Abstract
To date, cross-species comparisons of genetic interactomes have been restricted to small or functionally related gene sets, limiting our ability to infer evolutionary trends. To facilitate a more comprehensive analysis, we constructed a genome-scale epistasis map (E-MAP) for the fission yeast Schizosaccharomyces pombe, providing phenotypic signatures for ~60% of the non-essential genome. Using these signatures, we generated a catalogue of 297 functional modules, and assigned function to 144 previously uncharacterised genes, including mRNA splicing and DNA damage checkpoint factors. Comparison with an integrated genetic interactome from the budding yeast Saccharomyces cerevisiae revealed a hierarchical model for the evolution of genetic interactions, with conservation highest within protein complexes, lower within biological processes, and lowest between distinct biological processes. Despite the large evolutionary distance and extensive rewiring of individual interactions, both networks retain conserved features and display similar levels of functional cross-talk between biological processes, suggesting general design principles of genetic interactomes.
Introduction
Epistasis is a biological phenomenon where the phenotype of one gene is affected by the presence or absence of another gene. Such relationships are broadly termed genetic (or epistatic) interactions (GIs). Unlike protein-protein interactions (PPIs), which are limited to gene products that interact physically, GIs report on functional relationships, and reveal how groups of proteins and complexes work together to carry out higher level biological functions and describe the cross-talk between pathways and processes (Beltrao et al., 2010). Thus, GI networks are a natural complement to PPI maps and integrating these two types of information has proven to be extremely powerful in understanding complex biological phenomenon in a variety of systems (Kelley and Ideker, 2005; Keogh et al., 2005; Collins et al., 2007; Bandyopadhyay et al., 2008; Wilmes et al., 2008; Hannum et al., 2009). Genetic interactions serve as a bridge between genotype and phenotype and are instrumental in revealing functional redundancies in biological networks. For example, in S. cerevisiae, only ~1,100 out of ~6,000 possible individual gene deletions are lethal in rich medium (Giaever et al., 2002) while ~11,000 pairwise deletions have been reported to cause cell death (Stark et al., 2011). Furthermore it has been suggested that genetic interactions are vital to understanding the causes of human disease (Lehner, 2007) and may account for the “missing heritability” of complex trait studies (Carlborg and Haley, 2004; Hannum et al., 2009; Manolio et al., 2009; Zuk et al., 2012).
Genetic interactions can be divided into three broad categories: 1) aggravating (negative), whereby the double-mutant phenotype is stronger than is expected from the phenotypes associated with the single mutants; 2) alleviating (positive), whereby the double-mutant phenotype is weaker than anticipated and 3) neutral, where the measured phenotype is as expected (Phillips, 2008; Beltrao et al., 2010). Frameworks for modeling and scoring genetic interactions are normally centered at zero (i.e. a neutral gene pair) (Schuldiner et al., 2005; Collins et al., 2006, 2010; Baryshnikova et al., 2010; Horn et al., 2011) and have been developed to capture a continuous spectrum of phenotype strengths. The bulk of the available data has been generated in the budding yeast, S. cerevisiae, where fitness (derived from colony size) is most commonly used as a phenotypic readout. Several methodologies have been developed to quantify these relationships in a variety of other organisms, including E. coli (Butland et al., 2008; Typas et al., 2008), S. pombe (Roguev et al., 2007; Dixon et al., 2008), C. elegans (Lehner et al., 2006; Byrne et al., 2007) and D. melanogaster (Horn et al., 2011), by either deleting, mutating or knocking down expression of genes in a pair-wise fashion.
To date, genome-wide epistasis data has only been available for S. cerevisiae (Costanzo et al., 2010). In other organisms, the available datasets are either small in scale or focused on specific processes or pathways, including an analysis of chromatin function in S. pombe (Roguev et al., 2008), cell envelope biogenesis in E. coli (Babu et al., 2011), and signaling networks in D. melanogaster (Horn et al., 2011) and C. elegans (Lehner et al., 2006; Byrne et al., 2007). Therefore, the extent to which genetic interactions are conserved across species remains an open question. While earlier work has reported specific trends relating to the conservation and evolution of GIs (Byrne et al., 2007; Dixon et al., 2008; Roguev et al., 2008; Tischler et al., 2008), it is not clear how much of the knowledge gathered in one species can be applied to others and which individual interactions and network features are likely to be conserved. In this study, we present a genome-wide, quantitative genetic interaction map (or E-MAP (epistatic miniarray profile)) for the fission yeast, S. pombe. Fission yeast is estimated to be separated from S. cerevisiae by more than 400 million years of evolution (Sipiczki, 2000), and is in many ways more similar to metazoans, including aspects of mRNA splicing (due to the extensive presence of introns), gene expression controlled in part by the RNAi machinery, metazoan-like epigenetic mechanisms, and cell cycle regulation by the G2/M transition control (Wood, 2006). Our data allow for a comprehensive functional interrogation of these (and other) biological processes and facilitate the creation of a global S. pombe map of functional modules and assignment of specific function to many previously uncharacterised genes. Finally, analysis of these data in conjunction with our consolidated GI map from S. cerevisiae enables an unprecedented comparison of the genetic architecture of two organisms, revealing global trends that arguably exist in all eukaryotic species.
Results and Discussion
A global genetic interaction map in S. pombe
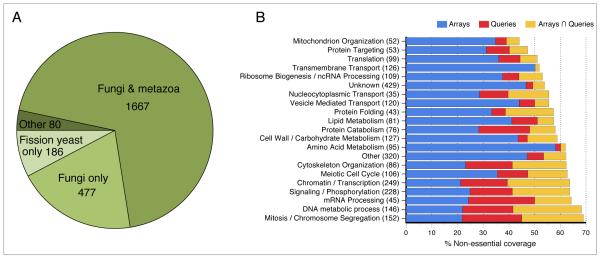
Using the PEM (Pombe Epistasis Mapper) system our group developed (Roguev et al., 2007), we screened 953 alleles (Table S1) of 876 genes against a fission yeast mutant library containing more than 2000 deletions (Table S1), resulting in an E-MAP containing ~1.6 million pairwise measurements (Datasets S1, S2). The majority of the genes screened are broadly conserved across eukaryotes, with subsets that are fungal- and fission yeast-specific (Figure 1A; Table S1). We obtained genetic interaction profiles for ~50% of the genome, resulting in representation of over half of the non-essential components of virtually every major biological process (Figure 1B; Table S1). Both internal and external validation showed the data to be of high quality and reproducibility (Supplemental Methods, Figure S1). All genetic interaction data are available online at (http://interactomecmp.ucsf.edu/pombe2012).
Figure 1. Overview of the genes included in this study.
(A) Species distribution of the genes in the fission yeast E-MAP. Species distribution data obtained from Pombase. For a complete list of the genes in each category, see Table S1. (B) Coverage of the non-essential genes with respect to different biological processes. Shown are genes present on the library array from Bioneer (http://pombe.bioneer.co.kr/) only (blue), as queries only (red) and present as both arrays and queries (orange). For each process, the total number of non-essential genes present in the E-MAP is given as the figure in brackets. For a full assignment of genes to different biological processes, see Table S1.
A global map of functional modules in S. pombe
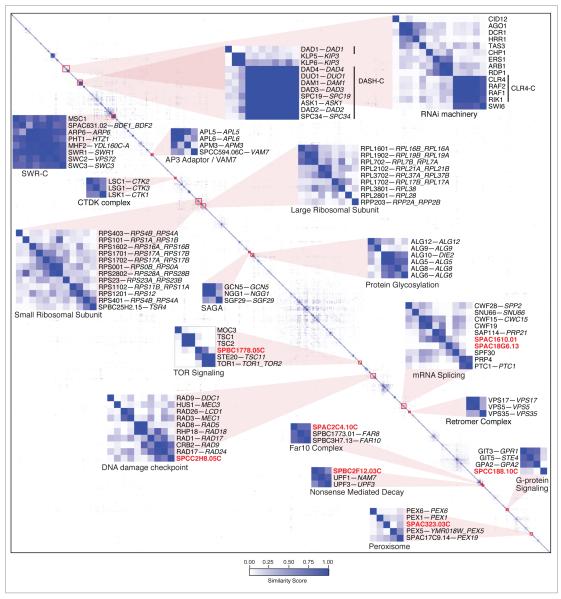
We previously reported that pairs of genes with similar genetic interaction profiles frequently encode proteins that belong to the same protein complex or work in the same functional pathway in fission yeast (Roguev et al., 2008), a network feature also observed in S. cerevisiae (Tong et al., 2004; Schuldiner et al., 2005; Collins et al., 2007; Beltrao et al., 2010). In an attempt to represent the entire dataset in an intuitive fashion, the profile from each mutant was compared to the profiles of all other mutants on the E-MAP and a similarity score was generated for each pair of mutants (Dataset S3, Supplemental Methods). These similarity scores were then subjected to hierarchical clustering, grouping genes that have similar genetic interaction profiles, suggesting that they are functionally related (Figure 2). Many known protein complexes were recapitulated from this matrix, including the SWR-C chromatin-remodelling complex (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004), CTDK-C (Sterner et al., 1995) and the GCN5 module of SAGA (Helmlinger et al., 2008) complexes that regulate transcription by RNA polymerase II, the retromer complex (Seaman et al., 1998; Iwaki et al., 2006), and the large and small subunits of the ribosome (Figure 2). Protein complexes containing components essential in S. cerevisiae, and thus difficult to genetically interrogate in that organism, were also identified, including the chromosome segregation complex, DASH-C (Figure 2). Interestingly, subunits of DASH-C clustered with the kinesins klp5 and klp6, whose protein products form a heterocomplex (Garcia et al., 2002) which functionally overlaps DASH-C in establishing bipolar chromosome attachment during mitosis (Sanchez-Perez et al., 2005). dad1 has a lower similarity score to other members of DASH-C (Figure 2), consistent with its unique role as a constitutive component of the kinetochore (Sanchez-Perez et al., 2005).
Figure 2. Hierarchical clustering of genetic interaction profiles.
Genes are grouped based on the similarity of their genetic interaction profiles. Modules discussed in the text are magnified and labeled and uncharacterised genes within these modules are highlighted in bold red. Genes are labeled using their S. pombe common name, followed by the common names of their S. cerevisiae orthologs if present (with paralogs separated by underscores). Only genes with at least one similarity score ≥ 0.1 are included in this representation (a complete dataset is provided in Dataset S3.
As genetic data allow for the grouping of factors that act together but are not necessarily physically associated, we were also able to identify several previously characterised functional pathways. These included components of the RNAi pathway, the AP3 adaptor complex with vam7 (Angers and Merz, 2009), components of the DNA damage checkpoint pathway and factors involved in protein glycosylation and TOR signaling (Figure 2). The TOR pathway in fission yeast, like that in higher eukaryotes, contains a tuberous sclerosis complex (TSC) composed of tsc1 and tsc2 that acts as a regulator for TOR signaling. In contrast to its regulatory role on TOR Complex 1 where the TSC negatively regulates TOR via GTPase RHEB, the TSC has been shown to be necessary for activation of TOR Complex 2 in mammalian cells (Huang et al., 2008). Consistent with this role, tsc1 and tsc2 group together with members of the TORC2 complex, including tor1 and ste20 (Figure 2). Furthermore, within the TORC2 group is the uncharacterised gene, SPBC1778.05c, which shows high sequence similarity (39%) (Figure S2A) with the human gene LAMTOR2, a factor known to regulate the Tor pathway (Sancak et al., 2010). This high sequence similarity together with our genetic evidence linking SPBC1778.05c to the TOR pathway, suggest that this gene is the S. pombe ortholog of LAMTOR2. Additional previously uncharacterised genes were also linked to specific function based on the hierarchical clustering, including a component of the Far8/Far10 complex (SPAC2C4.10c); a gene involved in peroxisome regulation (SPAC323.03c); a factor involved in the function of the UPF1/NAM7 nonsense mediated decay complex (SPBC2F12.03c), and a component of the G-protein signalling machinery (SPCC188.10c) (Figure 2).
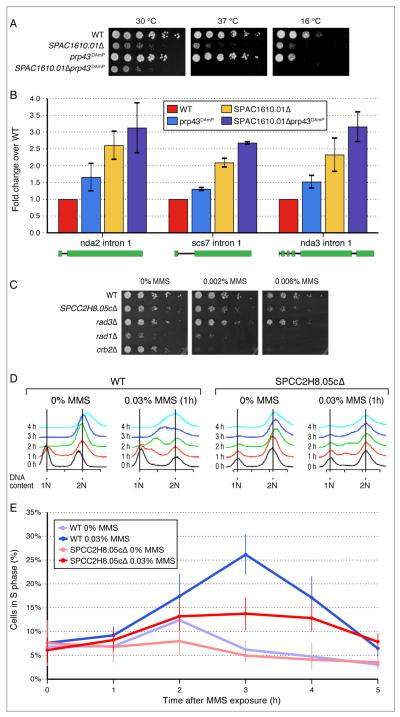
By applying a threshold to similarity metric used to generate the hierarchical clustering in Figure 2 (Figure S2B, Supplemental Methods), we were able to identify 297, non-overlapping, distinct functional modules with a minimum average similarity score of 0.1. These modules range in size from 2 to 26 genes (Table S2). In total, we were able to assign function to 144 previously uncharacterised genes by their inclusion in specific modules. For example, in module 289, which contains several genes involved in mRNA splicing, we found two previously uncharacterised genes; SPAC1610.01 and SPAC18G6.13. Deletion of one of them (SPAC1610.01) resulted in strong negative interaction with the splicing factor prp43 (Figure 3A), as well as increased level of intron accumulation of several genes (Figure 3B, Figure S3A), an effect exacerbated in a SPAC1610.01Δ prp43-DAmP double mutant. The S. cerevisiae ortholog of this gene, YKL183W, while functionally uncharacterised, is known to physically interact with the splicing factor Smd1 in S. cerevisiae (Yu et al., 2008). Furthermore, SPAC1610.01 belongs to the same protein family (ICln_channel) as the human methylosome subunit pICln which has been implicated in snRNA biogenesis (Pu et al., 1999), consistent with our observations in S. pombe.
Figure 3. Functional Characterization of SPAC1610.01 and SPCC2H8.05c.
(A) Temperature sensitive phenotype of SPAC1610.01Δ. Serial dilutions of wild type (WT), SPAC1610.01Δ and SPAC1610.01Δ prp43-DAmP mutants grown at 30°C, 37°C and 16°C. (B) Intron accumulation in SPAC1610.01Δ measured by qRT-PCR expressed as mean fold change over WT. Error bars indicate standard errors derived from at least two replicate experiments. See Figure S3A for a semi-quantitative PCR experiment. (C) SPCC2H8.05cΔ results in sensitivity to the DNA damaging agent, MMS (Methyl methanesulfonate). (D) MMS-induced S-phase delay in SPCC2H8.05cΔ. (E) Quantification of the S-phase distribution from (D). 3 hours after 0.03% MMS exposure, significantly fewer (p<0.01) SPCC2H8.05cΔ cells are in S-phase compared to WT. Means and standard errors (shown as vertical lines) were derived from five independent experiments.
We also found the uncharacterised gene SPCC2H8.05c as a part of module 203 (Table S2), which contains several well-characterised DNA damage checkpoint regulators, including rad9, rad17 and crb2. Further experiments showed that deletion of SPCC2H8.05c results in sensitivity to MMS (Figure 3C), as well as an S-phase delay in the cell cycle after exposure to MMS (Figure 3D, E), suggesting that this protein plays a role in regulating the DNA damage checkpoint pathway. Interestingly, SPCC2H8.05c has moderate sequence similarity (25%) (Figure S3B) and shows similar phenotypes to the human protein RHINO, a recently discovered DNA damage response factor (Cotta-Ramusino et al., 2011). A complete list of all functional modules and the proteins contained within them is presented in Table S2 and is also available in a searchable format on the web (http://interactomecmp.ucsf.edu/pombe2012/modules).
Identification of evolutionarily conserved functional modules
To date, large-scale, quantitative genetic interaction data has only been collected in S. cerevisiae. The S. pombe dataset described in this study is the largest genetic interaction map generated in another species, allowing us to carry out an extensive evolutionary analysis of the GI network architecture of two eukaryotic species. To facilitate this comparative cross-species analysis, we developed an algorithm to integrate the majority of existing quantitative genetic interaction data from S. cerevisiae into a single dataset, including data from a recent genome wide screen (Costanzo et al., 2010) and several smaller scale functionally focused E-MAP screens (Dataset S4) (Schuldiner et al., 2005; Collins et al., 2007; Wilmes et al., 2008; Fiedler et al., 2009; Aguilar et al., 2010; Bandyopadhyay et al., 2010; Zheng et al., 2010; Hoppins et al., 2011; unpublished data). The scoring system used to generate the genome wide dataset (SGA-score (Baryshnikova et al., 2010)) differs from that used to generate the functionally focused E-MAP datasets (S-score (Collins et al., 2010)), although both methods attempt to model the same biological phenomena. We first verified that the genome wide data were of similar quality to the functionally focused screens in terms of internal reproducibility (Figure S4A), ability to predict known genetic interactions (Figure S4B) and ability to predict protein-protein interactions (Figure S4C). We then verified that the genetic interaction scores from both methods were highly correlated (Figure S5A). Despite this high correlation, the range and distribution of interaction scores from both methods were significantly different (Figure S5B). To overcome this, a non-linear scaling method was applied to the genome-wide data (Figure S5C-E, Supplemental Methods) and the smaller scale E-MAP datasets before all S. cerevisiae data were merged into a final dataset (Dataset S4).
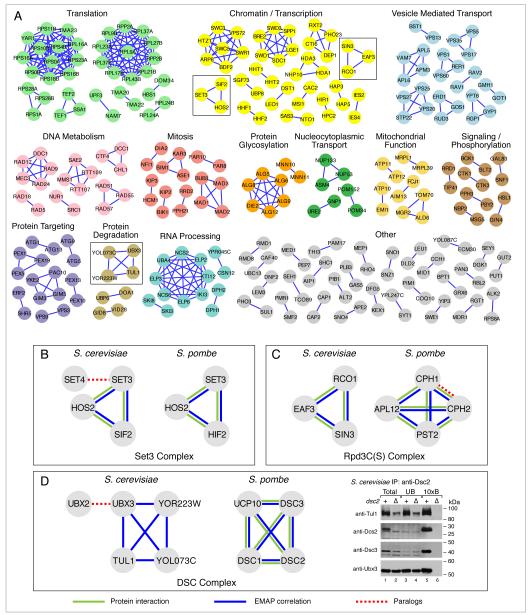
The identification of conserved biological sub-networks is a growing field of research (Sharan and Ideker, 2006). For example, methods have been developed to identify conserved linear pathways (Kelley et al., 2003) or protein complexes (Sharan et al., 2005) from protein interaction networks, or conserved co-regulated modules from gene expression (Stuart et al., 2003) or chromatin immunoprecipitation data (Tan et al., 2007). We developed a clustering procedure designed specifically to identify conserved functional modules from genetic interaction data (see Supplemental Methods for full details) and used it to identify 105 evolutionarily conserved functional modules present in both species (Figure 4A, Table S2). Gene Ontology (GO) analysis indicated that 61 of them are significantly enriched for known complexes, including the mitotic checkpoint complex (mad1, mad2, mad3, bub3) (Fraschini et al., 2001), or for pathways, such as the alg genes involved in oligosaccharyl synthesis (alg5, alg6, alg8, alg9, alg12, die2) (Jakob, 1998). A literature survey of the remaining 44 modules revealed that, although not documented in the Gene Ontology, many of them belong to the same pathway or complex, including the Tma20/Tma22 translation complex (Fleischer et al., 2006) and Aim13/Fcj1 (Figure 4A), which is part of the recently discovered MitOS complex (Hoppins et al., 2011).
Figure 4. Conserved Functional Modules.
(A) Groups of genes with highly correlated genetic interaction profiles in both S. pombe and S. cerevisae are shown. S. cerevisiae gene names were used for labeling, as many of the S. pombe orthologs lack common names. Modules are manually grouped and colored according to the biological process they are involved in. Modules from the insets are boxed and correspond to the Set3 complex (B), the Rpd3C(S) (C) and the DSC complex (D). A full list of the modules identified, and their S. pombe counterparts, is given in Table S2. Blue edges correspond to pairs of genes that have high E-MAP similarity scores, green edges represent pairs of factors that are physically associated from previous studies whereas dashed red edges represent paralogs within one species. For the immunoprecipitation assay in (D), Dsc2 binding proteins were immunopurified from detergent lysates of wild-type and dsc2Δ cells using anti-Dsc2 affinity purified polyclonal antibody. Equal amounts of total (lanes 1 and 2) and unbound fractions (lanes 3 and 4) along with 10× bound fractions (lanes 5 and 6) were immunoblotted using the indicated HRP-conjugated antibodies. See Figure S6 for an additional experiment confirming that Dsc2 binds to Ubx3, but not Ubx2.
For many of the identified modules, experimental support for their existence was previously present only in one species; evidence in the other species was either absent or based on sequence similarity alone (e.g. prefoldin and elongator in S.pombe). Furthermore, the exact ortholog mapping between these two species has been complicated in many cases by gene duplications prior to, or following their divergence more than 400 million years ago (Sipiczki, 2000). In these cases, it is unclear which of the several possible paralogs are part of the same functional module in the two modern organisms. In such instances, the E-MAP phenotypic signatures can be used to identify the correct functional orthology relationship. For example, in S. cerevisiae, there exist two orthologs of S. pombe set3 (SET3 and SET4), a putative methyltransferase in the Set3-C chromatin remodelling complex. This complex also contains Hos2, a histone deacetylase, and Sif2 (Pijnappel et al., 2001; Krogan et al., 2006). In conserved module 1, we find all three known components (SET3, HOS2 and SIF2) (Figure 4B). In budding yeast, SET4 displays a genetic interaction pattern distinct from the rest of the Set3-C, suggesting that it has a role outside the Set3-C. Consistent with this, Set4 has not been shown to physically associate with the Set3-C (Pijnappel et al., 2001; Krogan et al., 2006). The converse can be observed in another example in S. pombe: there are two orthologs of S. cerevisiae RCO1, both of which belong to conserved module 41, which corresponds to the Rpd3C(S) histone deacetylase complex (Figure 4C) involved in suppressing spurious transcription in coding regions of genes (Carrozza et al., 2005; Keogh et al., 2005). These data suggests that both of the proteins (Cph1 and Cph2) are physically part of the Rpd3C(S) complex in fission yeast, a prediction that is supported by protein-protein interaction studies (Shevchenko et al., 2008).
We hypothesized that conserved profile similarity likely reflects conserved co-pathway or co-complex membership. To test this, we focused on the DSC complex, which was recently identified in S. pombe and is required for cleavage of the membrane bound hypoxic transcription factor Sre1 in that organism (Stewart et al., 2011a). It has been suggested that the complex, which has functional links to the proteasome, may be involved in Golgi protein quality control (Stewart et al., 2011b). Initially, only four subunits of the complex were described (Dsc1, Dsc2, Dsc3, Dsc4), however a fifth has recently been reported (Ucp10/Dsc5) (Stewart et al., 2011a). S. cerevisiae has orthologs for dsc1 (TUL1), dsc2 (YOL073C) and dsc3 (YOR223W) but not dsc4, as well as a duplication of the ucp10 gene (UBX2 and UBX3). Consequently, it is not clear from sequence alone whether the complex is conserved, and how the paralogs should be annotated. In our analysis, we identified a conserved functional module (module 61) corresponding to four members of the S. pombe complex (Ucp10, Dsc1, Dsc2 and Dsc3) with S. cerevisiae orthologs (Figure 4D). UBX2, the paralog of UBX3, is not a part of the S. cerevisiae module, suggesting it is functionally and physically distinct from the DSC complex in budding yeast. In order to test this prediction, S. cerevisiae Yol073c (Dsc2) was immunoprecipitated using an antibody, and Tul1, Yor223w and Ubx3 were shown to be physically associated (Figure 4D), confirming that this complex does exist in budding yeast. Ubx2 was shown not to be physically associated with Dsc2 (Figure S6), consistent with our prediction.
Network feature conservation
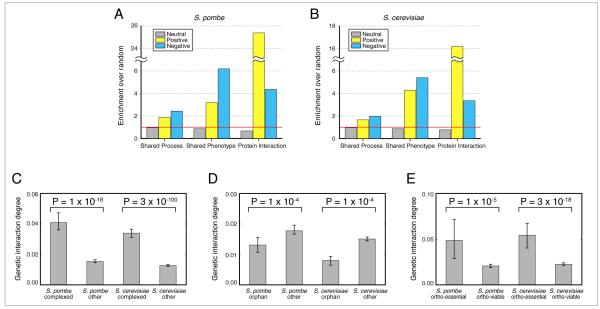
We next explored the conservation of global trends using the budding and fission yeast genetic interaction maps. By comparing genetic interaction data derived from S. cerevisiae to other, orthogonal datasets, several interesting observations have been previously reported. For example, pairs of genes that display strong genetic interactions are significantly more likely than random gene pairs to share other biological features, including similar deletion phenotypes (Tong et al., 2004), membership of the same biological process (Wilmes et al., 2008) and, particularly in the case of positive interactions, membership of the same protein complex (Schuldiner et al., 2005; Collins et al., 2007). We were able to confirm these observations in both S. pombe (Figure 5A) and S. cerevisiae (Figure 5B) on a global scale, suggesting they will also be present in other eukaryotic species. Additionally, genes whose products are members of protein complexes display a disproportionally high number of genetic interactions overall (Michaut et al., 2011), a network topology feature we find conserved in both S. cerevisiae and S. pombe (Figure 5C).
Figure 5. Conserved Network Features.
Pairs of genes which interact genetically in S. pombe (A) and S. cerevisiae (B) are more likely to display the same knockout phenotype and share membership of the same biological process and protein complex. Data are expressed as fold change over all gene pairs in the E-MAP. S. pombe and S. cerevisiae process annotations are presented in Table S1. (C) Members of known protein complexes have more genetic interactions. Bar height represents the median normalized genetic interaction degree for different categories of genes. (D) Sequence orphans have fewer genetic interactions (E) Non-essential S. pombe genes, whose S.cerevisiae ortholog is essential, have more genetic interactions than genes whose ortholog is non-essential. The same applies for S. cerevisiae. For (C), (D) and (E), the normalized genetic interaction degree for a gene is the number of significant genetic interactions for that gene, divided by the total number of measured interactions involving that gene. Error bars are calculated using 1000-fold bootstrap resampling. P-values are calculated using a two-sided Mann-Whitney U test.
Two classes of genes are especially interesting when trying to understand how genetic interactomes evolve. These are sequence orphans (genes with no identifiable orthologs in any other species) and ortho-essential genes (non-essential genes whose ortholog is essential). We find that in both species, sequence orphans have significantly fewer genetic interactions when compared to other genes (Figure 5D). These results are consistent with two of the predominant interpretations for the existence of sequence orphans: (i) sequence orphans may be rapidly evolving (Schmid and Aquadro, 2001), preventing the identification of a sequence ortholog, and the lack of genetic interactions represents a lack of functional constraints imposed by other genes and (ii) sequence orphans have arisen de novo from non-coding regions (Tautz and Domazet-Lošo, 2011) and the lack of interactions reflects incomplete integration into the cellular network. The latter theory is consistent with observations from protein-protein interaction networks (Capra et al., 2010).
Finally, in the two yeast species, 83% of the one–to-one orthologs have conserved dispensability, i.e. they are either essential or non-essential in both species (Kim et al., 2010). The remaining 17% (ortho-essential) genes have differing essentiality between the two species. We find that ortho-essential genes in both S. cerevisiae and S. pombe have ~2.5 times more genetic interactions than non-essential genes with non-essential orthologs (Figure 5E). These results suggest that although not essential for growth under standard laboratory conditions, these genes still contribute significantly to the robustness of the cell. The interpretation here depends primarily on whether one assumes that an ortho-essential gene was essential in the last common ancestor of the two species. If it was essential, and became non-essential in the modern organism, this may have happened through the accumulation of buffering relationships with other genes, also reflected by the high genetic interaction degree. On the converse, if it was non-essential in the ancestral species, but had a high number of buffering relationships with other genes, then a perturbation to any of these partners could render the gene essential in the modern organism.
Our cross-species analyses confirm that the presence of epistatic interactions generally reflects close functional associations among genes. It further suggests that genes that are evolving new or altered functions (i.e. sequence orphans) show delayed integration into the genetic interaction network, while genes with an essential ortholog are heavily integrated into the network. Since we have observed these network feature trends in two very divergent organisms, we suggest that they will be present throughout all eukaryotic species.
Hierarchical modularity of genetic interactions
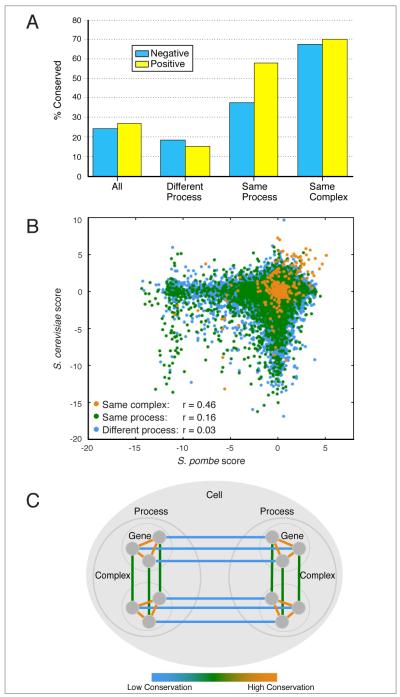
Previous work has shown that the genetic interactions between genes encoding components of the same protein complex, especially the positive ones, are highly conserved between budding and fission yeast (Roguev et al., 2008), suggesting that these functional modules are conserved across species. The data presented here support and expand these observations. To make our conservation estimates as accurate as possible, they were adjusted to take into account the reproducibility of different categories of interactions (Supplemental Methods). In addition to high conservation of positive genetic interactions within protein complexes (70%) (S-score > 1.8), we find a high degree of conservation for negative interactions (68%) (S-score < -2.3) (Figure 6A). This finding suggests that not only the dependencies, but also the buffering relationships within complexes are highly conserved.
Figure 6. Hierarchical Conservation of Genetic Interactions.
(A) Calculated percentage of conserved genetic interactions for different categories of gene pairs. Estimates were derived by comparing the observed cross-species conservation of genetic interactions to the within-species reproducibility of genetic interactions in the same category. See Supplemental Methods for full details. (B) A scatter plot of S. pombe and S. cerevisiae genetic interaction scores for pairs of genes belonging to different categories. r values are calculated using Pearson’s correlation coefficient. (C) A model for the evolution of genetic interactions with different colors representing the level of conservation. Genetic interactions between gene pairs whose products are co-complexed are highly conserved (orange), those between genes participating in the same biological process are less conserved (green), while interactions between genes involved in distinct biological processes are poorly conserved (blue).
However, biological systems do not exhibit just one level of modularity, since groups of complexes and pathways function together to carry out highly orchestrated and complex cellular processes such as translation or mitosis. Indeed, careful scrutiny of the data presented in Figure 2 reveals many instances of such hierarchical modularity. For example, two distinct clusters corresponding to the large and small ribosomal subunits can be distinguished. These are ultimately united in a single ribosomal subtree (Figure S2B). Higher up the tree, a larger cluster encompassing many genes involved in translation regulation and ribosome biogenesis is apparent (Figure S2B).
Interestingly, using the interaction strength cut-offs described above and process definitions obtained from the gene ontology (Supplemental Methods, Table S1), we find that interactions between genes belonging to the same biological process are less conserved than interactions within complexes (positive interactions: 58%; negative interactions: 38%), but significantly more conserved than interactions between genes functioning in separate processes (positive interactions: 19%; negative interactions: 15%) (Figure 6A). Analysis of the complete dataset is consistent with these observations: the genetic interactions between the two species become less conserved as larger modules are considered (same complex: r=0.46; same process: r=0.16; different process: r=0.03) (Figure 6B). These observations, combined with the fact that genes within the same complex or process are significantly more likely to interact than random gene pairs, suggests that biological systems exhibit multiple hierarchical levels of modularity and that the extent of rewiring of genetic interactions is dependent on the specificity of the module they belong to (Figure 6C).
Global connectivity of biological processes
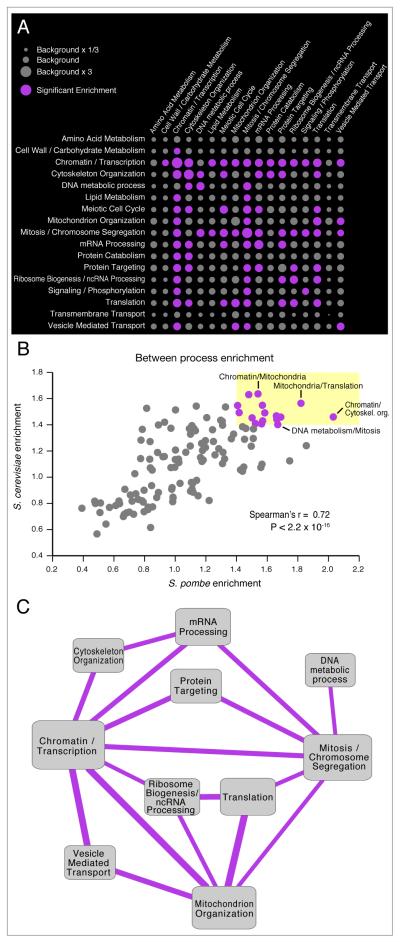
We next analyzed the functional connectivity between the different processes in the two organisms, identifying pairs of processes that are enriched (or depleted) for genetic interactions in fission yeast (Figure 7A, Table S3). Consistent with Figure 5A, we find that genes within the same process tend to be enriched in genetic interactions (large circles along the diagonal on Figure 7A). Interestingly, we also see significant enrichment between distinct biological processes, (large circles off the diagonal on Figure 7A). There is a clear indication of the existence of ‘hub processes’ – central processes that interact with many diverse functions, such as Chromatin/Transcription, Mitosis and Mitochondrion Organization. The role of chromatin as a ‘hub process’ has previously been identified in a genome wide S. cerevisiae genetic interaction map (Costanzo et al., 2010) and is also supported by smaller scale screens from C. elegans, suggesting that it may be a common feature of eukaryotic genetic interaction networks (Lehner et al., 2006). Conversely, we see that some processes, such as Amino Acid Metabolism and Transmembrane Transport, have very few genetic interactions (Figure 7A), suggesting a high degree of functional independence among these modules, with less impact on other cellular processes than hub modules, at least under the conditions used to collect the data
Figure 7. Conservation of Functional Cross-talk Between Biological Processes.
(A) Genetic cross-talk between distinct biological processes in S. pombe. The size of each circle represents the fraction of significant interactions between two processes compared to the fraction of significant interactions between all annotated genes. Purple circles represent significant enrichment of interactions between processes. Significance is assessed using a two-tailed binomial test, and the Bonferonni method is used to correct for multiple testing. Enrichment values and p-values are given in Table S3. For the S. cerevisiae enrichment map and data see Figure S7 and Table S3. (B) Enrichments observed in S. pombe are highly correlated with the ones in S. cerevisiae. Pairs of processes that are highly connected in both species are colored purple, highlighted inside the yellow box and are drawn as a network diagram in (C). Within process enrichments (i.e. the diagonal in Figure 7A) are not shown. (C) Conserved links between biological processes. Links represent pairs of processes that are linked by at least 1.4 times the background rate of genetic interactions in both species.
In order to analyze the evolutionary conservation of high-level inter-process connectivity, we created an analogous map for S. cerevisiae (Figure S7, Table S3). Comparison of the two maps (Figure 7B) shows that at a global level, both organisms share remarkable similarities and the level of cross-talk between distinct biological processes is highly conserved. This appears to happen independently of the extensive rewiring of individual interactions as in both species genes involved in Chromatin/Transcription and genes involved in Mitosis / Chromosome Segregation are significantly more likely to interact with each other than random gene pairs (>1.4 fold enrichment in both species) however, only ~25% of the individual interactions between these two processes are conserved. This suggests that although there is flexibility in terms of the implementation (the specific interactions between individual genes), there may be design requirements that must be met by all eukaryotic systems (the strong links between particular processes). For example, many cellular perturbations (including gene deletions (Hughes et al., 2000)) require an increase in transcription of specific genes, which offers an explanation for the tendency of genes in Chromatin / Transcription to act as genetic interaction hubs. This requirement for specific transcription is likely to be maintained across species, however the exact manner in which it is achieved, and which components are involved may be under less selective pressure.
Several of the processes that show conserved genetic links are not surprising, including DNA metabolism with Mitosis / Chromosome Segregation and Translation with Ribosome Biogenesis / ncRNA Processing. However, more intriguing connections also exist, including a link between Mitosis/Chromosome Segregation and mRNA Processing (Figure 7C) (Murakami et al., 2007; Tang et al., 2011). While further work will be required to understand the molecular mechanisms that link these different processes, the evolutionary conservation between both S. pombe and S. cerevisiae suggests that these links are likely to exist in other eukaryotic organisms.
Perspective
The availability of large-scale, genome-wide quantitative genetic interaction maps in the two model organisms, S. pombe and S. cerevisiae, has provided an opportunity for an unprecedented evolutionarily analysis of genetic interactomes across eukaryotic species. Additionally these data suggest ways to improve the design of similar experiments in more complex organisms. Genetic interaction mapping efforts can be broadly divided into large-scale unbiased screens (Tong et al., 2004; Costanzo et al., 2010), and those more focused on specific biological pathways or processes (Schuldiner et al., 2005; Collins et al., 2007; Roguev et al., 2008; Wilmes et al., 2008; Fiedler et al., 2009; Zheng et al., 2010; Babu et al., 2011; Horn et al., 2011). While both approaches have provided rich and unique biological insights, unbiased studies offer a number of advantages. Because the genes studied are not selected based on prior knowledge (e.g. sub-cellular localization, co-expression, common function), there is a greater chance to functionally annotate uncharacterised genes, such as the 144 we have assigned to functional modules in this study. Furthermore, unbiased gene selection increases the probability for identification of systems level trends, such as the connection reported here between essentiality in one species and genetic interaction degree in another. However, a major disadvantage of unbiased screens is the significant labour and cost involved in data collection, at least using the current approaches.
By contrast, focused screens can be carried out with more limited resources, are the method of choice for high-resolution, quantitative interrogation of distinct biological functions and are often associated with more specific, hypothesis-driven questions. Indeed, it is possible to saturate the interaction space within specific processes such as the early secretory pathway (Schuldiner et al., 2005), chromosome biology (Collins et al., 2007) and mitochondrial function (Hoppins et al., 2011). In addition to obtaining a detailed view of a particular process, these studies are beneficial in a number of other ways. Genes involved in the same process are more likely to genetically interact, resulting in a greater ratio of significant interactions discovered. Furthermore, in this study, we show that the interactions within biological processes are significantly more likely to be conserved across species, making them of potentially greater utility.
Both focused and unbiased screens currently share a common handicap in their inability to generate comprehensive datasets. Indeed, after over ten years of experiments in the budding yeast S. cerevisiae, only approximately six of a possible eighteen million pairwise interactions have been measured. Although this is a monumental achievement, it corresponds to only ~2% of the interactions that would need to be measured to obtain a complete mammalian genetic interactome, even without considering the complexities of different cell types. Furthermore, this does not take into account the generation of condition specific genetic interaction studies, for example using the differential E-MAP (or dE-MAP) approach (Bandyopadhyay et al., 2010), or genetically analysing multifunctional genes by mutating specific domains or individual amino acids (our unpublished data), both of which increase the potential screening space exponentially.
The issue of rational screen design is likely to become increasingly important as further genetic interaction detection methods are developed in metazoans (Lehner et al., 2006; Horn et al., 2011; Lin et al., 2012). We have previously proposed two possible solutions – an iterative experimental approach based on information theory (Casey et al., 2008), and an approach to exploit the overlap between smaller scale screens (Ryan et al., 2011). Our analysis suggests the additional possibility of exploiting the observations that in distantly related organisms certain categories of genes comprise genetic interaction hubs and certain pairs of processes are densely connected. Furthermore, we find that information collected from model systems about connections between individual genes may not be as useful as inferences derived from functional module definitions and the level of cross-talk between different processes.
These observations are also likely to be helpful in the search for epistasis in genome wide association studies. Genetic interactions are believed to account for a significant amount of the “missing heritability” of complex diseases (Moore, 2003; Carlborg and Haley, 2004; Zuk et al., 2012). Since in genome wide association studies testing every possible pair-wise interaction is computationally expensive and results in a significant loss of statistical power (Hirschhorn and Daly, 2005; Cordell, 2009), testing for interactions between logically selected subsets of interactions is likely to result in significant gains in the search for the cause of complex diseases (Pattin and Moore, 2008; Hannum et al., 2009).
Materials and Methods
Data collection
Genetic crosses were performed in high density (1536 format) on a Singer RoToR station using the PEM system and applying a previously published protocol. For a full list of strains see Tables S1. Data was collected in batches of 25-35 queries and colony sizes were measured using the Colony Measure Program (http://sourceforge.net/projects/ht-col-measurer/).
Scoring of genetic interactions
Raw data was scored using a published software toolbox. Individual batches were normalized and scored separately thus minimizing systematic experimental biases and batch-to-batch variation.
For a detailed description of methods related to characterization of SPAC1610.01, SPCC2H8.05c and the DSC complex as well as computational methods used see Supplementary Methods.
Supplementary Material
Highlights.
Genetic interaction map in fission yeast covering 60% of the non-essential genome
~300 functional modules identified, providing function for uncharacterized genes
Individual interactions between genes in different processes are poorly conserved
Overall levels of genetic cross-talk between pairs of processes are highly conserved
Acknowledgements
The authors wish to thank members of the Krogan lab and J. E. Haber for helpful discussion. We are grateful to Patrick Kemmermen for support with the online database and the Pombase team for responding to technical queries. This work was supported by grants from QB3@UCSF, the NIH (GM084448, GM084279, GM081879 and GM098101 to NJK; GM085764 and GM084279 to TI; GM21119 to CG; HL077588 to PE; ES019966 and CA013330 to WE and MCK) and the Science Foundation Ireland (Grant No. 08/SRC/I1407 to PC, DG and GC). CR is supported by the IRCSET-funded Ph.D. program in Bioinformatics and Systems Biology. PB is supported by the Human Frontiers Science Program. PE is an Established Investigator of the American Heart Association. TI is a David and Lucille Packard Fellow. CG is an American Cancer Society Research Professor of Molecular Genetics. NJK is a Searle Scholar and a Keck Young Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar PS, Fröhlich F, Rehman M, Shales M, Ulitsky I, Olivera-Couto A, Braberg H, Shamir R, Walter P, Mann M, et al. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nature Structural & Molecular Biology. 2010;17:901–908. doi: 10.1038/nsmb.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers CG, Merz AJ. HOPS Interacts with Apl5 at the Vacuole Membrane and Is Required for Consumption of AP-3 Transport Vesicles. Molecular Biology of the Cell. 2009;20:4563–4574. doi: 10.1091/mbc.E09-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Díaz-Mejía JJ, Vlasblom J, Gagarinova A, Phanse S, Graham C, Yousif F, Ding H, Xiong X, Nazarians-Armavil A, et al. Genetic Interaction Maps in Escherichia coli Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways. PLoS Genetics. 2011;7:e1002377. doi: 10.1371/journal.pgen.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional Maps of Protein Complexes from Quantitative Genetic Interaction Data. PLoS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung M-K, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn J-YY, Ou J, San Luis B-JJ, Bandyopadhyay S, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nature Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Babu M, Díaz-Mejía JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, et al. eSGA: E. coli synthetic genetic array analysis. Nature Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- Byrne A, Weirauch M, Wong V, Koeva M, Dixon S, Stuart J, Roy P. A global analysis of genetic interactions in Caenorhabditis elegans. Journal of Biology. 2007;6 doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Pollard KS, Singh M. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biology. 2010;11:R127–16. doi: 10.1186/gb-2010-11-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nature Reviews. Genetics. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Casey FP, Cagney G, Krogan NJ, Shields DC. Optimal stepwise experimental design for pairwise functional interaction studies. Bioinformatics (Oxford, England) 2008;24:2733–2739. doi: 10.1093/bioinformatics/btn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ, Weissman Jonathan, Guthrie Christine, Fink Gerald R. Quantitative Genetic Interaction Mapping Using the E-MAP Approach. Academic Press; 2010. pp. 205–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biology. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nature Reviews Genetics. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. The Genetic Landscape of a Cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science (New York, N.Y.) 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JLY, Prasad TSK, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe K-L, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes & Development. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. The EMBO Journal. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two Kinesin-like Kin I Family Proteins in Fission Yeast Regulate the Establishment of Metaphase and the Onset of Anaphase A. Current Biology. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hannum G, Srivas R, Guénolé A, van Attikum H, Krogan NJ, Karp RM, Ideker T. Genome-wide association data reveal a global map of genetic interactions among protein complexes. PLoS Genetics. 2009;5:e1000782. doi: 10.1371/journal.pgen.1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Gygi SP, Bähler J, Winston F. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes & Development. 2008;22:3184–3195. doi: 10.1101/gad.1719908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews. Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, DeVay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. The Journal of Cell Biology. 2011 doi: 10.1083/jcb.201107053. jcb.201107053–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nature Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Molecular and Cellular Biology. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Iwaki T, Hosomi A, Tokudomi S, Kusunoki Y, Fujita Y, Giga-Hama Y, Tanaka N, Takegawa K. Vacuolar protein sorting receptor in Schizosaccharomyces pombe. Microbiology (Reading, England) 2006;152:1523–1532. doi: 10.1099/mic.0.28627-0. [DOI] [PubMed] [Google Scholar]
- Jakob CA. Degradation of Misfolded Endoplasmic Reticulum Glycoproteins in Saccharomyces cerevisiae Is Determined by a Specific Oligosaccharide Structure. The Journal of Cell Biology. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, Ideker T. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proceedings of the National Academy of Sciences. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nature Biotechnology. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M-C, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Violations of robustness trade-offs. Molecular Systems Biology. 2010;6:384. doi: 10.1038/msb.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh M-C, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Molecular Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lehner B. Modelling genotype-phenotype relationships and human disease with genetic interaction networks. The Journal of Experimental Biology. 2007;210:1559–1566. doi: 10.1242/jeb.002311. [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Lin Y.-yi, Kiihl S, Suhail Y, Liu S-Y, Chou Y.-hsuan, Kuang Z, Lu J.-ying, Khor CN, Lin C-L, Bader JS, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science (New York, N.Y.) 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Human Heredity. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PloS One. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattin KA, Moore JH. Exploiting the proteome to improve the genome-wide genetic analysis of epistasis in common human diseases. Human Genetics. 2008;124:19–29. doi: 10.1007/s00439-008-0522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews. Genetics. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Séraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes & Development. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Molecular and Cellular Biology. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park H-O, Hayles J, et al. Conservation and Rewiring of Functional Modules Revealed by an Epistasis Map in Fission Yeast. Science. 2008:1162609. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Meth. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Ryan C, Greene D, Guenole A, van Attikum H, Krogan NJ, Cunningham P, Cagney G. Improved functional overview of protein complexes using inferred epistatic relationships. BMC Systems Biology. 2011;5:80. doi: 10.1186/1752-0509-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JBA. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. The EMBO Journal. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Aquadro CF. The Evolutionary Analysis of “Orphans” From the Drosophila Genome Identifies Rapidly Diverging and Incorrectly Annotated Genes. Genetics. 2001;159:589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epistatic Miniarray Profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. The Journal of Cell Biology. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nature Biotechnology. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan N, Shevchenko A, Stewart AF. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biology. 2008;9 doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biology. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. Reviews 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Research. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Molecular and Cellular Biology. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EV, Lloyd SJ-A, Burg JS, Nwosu CC, Lintner RE, Daza R, Russ C, Ponchner K, Nusbaum C, Espenshade PJ. Yeast SREBP cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. The Journal of Biological Chemistry. 2011a;287:672–681. doi: 10.1074/jbc.M111.317370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EV, Nwosu CC, Tong Z, Roguev A, Cummins TD, Kim D-U, Hayles J, Park H-O, Hoe K-L, Powell DW, et al. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Molecular Cell. 2011b;42:160–171. doi: 10.1016/j.molcel.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Tan K, Shlomi T, Feizi H, Ideker T, Sharan R. Transcriptional regulation of protein complexes within and across species. Proceedings of the National Academy of Sciences. 2007;104:1283–1288. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luca M, Portillio J, Ngo B, Chang C, Wen T, Murray J, Carr A. LAMMER kinase Kic1 is involved in pre-mRNA processing. Experimental Cell Research. 2011;317:2308–2320. doi: 10.1016/j.yexcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nature Reviews Genetics. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat Genet. 2008;40:390–391. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global Mapping of the Yeast Genetic Interaction Network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nature Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Molecular Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V. In: Schizosaccharomyces pombe comparative genomics; from sequence to systems. Sunnerhagen P, Piskur J, editors. Springer; Berlin / Heidelberg: 2006. pp. 233–285. [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yıldırım MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Benschop JJ, Shales M, Kemmeren P, Greenblatt J, Cagney G, Holstege F, Li H, Krogan NJ. Epistatic relationships reveal the functional organization of yeast transcription factors. Molecular Systems Biology. 2010;6:420. doi: 10.1038/msb.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences. 2012;109(4):1193–8. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.