Abstract
A high-throughput pipeline to engineer bacterial artificial chromosomes (BACs) expressing tagged genes of higher eukaryotes allows large-scale protein localization and interaction studies.
DNA engineering technology, allowing one to assemble DNA fragments like Lego pieces, has revolutionized biology. In this issue, Poser, Sarov and colleagues describe the BAC TransgeneOmics pipeline—a generic, high-throughput methodology that allows for the rapid generation of large DNA constructs, which will facilitate systematic characterization of protein function in mammals1.
Since its advent more than three decades ago2, recombinant DNA technology has completely changed our way of approaching biological questions and has provided invaluable tools for in-depth exploration and understanding of living systems. All DNA engineering methodologies are conceptually the same in that they aim to build new, artificial DNA molecules by stitching together pre-existing pieces. Two main technological branches have emerged: the classical restriction and ligation–based (copy and paste) strategies, and a set of advanced in vivo methods, referred to as ‘recombineering’3.
Classical copy-and-paste techniques are suitable for engineering relatively small and simple constructs that can comfortably accommodate entire genes from primitive genomes. These strategies, together with the use of generic DNA modules (for example, affinity tags), applied to yeast have yielded some of the most detailed and rich interactome4–6 and localization7 maps to date.
The classical techniques, however, are dependent on the availability of appropriate restriction sites and on the size of the fragments to be combined. This makes their implementation in higher organisms difficult, as those organisms tend to have longer genes with more complex structure and regulation and generally require larger, more sophisticated constructs that often require multiple cloning steps. And even though simpler organisms like yeast and bacteria make our wine, bread and cheese, we ultimately want to understand mice and men; recombineering now makes this possible.
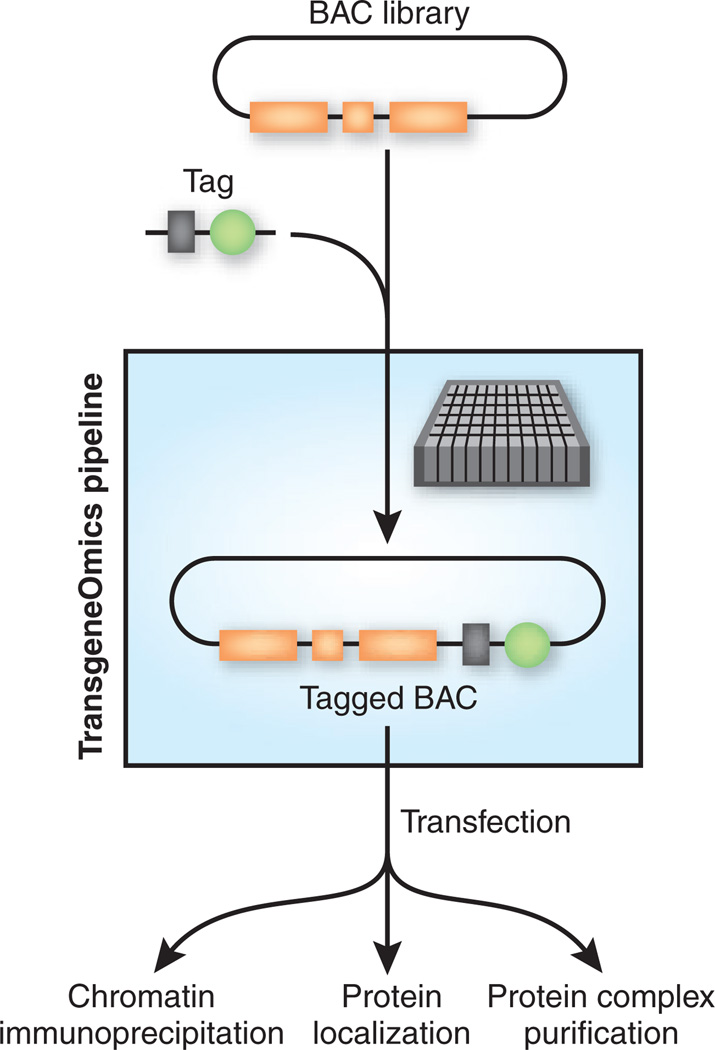
The generic pipeline that Poser, Sarov and colleagues describe—called BAC TransgeneOmics—allows for simple, rapid (and hence inexpensive) high-throughput generation of large DNA constructs, permitting the systematic exploration of interactomes in higher organisms (Fig. 1). The method uses a high-fidelity phage protein–based in vivo recombineering strategy8,9 for modifying BACs. BACs are the vectors of choice in metazoans as they are stable and can harbor very large DNA inserts that encompass complete transcription units together with their regulatory elements. This, in turn, allows a transgene to be expressed at near-physiological levels and largely preserves its in vivo regulation. High-quality BAC libraries compatible with the TransgeneOmics pipeline are readily available for several organisms.
Figure 1.
A schematic of the BAC TransgeneOmics pipeline. A BAC library is rapidly modified (a modular tag is added in this case). After introduction into cells, protein function can be interrogated in high throughput.
To validate their methodology, the authors used a modified version of the LAP tag10, a modular protein tag containing enhanced GFP (for protein localization and affinity purification) and S-peptide (for affinity purification), and successfully generated a large number of BAC transgenes. They placed particular emphasis on tailoring the system for liquid handling, thus making it more amenable for high-throughput studies. Indeed, using the BAC TransgeneOmics pipeline, complex constructs can be generated in less than a week. Creation of a whole-genome your favorite tag (YFT)-tagged mouse or human set is now feasible in a matter of months. As recombineering is essentially DNA context–independent, one can now conceive any construct architecture. The power of biochemistry (affinity purification) as well as genetics (dominant-negative, null or overexpression alleles) can be unleashed for large-scale protein characterization in higher eukaryotes. Moreover, as the endogenous regulation is effectively preserved, it now becomes possible to address questions with respect to specific cell types and tissues with relative ease.
Recombineering may represent a paradigm shift in our ways of manipulating DNA. And even though custom gene synthesis is now gaining momentum and becoming more accurate and affordable, recombineering will be important in functional genomics in mammalian systems over the next several years as it provides unparalleled flexibility, robustness and fidelity at a reasonable price.
Archimedes once said, “Give me a lever long enough and a fulcrum on which to place it, and I shall move the world.” With the recent advances in genome sequencing and the exponential increase of available genomes, we may already have the fulcrum. We now need the lever, and Poser, Sarov and colleagues have made a big leap in that direction. Simple, yet powerful and inexpensive techniques like the BAC TransgeneOmics pipeline should be easy to implement. Therefore, in the near future, interactome maps from higher eukaryotes will become available that will rival the quality and coverage of ones that we now have for simpler model organisms.
References
- 1.Poser I, et al. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen SN, Chang AC, Boyer HW, Helling RB. Proc. Natl. Acad. Sci. USA. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Court DL, Sawitzke JA, Thomason LC. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AC, et al. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 5.Krogan NJ, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 6.Collins SR, et al. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 7.Huh WK, et al. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 9.Sarov M, et al. Nat. Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 10.Cheeseman IM, Desai A. Sci. STKE. 2005 doi: 10.1126/stke.2662005pl1. pl1. [DOI] [PubMed] [Google Scholar]