Abstract
Controlled activation of the complement system, a key component of innate immunity, enables destruction of pathogens with minimal damage to host tissue. Complement factor H (CFH), which inhibits complement activation, and five CFH-related proteins (CFHR1–5) compose a family of structurally related molecules. Combined deletion of CFHR3 and CFHR1 is common and confers a protective effect in IgA nephropathy. Here, we report an autosomal dominant complement-mediated GN associated with abnormal increases in copy number across the CFHR3 and CFHR1 loci. In addition to normal copies of these genes, affected individuals carry a unique hybrid CFHR3–1 gene. In addition to identifying an association between these genetic observations and complement-mediated kidney disease, these results provide insight into the protective role of the combined deletion of CFHR3 and CFHR1 in IgA nephropathy.
Polymorphic variation within the CFH-CFHR gene locus influences susceptibility to diverse pathologies including age-related macular degeneration (AMD),1 meningococcal sepsis,2 and thrombotic and inflammatory kidney disease such as atypical hemolytic uremic syndrome (aHUS) and C3 glomerulopathy, respectively.3 Recently, a common copy number polymorphism that results in the deletion of both CFHR3 and CFHR1 genes (ΔCFHR3-1) has been associated with protection against the most common form of GN worldwide, IgA nephropathy, a pathology associated with complement deposition within the kidney.4 Although we have a detailed understanding of CFH biology, the biological role of the five CFHR proteins is unclear. It is likely that CFHR5 has a role in the processing of complement within the kidney because it has complement regulatory activity in vitro,5 it co-localizes with complement deposits within the kidney,6 and a CFHR5 mutation is associated with familial complement-mediated GN.7 Although CFHR1 and CFHR3 have complement regulatory activity in vitro,8,9 both have been considered biologically redundant because their complete absence in healthy populations is common. For example, the ∆CFHR3–1 homozygous deletion frequency is 4.7% in European-American and 16% in African-American populations.10 In fact, ∆CFHR3–1 is actually associated with disease protection. ∆CFHR3–1 is protective in both AMD10,11 and IgA nephropathy,4 pathologies associated with complement deposition in affected tissues. On the basis of a protective role for the ∆CFHR3–1 in IgA nephropathy, we developed the hypothesis that CFHR1 and CFHR3 impair complement processing within the kidney. This hypothesis would predict that an increase in CFHR1 and CFHR3 copy number would enhance susceptibility to complement-mediated kidney injury. Here, we report a novel CFHR3–1 hybrid gene located on an allele that also contained intact copies of the CFHR1 and CFHR3 genes. The hybrid gene was detected exclusively in eight affected individuals among a large kindred with familial complement-mediated GN.
The clinical description of this family has previously been reported12,13 and the clinical progress of the affected individuals is described in Table 1. The kindred contains eight individuals with the condition over three generations (Figure 1). Renal biopsies were performed in six affected individuals. In the remaining two affected individuals, disease positivity was deduced from abnormal urinalysis (Figure 1). Renal biopsy showed a distinct pattern of glomerular inflammation termed membranoproliferative GN type III (MPGN type III). Evidence of glomerular immunostaining for C3 in the absence of Igs was present. These features indicated Ig-independent complement activation through the complement alternative pathway and are the defining features of C3 glomerulopathy, a term that includes MPGN type III, C3 GN, and dense deposit disease.3 AMD-like lesions may develop in C3 glomerulopathy but ophthalmologic assessment in individuals 105, 212, 213, and 214 did not reveal any evidence of ocular drusen. C3 glomerulopathy has been associated with genetic and acquired defects in complement regulation.3 No evidence of anti-CFH autoantibodies or C3 nephritic factors, acquired causes of C3 glomerulopathy, were detected in the affected individuals and plasma C3 and CFH levels were normal (Table 2). Previous linkage data indicated that the causative defect was within a 22-cM region at chromosome 1q31–32 (maximum logarithm [base 10] of odds score = 3.86), which included the CFH-CFHR locus.13 Using Sanger sequencing, we did not identify any mutations in the CFH or the CFHR genes in the index case (pedigree number 105; Figure 1, data not shown). We next examined copy number variation (CNV) across the CFH-CFHR locus using multiplex ligation-dependent probe assay and TaqMan copy number assays (Supplemental Figure 1, A and B). Combined data from these approaches demonstrated the presence of three copies of two genomic segments within the CFHR locus. These segments included exons 1, 2, and 3 of CFHR3 and exons 3, 4, 5, and 6 of CFHR1 (Figure 2A). Abnormal rearrangement within the CFH-CFHR gene cluster can also impair the surface recognition domains of CFH and predispose individuals to familial aHUS.14,15 Importantly, in affected individuals, there was no evidence of structural mutations affecting the CFH gene, serum CFH levels were normal, and anti-CFH antibodies were not detected.
Table 1.
Updated clinical summaries of affected individuals
| Pedigree No. | Presentation | End Stage Renal Failure | Renal Transplantation | Ocular Drusen | |||
|---|---|---|---|---|---|---|---|
| Age (yr) | Proteinuria (g/24 h) | Hematuria | Biopsy Diagnosis | ||||
| 103 | 25 | >3 | +++ | MPGN | Yes | Yes | NA |
| 105 | 51 | 3.06 | ++ | MPGN | Yes | Yes | Absent |
| 107 | 51 | +++a | +++ | NA | No | — | NA |
| 207 | 22 | +++a | + | NA | No | — | NA |
| 212 | 28 | 3.77 | + | MPGN | Yes | Yes | Absent |
| 213 | 4 | 5.0 | Negative | MPGN | Yes | Yes | Absent |
| 214 | 21 | 3.13 | + | MPGN | Yes | Yes | Absent |
| 223 | 16 | 0.3 | +++ | MPGN | No | - | NA |
MPGN, membranoproliferative GN; NA, not assessed.
Qualitative proteinuria.
Figure 1.
Pedigree with familial C3 glomerulopathy. Affected individuals indicate those with C3 glomerulopathy confirmed on renal biopsy. Probable affected indicates those with significant proteinuria (>300 mg/24 h or at least 3+ on urinalysis) or hematuria (at least 3+ on two occasions). Unaffected individuals are healthy and do not have abnormal urinalysis.
Table 2.
Complement profile
| Pedigree No. | Status | C3 | C4 | CH100 | AP100 | CFHa | CFIa | C3NeF | Anti-CFH |
|---|---|---|---|---|---|---|---|---|---|
| Normal range | 0.7–1.7 g/L | 0.16–0.54 g/L | 50%–150% NHP | 50%–150% NHP | % NHP | % NHP | n/a | AEU | |
| 103 | Affected | 0.93 | 0.39 | 158 | 109 | 102 | 152 | ND | ND |
| 105 | Affected | 0.84 | 0.29 | 155 | 39 | 77 | 87 | ND | ND |
| 212 | Affected | 1.16 | 0.54 | 126 | 108 | 119 | 123 | ND | ND |
| 213 | Affected | 1.12 | 0.40 | 172 | 32 | 84 | 96 | ND | ND |
| 214 | Affected | 0.79 | 0.26 | 145 | 64 | 86 | 103 | ND | ND |
| 215 | Unaffected | 1.12 | 0.23 | 155 | 74 | 77 | 96 | ND | ND |
NHP, normal human pool; C3NeF, C3 nephritic factor; AEU, arbitrary ELISA units; CFI, complement factor I; ND, not detected.
No validated reference range exists for CFH and CFI and results are thus expressed as the percentage of NHP. C3NeF was detected using a commercial assay (The Binding Site, UK).
Figure 2.
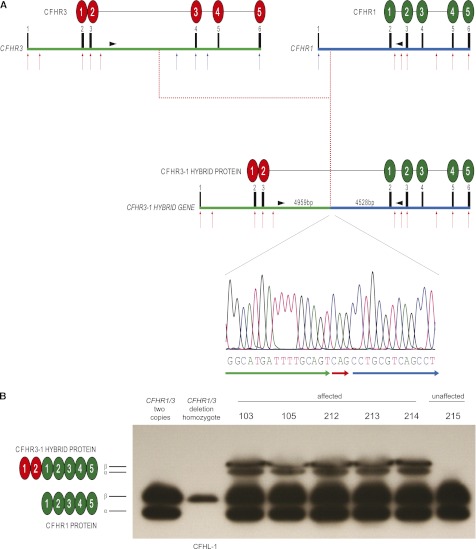
The CFHR3–1 hybrid gene structure, genomic breakpoint, and protein. (A) Schematic representation of the CFHR3–1 hybrid gene and protein. Red arrows indicate sites of probes giving three copy signals, whereas blue arrows depict probes giving two copy signals using copy number assays. Exons and protein domains are numbered. The intergenic distance between CFHR3 and CFHR1 is 24 kb. The genomic breakpoint (4959 bp into CFHR3 intron 3) was identified using paired-end mapping analysis of next-generation sequence and long-range genomic PCR and amplicon sequencing. The breakpoint sequence contained a CAG triplet not identified in the introns of either gene (red arrow). (B) Serum Western blot using polyclonal anti-CFH antibody demonstrated the presence of two additional protein bands of similar molecular mass only in sera from affected individuals. The two bands represent differentially glycosylated isoforms (designated α and β) comparable with that seen with CFHR1. CFHL-1, CFH-like protein 1, a protein derived from alternative splicing of the CFH gene that is visible on this gel only in the absence of CFHR1 protein (lane 2).
We therefore hypothesized that a complex rearrangement that resulted in a CFHR3-CFHR1 hybrid gene in addition to the normal diploid copies of both CFHR3 and CFHR1 genes had occurred, and that this was inherited in an autosomal dominant fashion in affected individuals. The region of interest (NCBI37/hg19: Chr1: 196621008–196978800) containing the CFHR locus was successfully targeted using the Agilent SureSelect target enrichment system. Deep sequencing with coverage of >1000× was performed in one affected individual (pedigree number 214) and one related unaffected individual (pedigree number 104). Two partial gene duplication events were identified by comparing sequence coverage discrepancies between individuals; the first starting at least 10 kb upstream of CFHR3′s transcription start site and terminating in intron 3 of CFHR3, the second starting in intron 1 of CFHR1 and terminating at least 13 kb downstream of CFHR1 (Supplemental Figure 1C). Analysis of the sequencing which compared atypical mapping of paired-end reads between these individuals revealed two genomic breakpoints (NCBI37/hg19:chr1:196754062 at CFHR3 intron 3 and NCBI37/hg19:chr1: 196790079 at CFHR1 intron 1) present only in the affected individual (Supplemental Figures 2 and 3).
To confirm the presence of these genomic breakpoints, primers located within the duplicated regions were used to generate a 6-kb amplicon. As expected, it was possible to generate this amplicon using genomic DNA from affected individuals only. Amplicon sequencing confirmed the genomic breakpoints identified using the paired-end mapping analysis. These data indicated a CFHR3-CFHR1 hybrid gene comprising exons 1, 2, and 3 from CFHR3 and exons 2, 3, 4, 5, and 6 from CFHR1 (Figure 2A). This hybrid gene was present on a CFH-CFHR allele that contained intact copies of the CFHR1 and CFHR3 genes. We next designed a genomic PCR that would amplify this breakpoint (757-bp amplicon) in addition to normal genomic sequence (366-bp amplicon). The abnormal amplicon was present in all affected members of the kindred but not in unaffected individuals (Supplemental Figure 4) or 450 Caucasian control samples (data not shown).
Analysis of the predicted hybrid protein cDNA sequence indicated that splicing from CFHR3 exon 3 to CFHR1 exon 2 would generate a novel protein containing 454 amino acids. The protein would comprise leader peptide sequence and the initial two protein domains, termed short consensus repeat domains, of CFHR3 (derived from CFHR3 exons 1, 2, and 3) and all five short consensus repeat domains of CFHR1 (derived from CFHR1 exons 2, 3, 4, 5, and 6; Figure 2A). Using a polyclonal anti-human CFH antibody we were able to demonstrate the presence of two aberrant protein bands of similar molecular weight in affected but not unaffected sera (Figure 2B). Two CFHR1 protein bands of similar molecular mass (37-kD CFHR1α and 43-kD CFHR1-β) are seen in sera and we postulated that the two aberrant protein bands represented differentially glycosylated isoforms of the CFHR3–1 hybrid protein (Figure 2B). Using heparin-purified sera from affected individuals, we were able to isolate and analyze the aberrant proteins by peptide mass fingerprinting (Supplemental Figure 5). This analysis identified seven peptides within the predicted hybrid protein sequence (Supplemental Figure 5), including a unique peptide sequence that spanned the junction between CFHR3 protein sequence and CFHR1 protein sequence (VTTFCDFPK) (Supplemental Figure 5).
It is important to note that linkage to the disease locus was reported in this family a decade ago13 and exon sequencing of the CFH-CFHR locus did not detect the structural variation. Paired-end mapping analysis and copy number assays across the locus successfully identified the hybrid CFHR3–1 gene. Hence, target enrichment followed by paired-end mapping analysis is an efficient method of detecting rearrangements and CNV in linkage peaks and this approach can be applied on a genome-wide level.16,17
The CFHR3–1 hybrid gene is a unique cause of C3 glomerulopathy. The association is robust because we were able to demonstrate complete segregation with disease in a large kindred. It is likely that this hybrid gene arose as a result of an abnormal crossover event during meiosis. Frequent interspersed repeat elements within the CFH-CFHR locus most likely predispose to such events.18 The association between an abnormal CFHR3–1 hybrid gene on an allele with intact copies of both CFHR3 and CFHR1 in familial C3 glomerulopathy, together with the observations that ΔCFHR3–1 is common among healthy individuals and protective in IgA nephropathy and AMD, suggested that the CFHR3–1 protein causes C3 glomerulopathy through a dominant mechanism. However, further studies are needed to demonstrate causation in this family. In a previous study, we identified a heterozygous internal duplication within CFHR5 in familial C3 glomerulopathy.7 Whether this represented a loss of function or dominant negative change remained uncertain because the consequences of complete CFHR5 deficiency, unlike complete deficiency of CFHR1 and CFHR3, were unknown. We speculate that the CFHR3–1 and CFHR5 mutant proteins mediate abnormal accumulation of C3 within the kidney through a common dominant mechanism most likely interfering with the C3 regulation by CFH and CFHR5. It may be the case that normal CFHR3 and CFHR1 proteins also interfere with this process, albeit to a lesser degree; consequently, the absence of these proteins, afforded by the ΔCFHR3–1 polymorphism, results in amelioration of complement-mediated renal injury in IgA nephropathy.
Concise Methods
Sequencing
Exon sequencing of CFH and CFHR1–5 genes was performed in two affected individuals (numbers 105 and 213). Amplicons that targeted the exons and at least 50 bp of contiguous intronic sequence were amplified by PCR using primers designed using Primer3 software. Sequencing reactions were performed using BigDye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Both PCR and sequencing reaction products were purified using Edge Biosystems Gel Filtration plates (Edge Biosystems, Gaithersburg, MD). The sequencing was run on a 3730 DNA Analyzer and analyzed using Sequencher DNA sequencing analysis software (Gene Codes Corp, Ann Arbor, MI).
Multiplex Ligation-Dependent Probe Amplification
Probes used targeted CFH exons 1, 2, 3, 4, 6, 11, 12, 13, 18, and 23; CFHR3 exons 1, 2, 3, 4, and 6; CFHR1 exons 1, 3, 5, and 6; CFHR2 exons 1, 2, 3, and 4; and CFHR5 exons 1, 2, and 3 (MLPA P236-A1; MRC Holland, www.mlpa.com). Genomic DNA was processed according to the manufacturer’s instructions. Assay readings were validated using control samples that included individuals with known heterozygous or homozygous polymorphic deletion of the CFHR1 and CFHR3 genes. Data were depicted as mean ± SD.
CNV with Real-Time PCR
TaqMan probes (Applied Biosystems) were used to further dissect CNV within the CFHR3 and CFHR1 genes. Genomic DNA was processed according to manufacturer’s instructions. Assay readings were normalized to control samples and values represent mean ± SD. All probes were validated using genomic DNA from controls with either heterozygous or homozygous polymorphic deletion of the CFHR1 and CFHR3 genes.
In-Solution Target Enrichment of the CFHR Locus and Next-Generation Sequencing
Extracted DNA from affected individual 214 and unaffected individual 104 were controlled using fluorescent dosage with picogreen as well as Nanodrop analysis. Illumina libraries were constructed according to the manufacturer’s SureSelect protocol, which involves replacement of one of the Illumina paired-end adapters with the SureSelect precapture adapter and precapture PCR. Library construction was carried out using the automated SPRI-TE system (Beckman Coulter Genomics). Libraries were controlled on the Agilent Bioanalyzer. In parallel, baits were designed on the CFHR locus defined as chr1:196621008–196978804 using the online Agilent eArray design tool. Default design parameters were used to generate a total of 2179 baits of 120-bp length, covering 150 kb of the 358-kb region of interest, essentially avoiding nonunique locations such as long interspersed nuclear element repeats and long terminal repeats with the default limited 20-bp allowance of bait overlap with nonunique sequence. This represents around 4% of bait space provided for custom bait libraries by the manufacturer. Expected actual extent covered by captured fragments is in general several tens of kilobases higher because the method also captures fragments having only partial overlaps with the 120-bp reads. The design was visually checked on the UCSC genome browser (University of California, Santa Cruz, California). Baits were ordered from the manufacturer; upon reception, recommended quantities of baits were mixed with denatured Illumina libraries and hybridization was carried out over 24 hours. After capture, noncaptured DNA was washed away, the hybridized molecules were eluted, and postcapture PCR was performed integrating the Illumina 3′ index adapters so as to tag the libraries with DNA barcodes. Library quality control and dosage was carried out so as to pool an equivalent number of molecules from the two libraries for sequencing in a single lane. Paired-end 2×50-bp sequencing was performed on the Illumina Genome Analyzer IIx, generating 22M read pairs for the affected and 18M read pairs for the unaffected individuals.
Breakpoint Analyses
Sequences were mapped on the human genome by the manufacturer’s Eland program, comprised in the Illumina CASAVA 1.8 software suite. The mapped reads were realigned using the SRMA 0.15 assembler. Resulting sequence coverage was examined using Integrative Genomics Viewer software. Subsequently, atypical pairs were isolated using a BAMtools 0.9 custom filter, specifying insert sizes >1 kb. These atypical pairs were provided as input to SVDetect 0.7. SVDetect was instructed to compare the two libraries so as to isolate locations with atypical read pairs present in the affected sequence set and absent from the unaffected sequence set. Finally, Circos 0.55 was configured to project the output from SVDetect onto a circular CFHR locus representation (Supplemental Figure 3A). The output from SVDetect was also uploaded onto the Santa Cruz UCSC genome browser so as to compare putative breakpoint locations against known transcripts (Supplemental Figure 3B).
PCR
We designed a genomic PCR that would generate a single 366-bp amplicon in individuals with at least one copy of the CFHR3 and CFHR1 genes using the following primers: forward, 5′CCTTGTTGACTTTCCATCTCG3′; and reverse, 5′AGGAAACCCATCTCATGTGC3′, which targets GCACATGAGATGGGTTTCCT in CFHR3 intron 3–4. In the presence of the CFHR3–1 hybrid gene, an additional 757-bp amplicon is generated using the same forward primer: 5′CCTTGTTGACTTTCCATCTCG3′ and the reverse primer 5′GGTGGCTTATGCCTGCAA3′, which targets TTGCAGGCATAAGCCACC in the forward strand of CFHR1 intron 1–2. PCR was performed using Qiagen HotStar Taq DNA polymerase with 10 ng of genomic DNA according to the manufacturer's protocol. Both reactions were multiplexed in the same reaction. Affected individuals had both a 366-bp and a 757-bp amplicon. In contrast, only the 366-bp control amplicon was seen in all unaffected individuals examined.
Western Blots
Western blot for CFHR proteins was performed using heparin-purified sera from healthy controls and an affected individual (Supplemental Figure 3A). Samples were enriched for CFHR proteins using heparin affinity chromatography and immunoprecipitation. Samples were incubated with heparin-coated sepharose beads (GE Healthcare). After centrifugation and washing, bound proteins were eluted with 500 mM NaCl, 20 mM TRIS-HCl, pH 7.5. Eluted proteins were immunoprecipitated with a mouse monoclonal anti-human CFHR1 antibody (a gift from Dr. Claire Harris, Cardiff University, Wales), which recognizes an epitope common to CFHR1, CFHR2, and CFHR5, and run on standard 12% SDS PAGE under nonreducing conditions and probed with a goat polyclonal anti-human CFH antibody (Quidel).
Peptide Mass Fingerprinting
After electrophoresis, a replicate gel was stained with Coomassie blue and the aberrant bands were manually excised and the gel fragment subjected to peptide mass fingerprinting. Protein samples were reduced and alkylated with iodoacetamide and then digested with trypsin. Peptide mass determination was performed using a Bruker Autoflex III MALDI TOF/TOF instrument by Alphalyse (www.alphalyse.com).
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank the patients and their families.
M.C.P. is a Wellcome Trust Senior Fellow in Clinical Science (WT082291MA) and T.H.M., E.G.d.J., and K.L.R. are funded by this fellowship. Additional support was obtained from the National Institute for Health Research Biomedical Research Centre Funding Scheme. K.A.V. is a Kidney Research UK Clinical Fellow (TF8/2009).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2012020166/-/DCSupplemental.
References
- 1.de Córdoba SR, de Jorge EG: Translational mini-review series on complement factor H: Genetics and disease associations of human complement factor H. Clin Exp Immunol 151: 1–13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davila S, Wright VJ, Khor CC, Sim KS, Binder A, Breunis WB, Inwald D, Nadel S, Betts H, Carrol ED, de Groot R, Hermans PW, Hazelzet J, Emonts M, Lim CC, Kuijpers TW, Martinon-Torres F, Salas A, Zenz W, Levin M, Hibberd ML, International Meningococcal Genetics Consortium : Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet 42: 772–776, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Fakhouri F, Frémeaux-Bacchi V, Noël LH, Cook HT, Pickering MC: C3 glomerulopathy: A new classification. Nat Rev Nephrol 6: 494–499, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McRae JL, Duthy TG, Griggs KM, Ormsby RJ, Cowan PJ, Cromer BA, McKinstry WJ, Parker MW, Murphy BF, Gordon DL: Human factor H-related protein 5 has cofactor activity, inhibits C3 convertase activity, binds heparin and C-reactive protein, and associates with lipoprotein. J Immunol 174: 6250–6256, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Murphy B, Georgiou T, Machet D, Hill P, McRae J: Factor H-related protein-5: A novel component of human glomerular immune deposits. Am J Kidney Dis 39: 24–27, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Gale DP, de Jorge EG, Cook HT, Martinez-Barricarte R, Hadjisavvas A, McLean AG, Pusey CD, Pierides A, Kyriacou K, Athanasiou Y, Voskarides K, Deltas C, Palmer A, Frémeaux-Bacchi V, de Cordoba SR, Maxwell PH, Pickering MC: Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet 376: 794–801, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Köhl J, Zipfel PF, Weber BH, Skerka C: An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet 19: 4694–4704, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Hälbich S, Mihlan M, Schlötzer-Schrehardt U, Zipfel PF, Skerka C: Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114: 2439–2447, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, Meri S, Bergeron J, Zernant J, Merriam J, Gold B, Allikmets R, Dean M, AMD Clinical Study Group : Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann Med 38: 592–604, 2006 [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U: A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet 38: 1173–1177, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Neary J, Dorman A, Campbell E, Keogan M, Conlon P: Familial membranoproliferative glomerulonephritis type III. Am J Kidney Dis 40: E1, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Neary JJ, Conlon PJ, Croke D, Dorman A, Keogan M, Zhang FY, Vance JM, Pericak-Vance MA, Scott WK, Winn MP: Linkage of a gene causing familial membranoproliferative glomerulonephritis type III to chromosome 1. J Am Soc Nephrol 13: 2052–2057, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Francis NJ, McNicholas B, Awan A, Waldron M, Reddan D, Sadlier D, Kavanagh D, Strain L, Marchbank KJ, Harris CL, Goodship TH: A novel hybrid CFH/CFHR3 gene generated by a microhomology-mediated deletion in familial atypical hemolytic uremic syndrome. Blood 119: 591–601, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, Pirson Y, Jackson MS, Hughes A, Wood KM, Goodship JA, Goodship TH: Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med 3: e431, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medvedev P, Stanciu M, Brudno M: Computational methods for discovering structural variation with next-generation sequencing. Nat Methods 6[Suppl]: S13–S20, 2009 [DOI] [PubMed] [Google Scholar]
- 17.Slade I, Stephens P, Douglas J, Barker K, Stebbings L, Abbaszadeh F, Pritchard-Jones K, Cole R, Pizer B, Stiller C, Vujanic G, Scott RH, Stratton MR, Rahman N, FACT collaboration : Constitutional translocation breakpoint mapping by genome-wide paired-end sequencing identifies HACE1 as a putative Wilms tumour susceptibility gene. J Med Genet 47: 342–347, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL: Complement factor H: Sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 37: 41–52, 2000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.