fig. 5B). For βcatenin-1(RNAi) animals, the penetrance of the bipolar phenotype in the population increased more gradually over time, as knockdown was effected over several feeding/regeneration cycles (Fig. 5A). To assess whether these PZQ and genetic (βcatenin-1 RNAi) pathways to bipolarity were independent, we performed an enhancement screen using a low dose of PZQ and a subpenetrant RNAi feeding cycle, either alone or in combination (Fig. 6). Low-dose PZQ or suboptimal βcatenin-1 RNAi produced a low percentage of two-headed worms after regeneration (9 ± 5% and 20 ± 7%, respectively; Fig. 6). However, in combination, the same treatments produced many bipolar worms (80 ± 4%; Fig. 6), a proportion ∼2.8-fold larger than simple additivity of bipolar percentages from the individual treatments. This synergism between PZQ and βcatenin-1 RNAi treatments implied mechanistic convergence in their actions. We conclude PZQ acts rapidly via inhibitory interactions with Wnt signaling events that control AP polarity through βcatenin-1 (Fig. 6), but not as a direct inhibitor of βcatenin-1 itself owing to the phenotypic divergence between the different bipolarity inducing treatments (Fig. 5B).
Figure 5.
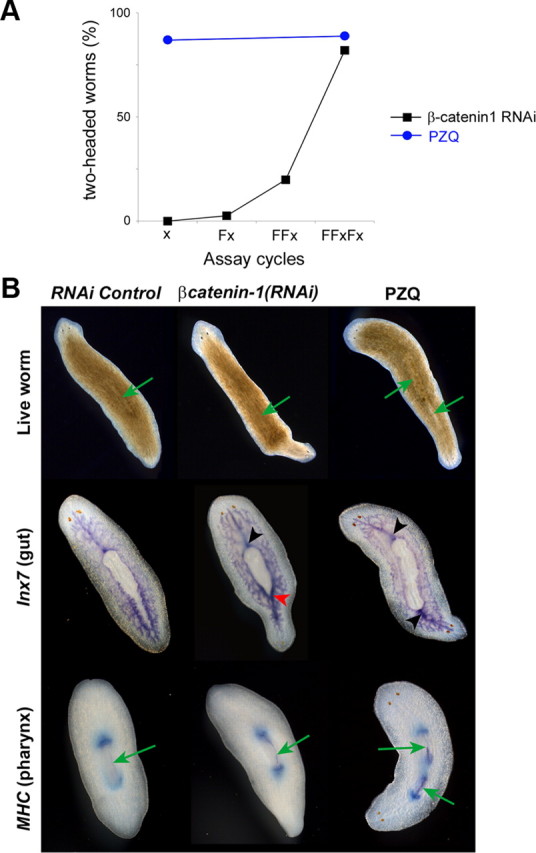
Comparison of genetic and pharmacological routes to bipolarity. A, Number of two-headed worms produced by pharmacological (70 μm PZQ, 24 h) or knockdown methods [(Dj-βcatenin-1(RNAi)] at indicated number of feeding (F) and regeneration (x) cycles from a representative experiment. B, Images of live worms (top) and in situ hybridization staining of gut (Inx7, middle) and pharyngeal (MHC, bottom row) markers in control (control RNAi, left) and PZQ-treated (70 μm for 24 h; imaged after 7 d; right) regenerating worms and worms subject to RNAi of βcatenin-1 (middle, 7 d). In βcatenin-1(RNAi) worms, the bipolar conversion occurred more slowly than with PZQ exposure. Newly anteriorized structures appeared to emerge from the posterior regenerative blastema without immediate remodeling of tissues within the trunk fragment. In situ hybridization of gut and pharyngeal marker after 14 d did not show clear evidence of full duplication unlike the effects elicited by PZQ. Rather, AP remodeling occurred over time in βcatenin-1(RNAi) worms subsequent to the emergence of head structures, in a manner reminiscent of intercalation events as seen in grafting experiments. Green arrows indicate the pharynx in live worms (top). Black arrowhead highlights a typical single anterior gut branch connecting with the pharynx. Red arrowhead indicates a gut organization intermediate between the normal anterior and posterior morphology.
Figure 6.
PZQ and βcatenin-1 knockdown act synergistically to anteriorize regeneration. Analysis of the interaction between drug (PZQ) and genetic [βcatenin-1(RNAi)] routes to bipolarity. Graph details the percentage of two-headed worms resulting from treatments with high- (70 μm) or low-dose PZQ (25 μm for 24 h, blue) compared with optimal (multiple feeding and regeneration cycles) and suboptimal RNAi of βcatenin-1 (two feedings and single regeneration cycle, red). Treatment of worms subject to suboptimal βcatenin-1(RNAi) with low-dose PZQ is shown in green. Images show phenotypes associated from drug treatments (blue), βcatenin-1 knockdown (red), and dual treatments (green). The two images for βcatenin-1(RNAi) are representative of results from the suboptimal (left) and optimal (right) RNAi protocol.
An alternative to analysis of dual anteriorizing cues is evaluation of antagonism between PZQ and posteriorizing signals. The intractability of transgenic methods in planarians precluded a gain of function approach (e.g., βcatenin-1 overexpression). Rather such analyses must be realized via RNAi of inhibitors of posteriorization circuits to potentiate “tail” signaling indirectly. In the context of Wnt signaling, APC—a physiological inhibitor of βcatenin stability—provides such a target. APC RNAi yielded two-tailed animals regenerating from trunk fragments (Fig. 7A), consistent with the logic that knockdown of APC elevates βcatenin-1 (Gurley et al., 2008). The two-tailed worms displayed impaired movement, and owing to the lack of CNS coordination of the feeding response, were viable for only ∼1 month. Cotreatment of APC RNAi worms with PZQ (70 μm for 24 h) resulted in a high proportion of two-tailed worms (94 ± 4% two-tailed worms, n = 3, Fig. 7A). This contrasted with PZQ exposure in Smed-six1 RNAi worms, or naive worms which produced a high proportion of two-headed worms under identical conditions (Fig. 7A). Lengthening the duration of PZQ exposure to 2 or even 3 d failed to inhibit the two-tailed phenotype observed in APC(RNAi) worms (Fig. 7A). These data suggest APC impacts anterior–posterior patterning downstream of the target of PZQ, as PZQ treatment is unable to overcome the effects of APC RNAi.
Figure 7.
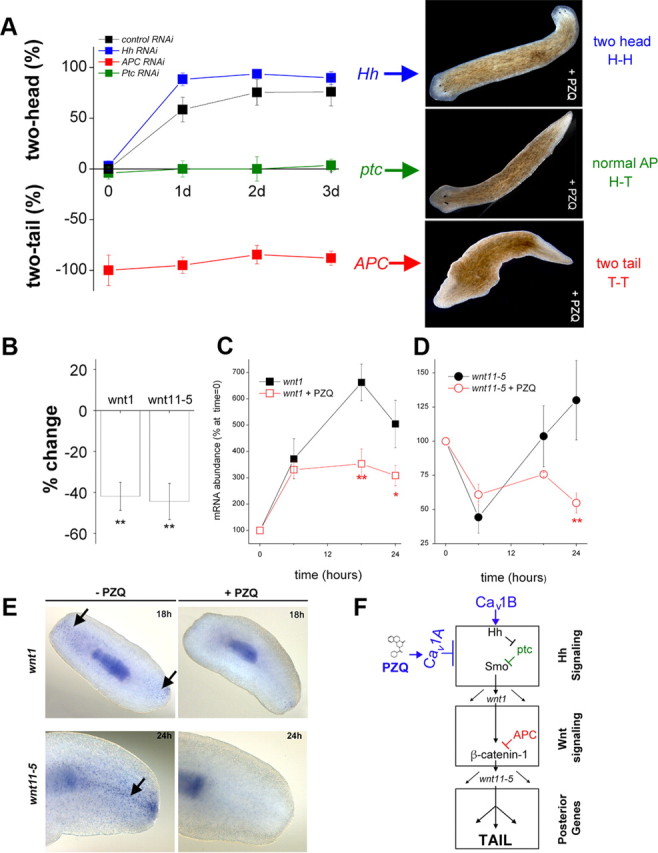
Inhibitory interaction of PZQ-evoked Ca2+ influx with Hh/Wnt signaling pathway. A, Left, Chemical genetic screen of PZQ efficacy in different cohorts of RNAi worms, including negative RNAi control (black), Hh (blue), APC (red), and Ptc (green) RNAi. The duration of PZQ exposure (90 μm) is shown on the abscissa, and the resulting bipolarity (two-headed, normal, two-tailed) is shown on the ordinate. Right, Representative images of dominant phenotype for Hh, Ptc, and APC RNAi worms exposed to PZQ. B, qPCR data of changes in wnt1 and wnt11-5 levels in regenerating trunk fragments exposed to PZQ (90 μm, 24 h) relative to untreated controls. C, qPCR analysis of wnt1 mRNA levels in the posterior blastema at indicated times after amputation (at t = 0) in the absence (black) and presence of PZQ (red squares, 90 μm). Asterisks indicate probability of similarity at p < 0.05 (*) and p < 0.01 (**). D, Similar qPCR analysis for wnt11-5 levels. E, In situ hybridization of wnt1 and wnt11-5 (arrowed) in the absence and presence of PZQ (90 μm) in regenerating trunk fragments at indicated times. F, Schematic of signaling modules involved in anterior–posterior specification. At least two distinct signal transduction pathways—Hedgehog (top) and Wnt signaling (middle) modules—control AP specification during regeneration as evidenced by RNAi of individual components of each module. These modules culminate to impact levels of βcatenin-1, which regulate a posterior fate circuit. Our data demonstrate an interaction of PZQ-evoked Ca2+ influx via Cav1A with Hh/Wnt signaling (top), localized upstream to APC within the Hh signaling module. Cav1B likely inhibits the trafficking/release of Hh from neurons (see Discussion). GenBank accession numbers: APC, HQ738520; βcatenin-1, HQ738521; wnt11-5, HQ738522; wnt1, HQ738523.
Recent experiments have implicated Hh signaling as an upstream transcriptional regulator of Wnt expression in planarians (Rink et al., 2009; Yazawa et al., 2009). If PZQ acts upstream of canonical Wnt signaling events, does PZQ-evoked Ca2+ entry impact Hh signaling? To test this, we applied similar logic (knockdown of an inhibitor of posteriorization) to examine the effects of knockdown of ptc, an endogenous inhibitor of the Hh signaling module. Knockdown of Dj-ptc posteriorized regeneration (35 ± 3%, worms with two-tails or inhibited head), a low penetrance compared with APC RNAi but consistent with modulation of an upstream modulator (Yazawa et al., 2009). Different from results with APC RNAi, PZQ treatment of ptc(RNAi) worms blocked the formation of two-tailed worms (Fig. 7A). In contrast, knockdown of Hh, the physiological ligand and upstream component of ptc, resulted in a small percentage of two-headed worms (3.1 ± 0.7%, n = 3 trials), and treatment of Hh RNAi worms with PZQ mimicked results seen with control cohorts at longer time periods (Fig. 7A). The ability of PZQ to suppress bipolar tail formation in ptc(RNAi) worms supports a modulation of Hh signaling components by voltage-operated Ca2+ influx, upstream of canonical Wnt signaling (Fig. 7F).
If the logic that PZQ impacts Hh signaling is correct, then PZQ should modulate the levels of mediators that serve as the output of Hh signaling events. Hh signaling regulates the transcription of Wnt genes—notably wnt1, a wound-induced Wnt that activates βcatenin-1 during tail regeneration, and the downstream effector wnt11-5 (Fig. 7F) (Adell et al., 2009; Petersen and Reddien, 2009; Gurley et al., 2010). Crucially, loss of Hh signaling activity (via RNAi) inhibits wnt1 expression (Rink et al., 2009; Yazawa et al., 2009). Therefore, if PZQ inhibits Hh signaling, wnt1 expression should be reduced.
Consequently, we performed qPCR analysis of wnt1 in trunk and posterior blastema samples. Relative to control samples, regenerating trunk samples exposed to PZQ showed a decrease in wnt1 and wnt11-5 levels (Fig. 7B). Resolution of the time course of changes in wnt1 and wnt11-5 in the posterior blastema after amputation revealed that PZQ exposure attenuated the early wound-induced increase in wnt1 expression (∼12–18 h, Fig. 7C) that occurs before determination of polarity (i.e., preceding changes in wnt11-5, Fig. 7D). Therefore, PZQ is impacting wnt levels at a time frame causative, rather than consequent, of polarity specification (Petersen and Reddien, 2009). Further, in situ hybridization patterns of wnt1 and wnt11-5 during trunk fragment regeneration were compared to samples treated with PZQ. wnt1 and wnt11-5 expression was reduced by PZQ with similar kinetics to qPCR results (Fig. 7E). As PZQ-evoked changes in wnt1 intensity precede changes in wnt11-5, these data are consistent with the conclusion that PZQ is impacting early events. Finally, as Wnt/βcatenin signaling maintains AP axis polarity in intact worms (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008), we were also interested in determining whether PZQ modulated Wnt expression during normal body homeostasis. In intact worms, wnt1 and wnt11-5 expression was also decreased by PZQ exposure (data not shown). Therefore, both qPCR and in situ hybridization approaches demonstrated that PZQ decreased wnt1 expression, consistent with early inhibition of the Hh signaling module by PZQ.
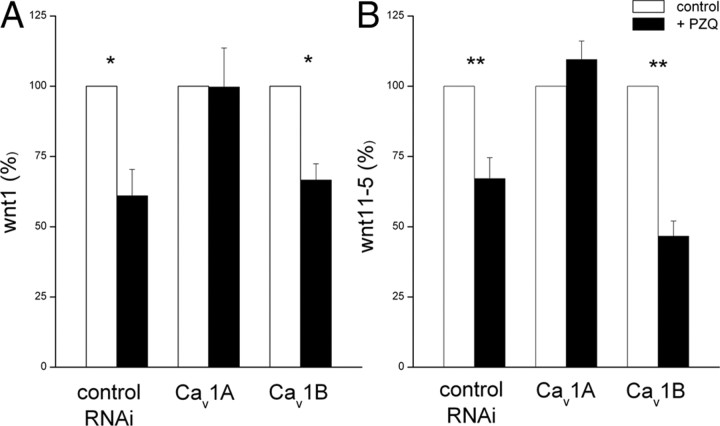
We have previously shown that PZQ-evoked Ca2+ entry is inhibited by RNAi of Cav1A (Fig. 4D). If PZQ-evoked changes in wnt1 were also dependent on Ca2+ entry via Cav1A, then RNAi of Cav1A channels should attenuate the PZQ-evoked decrease in wnt1. Therefore, we performed qPCR analysis of both wnt1 and wnt11-5 in Cav1A(RNAi) and Cav1B(RNAi) worms, compared to RNAi controls [Smed-six-1(RNAi)]. Whereas PZQ treatment resulted in decreased levels of wnt1 and wnt11-5 in the control RNAi cohort, knockdown of Cav1A prevented any PZQ-evoked decrease in either wnt1 or wnt11-5 (Fig. 8). In contrast, PZQ treatment was still effective at causing a decrease in wnt1 and wnt11-5 in Cav1B(RNAi) worms. We conclude that RNAi of Cav1A, but not Cav1B, prevents PZQ-evoked Ca2+ entry and PZQ-evoked inhibition of wnt1 and wnt11-5.
Figure 8.
RNAi of Cav1A blocks PZQ-evoked changes in wnt mediators. qPCR analysis of wnt1 (A) and wnt11-5 (B) levels in posterior blastema samples from different cohorts of RNAi worms [Smed-six-1 (RNAi) worms, Cav1A and Cav1B (RNAi) worms] isolated after 24 h of regeneration in the absence (open) or presence (solid) of PZQ (70 μm). Difference from controls is indicated at p < 0.01 (**) and p < 0.05 (*).
Nervous system expression of Cav channels and Hh signaling machinery
The conclusion that PZQ inhibited Hh signaling implied a spatial relationship between Cav channels and the Hh signaling machinery. In situ analysis of Cav1A and Cav1B localization in intact worms revealed Cav1A was predominantly expressed in brain as well as pharynx, whereas Cav1B was confined to the CNS with expression in the brain and ventral nerve cords (Fig. 9A). Expression of Cav1 isoforms in the nervous system was also evident from analysis of Cav1 staining in regenerating trunk fragments. Cav1A and Cav1B were detected in regenerating brain tissue at the anterior blastema by 18 h after amputation (Fig. 9B), at the same time point as the earliest known anterior markers. These data do not preclude a later role of Ca2+ channels in the development of anterior structures (Beane et al., 2011) in addition to their earlier role in modulating Hh signaling after injury (Figs. 7, 8). This neuronal localization of both Cav1 isoforms is therefore consistent with the cell physiological data resolving PZQ-evoked Ca2+ influx in a neuronally derived cell population (Fig. 4). Crucially, an antisense probe against Hh was also found to stain the CNS (Fig. 9A) (Rink et al., 2009; Yazawa et al., 2009). Therefore the demonstration that Cav1A, Cav1B, and Hh are all expressed within the planarian nervous system further supports the regulatory interplay between voltage-operated Ca2+ entry and Hh signaling.
Figure 9.
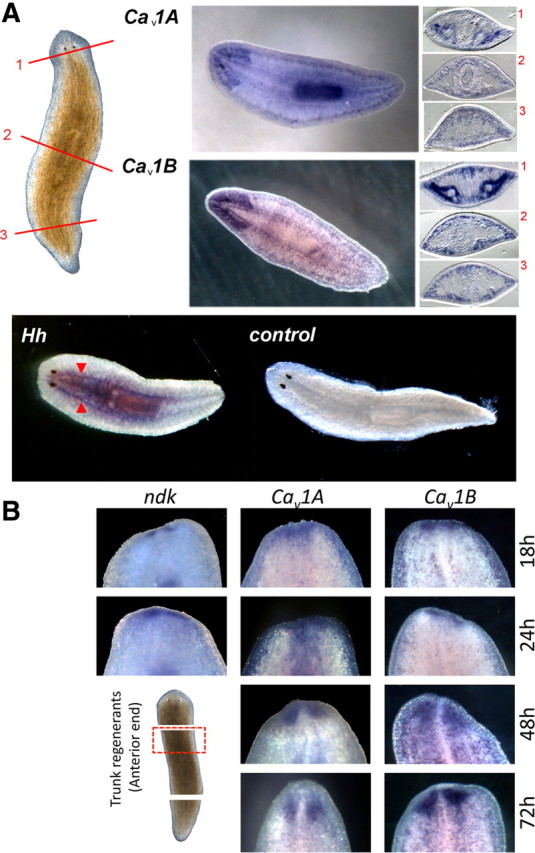
Localization of Cav1 channels with Hh in the planarian nervous system. A, Top, Cav1A and Cav1B localization in whole-mount and sectioned samples. Brightfield image of intact planarian shows location of cross sections: anterior (1, top), pharyngeal (2, middle), and postpharyngeal (3, bottom). Sections are orientated with the ventral side at the bottom. Bottom, Whole-mount in situ hybridization of Hh showing localization of mRNA within the ventral nerve cords (red arrows) compared with controls. B, Whole-mount in situ hybridization of ndk (a brain marker), Cav1A, and Cav1B in the anterior blastema during trunk fragment regeneration at the indicated times.
Discussion
The planarian model holds great appeal for neuroscientists interested in studying the wholesale regeneration of a nervous system (Cebrià, 2007). The experimental system is simple and increasingly tractable (Newmark and Sánchez Alvarado, 2002; Reddien et al., 2005; Robb et al., 2008; Chan and Marchant, 2011), yet the endpoint is complex in terms of structure (Morita and Best, 1966; Mineta et al., 2003), gene-expression profiles (Cebrià et al., 2002), neurotransmitter diversity (Ribeiro et al., 2005; Collins et al., 2010), and potential for behavioral insight (Kitamura et al., 2003; Raffa et al., 2003).
Our interest relates to the use of this system to identify small molecules efficacious in vivo at modulating stem cell behavior. Indeed, the utility of invertebrate models for studying conserved mechanisms of stem cell regulation is increasingly appreciated (Brand and Livesey, 2011). We previously discovered that agents disrupting cellular Ca2+ homeostasis anteriorized regeneration, with PZQ (a drug of unknown mechanism of action) being exceedingly effective at producing worms with dual, integrated nervous systems (Nogi et al., 2009). Here, by identifying a family of planarian Cavα subunits, we provide chemical genetic data that PZQ activates a specific Cav1 isoform (Cav1A) to miscue regeneration by inhibiting Hh signaling. Knockdown of Cav1A prevented PZQ-evoked anteriorization (Fig. 3), PZQ-evoked Ca2+ entry (Fig. 4), and the PZQ-evoked decrease in wnt1 (Fig. 8), the output of neuronally derived Hh signaling (Rink et al., 2009; Yazawa et al., 2009). The significance of these data is twofold. First, they establish a unique interplay between specific Cav channels isoforms and Hedgehog signaling in the control of stem cell differentiation that on the basis of recently published data appears also relevant to vertebrates. Second, they provide in vivo support for PZQ efficacy being dependent on neuronal Cav isoforms of discrete subunit composition.
Cav channels and neuronal stem cell differentiation
Unbiased in vitro screens have uncovered new, and existing, activators of voltage-operated Ca2+ influx that regulate the differentiation and proliferation of various multipotent stem cells (Schneider et al., 2008; Wang et al., 2009). In the context of neurogenesis, application of Cav1 agonists to proliferating neuronal stem cells in culture enhances neuronal fate (Deisseroth et al., 2004; D'Ascenzo et al., 2006; Diamandis et al., 2007; Schneider et al., 2008). By extending such findings to a model suited for studying pluripotent cells in vivo, our data demonstrate a novel role for voltage-operated Ca2+ entry in regulating wholesale nervous system regeneration. The observed anteriorization of regenerative responses by PZQ-evoked Ca2+ entry is reminiscent of proposed roles for voltage-operated Ca2+ entry in neurogenesis and neural induction in vertebrate models (Leclerc et al., 1997; Webb and Miller, 2003; Deisseroth et al., 2004; Whitaker, 2006), and the consequent Ca2+-dependent inhibition of Hh signals supportive of an emerging literature showing reciprocal interplay between morphogens and Cav1 channel activity. Examples include Wnts [which can both activate Cav1 channels (Panáková et al., 2010) and serve as transcriptional effectors of neuronal voltage-operated Ca2+ entry (Alvania et al., 2006)], as well as noggin and FGF signals that regulate Cav activity during neural induction (Moreau et al., 2008; Lee et al., 2009). Therefore, our findings underscore a fundamental and evolutionary conserved role for voltage-operated Ca2+ influx controlling stem cell differentiation, and the utility of basic invertebrate models for studying neural stem cell biology (Brand and Livesey, 2011).
Characterization of planarian Cav channels revealed a surprisingly diverse family of Cavα subunits compared to better studied invertebrate models (Fig. 1B). This diversity is likely a general characteristic of flatworms: first, four HVA Cavα subunits are predicted in the Schistosoma mansoni genome (Kohn et al., 2001b; Berriman et al., 2009); second, each of the subunits described here has a clearly identifiable homolog within the Schmidtea mediterranea genome (Robb et al., 2008). The Cav channel diversity was functionally significant, rather than reflecting redundant gene duplication, as the two Cav1 family isoforms differentially regulated regenerative outcomes. RNAi of Cav1A blocked bipolar regeneration, whereas Cav1B RNAi increased the number of two-headed regenerants whether in the absence of drug or from PZQ or K+ exposure (Fig. 3). The surprising different roles for Ca2+ influx through Cav1 isoforms (Cav1A vs Cav1B) explains previous observations that established activators and inhibitors of Cavs can both yield bipolar worms, albeit with different penetrances (Nogi et al., 2009). Differential selectivity of pharmacological agents for Cav1 isoforms with opposing roles likely underpins these observations, and highlights the possibility that small molecule neurogenics can encompass compounds acting as either selective activators (Cav1A) or inhibitors (Cav1B) of Cav channels in vivo. Data suggesting differences in pharmacophore binding profiles to specific neuronal Cav1 channel isoforms is significant in this regard for future development of Cav1 subtype-specific neurogenic compounds (Sinnegger-Brauns et al., 2009).
Finally, pharmacological profiling of the neuronal flatworm Cavα subunits is extremely important in the context of defining the molecular site of action of PZQ. PZQ is the mainstay therapeutic for combating schistosomiasis, a parasitic flatworm disorder that infects >200 million people worldwide (Day et al., 1992; Cioli and Pica-Mattoccia, 2003; Caffrey, 2007). However, the relevant in vivo target(s) of this clinically important drug have remained undefined for decades, hampering rational design of new antischistosomal agents that target the same vulnerable pathways in the parasite. Our data are significant in this regard by first, narrowing PZQ efficacy to Ca2+ channel complexes of specific Cavα composition (Cav1A), and second, suggesting a revised focus on a neuronal rather than a muscular site(s) of action of PZQ.
Ca2+ regulation of Hh signaling
During planarian regeneration, Hh is the most upstream activator of neoblast differentiative responses following wounding (Petersen and Reddien, 2009; Yazawa et al., 2009). Hh-evoked changes in wnt1 and wnt11-5 expression occur in a stem cell-independent manner (Petersen and Reddien, 2009; Yazawa et al., 2009), such that initial wnt1 and wnt11-5 expression occurs even in irradiated worms (Petersen and Reddien, 2009). In vertebrates also, the Hedgehog system acts as a paracrine regulator of stem cell behavior in normal proliferative scenarios, and as an aberrant pathway in cancer (Varjosalo and Taipale, 2007; Yauch et al., 2008; Traiffort et al., 2010). In the CNS, Shh release has been proposed to maintain an adult neurogenic niche and regulate the proliferation of neuronal precursors in different brain regions (Traiffort et al., 2010). Our demonstration of regulation of Hh signaling by PZQ-evoked voltage-operated Ca2+ entry establishes a largely unrecognized functional interaction between two signaling systems that individually are highly competent at nuclear reprogramming and crucial for CNS development (Whitaker, 2006; Greer and Greenberg, 2008; Traiffort et al., 2010). Is the regulatory interplay demonstrated between these signaling pathways in planaria conserved in vertebrate systems? Although PZQ efficacy is unique to the flatworm system (as is ideal for a selective therapeutic), the principle of Cav regulation of Hedgehog signaling appears conserved on the basis of two recently published studies.
First, cytoplasmic Ca2+ signals have been shown to act as downstream effectors of Sonic hedgehog (Shh) signaling in Xenopus embryonic neurons (Belgacem and Borodinsky, 2011). By imaging the neural tube of developing frog embryos, Ca2+ spike activity trended with the Shh gradient crucial for dorsoventral patterning of the spinal cord. Shh failed to increase Ca2+ spike activity when Cavs were blocked (Belgacem and Borodinsky, 2011). Such data place Ca2+ as a downstream effector of Hh activity, a coupling that may also exist in stem cells and other cell types (Osawa et al., 2006; Heo et al., 2007). This observation is entirely consistent with our data showing Cav1A regulation of the Hh signaling module at, or downstream of, Ptc (Fig. 7). Nonetheless, the specific Hh signaling components that are regulated by Ca2+, and the customization of this regulation between different systems, remain to be elucidated. However, the demonstration that PZQ activates a Cav1 channel to inhibit Hh transcriptional effects is highly reminiscent of the paradigm that Cav1 silencing is needed to support activity-dependent gene expression in certain vertebrate neurons (Chang and Berg, 2001).
Second, recent evidence from various neuronal cell types has shown that Shh is sorted to the regulatory secretory pathway in axons and is available for release by depolarization (Beug et al., 2011). The demonstration of a link between Cav channels and Hh signaling suggests an obvious connection between neuronal activity, Cav activation, and synaptic Hh secretion that may be important for both maintaining progenitors and regulating their proliferation. In planaria, Cav1B likely fulfills this role by regulating neuronal Hh release to ensure normal posterior patterning. As Hh is predominantly localized to the planarian nervous system, it is likely that neuronal damage on amputation releases Hh to the surrounding environment effecting the wnt1 wound response. Continued delivery of Hh to posterior wounds through the ventral nerve cords has been suggested (Yazawa et al., 2009) to stabilize posterior-specification mechanisms dependent on βcatenin-1 and wnt11-5. Loss of Cav1B function would therefore increase anteriorization outcomes (Fig. 3) by repression of depolarization-evoked Hh release.
In summary, our data establish a unique regulatory interplay between specific Cav1 isoforms and Hh signals that control planarian nervous system regeneration in vivo. This is demonstrated by activation of Cav1A by the antischistosomal drug PZQ, casting new light on the relevant in vivo mechanism of action of this important clinical agent.
Footnotes
References
- Adell T, Salò E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- Alvania RS, Chen X, Ginty DD. Calcium signals control Wnt-dependent dendrite growth. Neuron. 2006;50:813–815. doi: 10.1016/j.neuron.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Angelucci F, Basso A, Bellelli A, Brunori M, Pica Mattoccia L, Valle C. The anti-schistosomal drug praziquantel is an adenosine antagonist. Parasitology. 2007;134:1215–1221. doi: 10.1017/S0031182007002600. [DOI] [PubMed] [Google Scholar]
- Beane WS, Morokuma J, Adams DS, Levin M. A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem Biol. 2011;18:77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc Natl Acad Sci U S A. 2011;108:4482–4487. doi: 10.1073/pnas.1018217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JB, Noel J. Complex synaptic configurations in planarian brain. Science. 1969;164:1070–1071. doi: 10.1126/science.164.3883.1070. [DOI] [PubMed] [Google Scholar]
- Beug ST, Parks RJ, McBride HM, Wallace VA. Processing-dependent trafficking of Sonic hedgehog to the regulatory secretory pathway in neurons. Mol Cell Neurosci. 2011;46:583–596. doi: 10.1016/j.mcn.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. doi: 10.1016/j.neuron.2011.05.016. [DOI] [PubMed] [Google Scholar]
- Caffrey CR. Chemotherapy of schistosomiasis: present and future. Curr Opin Chem Biol. 2007;11:433–439. doi: 10.1016/j.cbpa.2007.05.031. [DOI] [PubMed] [Google Scholar]
- Cebrià F. Regenerating the central nervous system: how easy for planarians! Dev Genes Evol. 2007;217:733–748. doi: 10.1007/s00427-007-0188-6. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Kudome T, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Agata K. The expression of neural-specific genes reveals the structural and molecular complexity of the planarian central nervous system. Mech Dev. 2002;116:199–204. doi: 10.1016/s0925-4773(02)00134-x. [DOI] [PubMed] [Google Scholar]
- Chan JD, Marchant JS. Pharmacological and functional genetic assays to manipulate regeneration of the planarian Dugesia japonica. J Vis Exp. 2011:e3058. doi: 10.3791/3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KT, Berg DK. Voltage-gated channels block nicotinic regulation of CREB phosphorylation and gene expression in neurons. Neuron. 2001;32:855–865. doi: 10.1016/s0896-6273(01)00516-5. [DOI] [PubMed] [Google Scholar]
- Cioli D, Pica-Mattoccia L. Praziquantel. Parasitol Res. 2003;90:S3–S9. doi: 10.1007/s00436-002-0751-z. [DOI] [PubMed] [Google Scholar]
- Collins JJ, 3rd, Hou X, Romanova EV, Lambrus BG, Miller CM, Saberi A, Sweedler JV, Newmark PA. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ascenzo M, Piacentini R, Casalbore P, Budoni M, Pallini R, Azzena GB, Grassi C. Role of L-type Ca2+ channels in neuronal stem/progenitor differentiation. Eur J Neurosci. 2006;23:935–944. doi: 10.1111/j.1460-9568.2006.04628.x. [DOI] [PubMed] [Google Scholar]
- Day TA, Bennett JL, Pax RA. Praziquantel: the enigmatic antiparasitic. Parasitol Today. 1992;8:342–344. doi: 10.1016/0169-4758(92)90070-i. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Singla S, Toda H, Monje M, Palmer TD, Malenka RC. Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron. 2004;42:535–552. doi: 10.1016/s0896-6273(04)00266-1. [DOI] [PubMed] [Google Scholar]
- Diamandis P, Wildenhain J, Clarke ID, Sacher AG, Graham J, Bellows DS, Ling EKM, Ward RJ, Jamieson LG, Tyers M, Dirks PB. Chemical genetics reveals a complex functional ground state of neural stem cells. Nat Chem Biol. 2007;3:268–273. doi: 10.1038/nchembio873. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, Sánchez Alvarado A, Kim JK, Rajewsky N. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekar M, Salunkhe AM, Mallia AK, He YX, Kalyanasundaram R. Praziquantel affects the regulatory myosin light chain of Schistosoma mansoni. Antimicrob Agents Chemother. 2009;53:1054–1060. doi: 10.1128/AAC.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JS, Lee MY, Han HJ. Sonic hedgehog stimulates mouse embryonic stem cell proliferation by cooperation of Ca2+/protein kinase C and epidermal growth factor receptor as well as Gli1 activation. Stem Cells. 2007;25:3069–3080. doi: 10.1634/stemcells.2007-0550. [DOI] [PubMed] [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- King GF. Modulation of insect Cav channels by peptidic spider toxins. Toxicon. 2007;49:513–530. doi: 10.1016/j.toxicon.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Inden M, Sanada H, Takata K, Taniguchi T, Shimohama S, Orii H, Mochii M, Agata K, Watanabe K. Inhibitory effects of antiparkinsonian drugs and caspase inhibitors in a parkinsonian flatworm model. J Pharmacol Sci. 2003;92:137–142. doi: 10.1254/jphs.92.137. [DOI] [PubMed] [Google Scholar]
- Kohn AB, Anderson PAV, Roberts-Misterly JM, Greenberg RM. Schistosome calcium channel β subunits. Unusual modulatory effects and potential role in the action of the antischistosomal drug praziquantel. J Biol Chem. 2001a;276:36873–36876. doi: 10.1074/jbc.C100273200. [DOI] [PubMed] [Google Scholar]
- Kohn AB, Lea J, Roberts-Misterly JM, Anderson PAV, Greenberg RM. Structure of three high voltage-activated calcium channel α1 subunits from Schistosoma mansoni. Parasitology. 2001b;123:489–497. doi: 10.1017/s0031182001008691. [DOI] [PubMed] [Google Scholar]
- Kwok TCY, Hui K, Kostelecki W, Ricker N, Selman G, Feng Z-P, Roy PJ. A genetic screen for dihydropyridine (DHP)-resistant worms reveals new residues required for DHP-blockage of mammalian calcium channels. PLoS Genet. 2008;4:e1000067. doi: 10.1371/journal.pgen.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc C, Daguzan C, Nicolas M-T, Chabret C, Duprat A-M, Moreau M. L-type calcium channel activation controls the in vivo transduction of the neuralizing signal in the amphibian embryos. Mech Dev. 1997;64:105–110. doi: 10.1016/s0925-4773(97)00054-3. [DOI] [PubMed] [Google Scholar]
- Lee KW, Moreau M, Néant I, Bibonne A, Leclerc C. FGF-activated calcium channels control neural gene expression in Xenopus. Biochim Biophys Acta. 2009;1793:1033–1040. doi: 10.1016/j.bbamcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Mannini L, Rossi L, Deri P, Gremigni V, Salvetti A, Saló E, Batistoni R. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol. 2004;269:346–359. doi: 10.1016/j.ydbio.2004.01.042. [DOI] [PubMed] [Google Scholar]
- Marchant JS, Beecroft MD, Riley AM, Jenkins DJ, Marwood RD, Taylor CW, Potter BVL. Disaccharide polyphosphates based upon adenophostin A activate hepatic D-myo-inositol 1,4,5-trisphosphate receptors. Biochemistry. 1997;36:12780–12790. doi: 10.1021/bi971397v. [DOI] [PubMed] [Google Scholar]
- McTigue MA, Williams DR, Tainer JA. Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J Mol Biol. 1995;246:21–27. doi: 10.1006/jmbi.1994.0061. [DOI] [PubMed] [Google Scholar]
- Mineta K, Nakazawa M, Cebria F, Ikeo K, Agata K, Gojobori T. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci U S A. 2003;100:7666–7671. doi: 10.1073/pnas.1332513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Néant I, Webb SE, Miller AL, Leclerc C. Calcium signalling during neural induction in Xenopus laevis embryos. Philos Trans R Soc Lond B Biol Sci. 2008;363:1371–1375. doi: 10.1098/rstb.2007.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Best JB. Electron microscopic studies of planaria. III. Some observations on the fine structure of planarian nervous tissue. J Exp Zool. 1966;161:391–411. doi: 10.1002/jez.1401610307. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Nogi T, Zhang D, Chan JD, Marchant JS. A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel β subunits: subversion of flatworm regenerative polarity. PLoS Negl Trop Dis. 2009;3:e464. doi: 10.1371/journal.pntd.0000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novozhilova E, Kimber MJ, Qian H, McVeigh P, Robertson AP, Zamanian M, Maule AG, Day TA. FMRFamide-like peptides (FLPs) enhance voltage-gated calcium currents to elicit muscle contraction in the human parasite Schistosoma mansoni. PLoS Negl Trop Dis. 2010;4:e790. doi: 10.1371/journal.pntd.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa H, Ohnishi H, Takano K, Noguti T, Mashima H, Hoshino H, Kita H, Sato K, Matsui H, Sugano K. Sonic hedgehog stimulates the proliferation of rat gastric mucosal cells through ERK activation by elevating intracellular calcium concentration. BBRC. 2006;344:680–687. doi: 10.1016/j.bbrc.2006.03.188. [DOI] [PubMed] [Google Scholar]
- Panáková D, Werdich AA, MacRae CA. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel. Nature. 2010;466:874–878. doi: 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pax R, Bennett JL, Fetterer R. A benzodiazepine derivative and praziquantel: effects on musculature of Schistosoma mansoni and Schistosoma japonicum. Naunyn Schmiedebergs Arch Pharmacol. 1978;304:309–315. doi: 10.1007/BF00507974. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa RB, Stagliano GW, Umeda S. κ-opioid withdrawal in Planaria. Neurosci Lett. 2003;349:139–142. doi: 10.1016/s0304-3940(03)00814-0. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro P, El-Shehabi F, Patocka N. Classical transmitters and their receptors in flatworms. Parasitology. 2005;131:S19–S40. doi: 10.1017/S0031182005008565. [DOI] [PubMed] [Google Scholar]
- Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian hh signaling regulates regeneration polarity and links hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb SMC, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucl Acids Res. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Schneider JW, Gao Z, Li S, Farooqi M, Tang TS, Bezprozvanny I, Frantz DE, Hsieh J. Small-molecule activation of neuronal cell fate. Nat Chem Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- Sinnegger-Brauns MJ, Huber IG, Koschak A, Wild C, Obermair GJ, Einzinger U, Hoda JC, Sartori SB, Striessnig J. Expression and 1,4-dihydropyridine-binding properties of brain L-type calcium channel isoforms. Mol Pharmacol. 2009;75:407–414. doi: 10.1124/mol.108.049981. [DOI] [PubMed] [Google Scholar]
- Tallima H, El Ridi R. Praziquantel binds Schistosoma mansoni adult worm actin. Int J Antimicrob Agents. 2007;29:570–575. doi: 10.1016/j.ijantimicag.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Traiffort E, Angot E, Ruat M. Sonic Hedgehog signaling in the mammalian brain. J Neurochem. 2010;113:576–590. doi: 10.1111/j.1471-4159.2010.06642.x. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog signaling. J Cell Sci. 2007;120:3–6. doi: 10.1242/jcs.03309. [DOI] [PubMed] [Google Scholar]
- Wang W, Walker JR, Wang X, Tremblay MS, Lee JW, Wu X, Schultz PG. Identification of small-molecule inducers of pancreatic β-cell expansion. Proc Natl Acad Sci U S A. 2009;106:1427–1432. doi: 10.1073/pnas.0811848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Calcium signaling during early embryonic development. Nat Rev Mol Cell Biol. 2003;4:539–551. doi: 10.1038/nrm1149. [DOI] [PubMed] [Google Scholar]
- Whitaker M. Calcium at fertilization and in early development. Physiol Rev. 2006;86:25–88. doi: 10.1152/physrev.00023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest PM, Li Y, Olds GR, Bowen WD. Inhibition of phosphoinositide turnover by praziquantel in Schistosoma mansoni. J Parasitol. 1992;78:753–755. [PubMed] [Google Scholar]
- Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci U S A. 2009;106:22329–22334. doi: 10.1073/pnas.0907464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Ng S, Zhang M, Bouhours M, Wang Y, Wang M, Hung W, Aoyagi K, Melnik-Martinez K, Li M, Liu F, Schafer WR, Zhen M. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6:e55. doi: 10.1371/journal.pbio.0060055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Feng G, Ren D, Eberl DF, Hannan F, Dubald M, Hall LM. Cloning and characterization of a calcium channel alpha 1 subunit from Drosophila melanogaster with similarity to the rat brain type D isoform. J Neurosci. 1995;15:1132–1143. doi: 10.1523/JNEUROSCI.15-02-01132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]