Abstract
Insulin is susceptible to thermal fibrillation, a misfolding process that leads to nonnative cross-b assembly analogous to pathological amyloid deposition. Pharmaceutical formulations are ordinarily protected from such degradation by sequestration of the susceptible monomer within native protein assemblies. With respect to the safety and efficacy of insulin pumps, however, this strategy imposes an intrinsic trade-off between pharmacokinetic goals (rapid absorption and clearance) and the requisite physical properties of a formulation (prolonged shelf life and stability within the reservoir). Available rapid-acting formulations are suboptimal in both respects; susceptibility to fibrillation is exacerbated even as absorption is delayed relative to the ideal specifications of a closed-loop system. To circumvent this molecular trade-off, we exploited structural models of insulin fibrils and amyloidogenic intermediates to define an alternative protective mechanism. Single-chain insulin (SCI) analogs were shown to be refractory to thermal fibrillation with maintenance of biological activity for more than 3 months under conditions that promote the rapid fibrillation and inactivation of insulin. The essential idea exploits an intrinsic incompatibility between SCI topology and the geometry of cross-b assembly. A peptide tether was thus interposed between the A- and B-chains whose length was (a) sufficiently long to provide the “play” needed for induced fit of the hormone on receptor binding and yet (b) sufficiently short to impose a topological barrier to fibrillation. Our findings suggest that ultrastable monomeric SCI analogs may be formulated without protective self-assembly and so permit simultaneous optimization of pharmacokinetics and reservoir life.
Keywords: amyloid, closed-loop system, insulin pump, intraperitoneal pump, pump reservoir
Introduction
Insulin is susceptible to physical and chemical degradation. Physical degradation refers to an irreversible change in physical state without change in covalent structure; the latter is designated chemical degradation. The major barrier to the storage and use of insulin above room temperature (such as in an intraperitoneal pump) is fibrillation, a universal property of polypeptides leading to cross-b linear polymers.1 Studies of fibrillation provide a foundation for the design of ultrastable and ultrafast insulin analogs with potential application to the artificial pancreas project.2,3
Insulin formulations are shipped and stored at 4–8 °C; shelf life is typically 2 years. Degradation accelerates rapidly with increasing temperature. Vials, once used, must be kept below 30 °C and discarded after 30 days (15 days in the case of diluted solutions of Humalog® or Novolog®). Mechanisms of chemical degradation (such as scission of disulfide bonds leading to covalent polymers) are well characterized.4–9 Studies of insulin analogs suggest that flexible regions are particularly susceptible to chemical degradation, and so stabilizing substitutions protect from such processes.10 By contrast, physical degradation is not well understood.1,11 Despite the importance of fibrillation in biotechnology and medicine, how globular proteins form fibrils remains enigmatic. Although molecular models have been proposed,12,13 the structure of an insulin fibril has not been elucidated at atomic resolution.
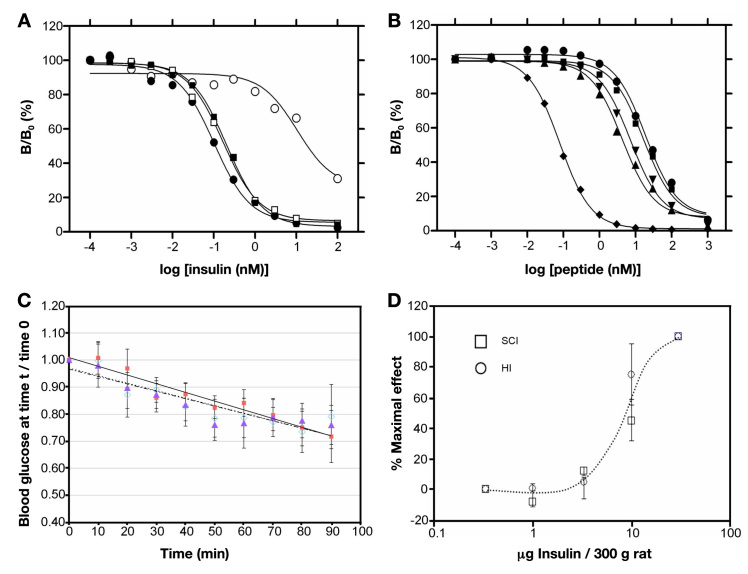
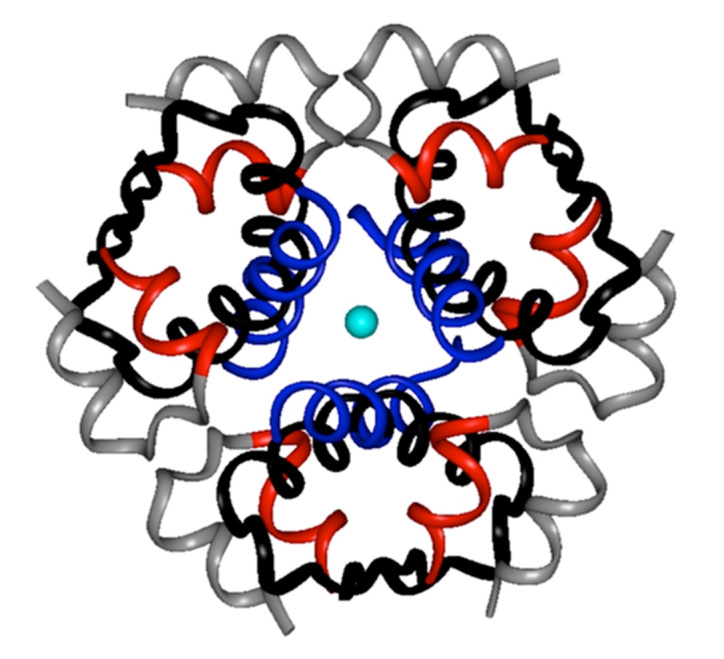
The present study exploited general structural features of an insulin fibril as a basis for rational design. It has been established, for example, that it is the monomeric polypeptide (as distinct from native self-assemblies) that mediates aggregation-coupled misfolding. Accordingly, conditions that favor (or disfavor) native self-assembly are associated with delayed (or accelerated) fibrillation.9 Insulin fibrillation is thus promoted by factors that destabilize its classical pathway of assembly, presumably by increasing the availability of the monomer.14 Fibrillation is further accelerated by partial unfolding as induced by agitation or elevated temperatures.15 The zinc insulin hexamer (depicted in Figure 1) has long provided a vehicle for pharmaceutical formulation due to its stability. Sequestration of the insulin monomer by native self-assembly forestalls partial unfolding and nonnative aggregation (Figure 2).
Figure 1.
Structure of insulin as an R6 hexamer [Protein Data Bank (PDB) entry 1 TRZ]. The two axial zinc ions (center) are shown as over-lying light blue spheres. The A-chains are shown in red (residues A1–A8) and gray (A9–A21); the B-chains are shown in blue (R-specific B1–B8 α-helix) and black (B9–B30). Phenolic ligands are not shown.
Figure 2.
Mechanism of insulin fibrillation via partial unfolding of monomer.28 The native hexamer is protected by self-assembly (far left). Disassembly leads to an equilibrium between native and partially folded monomers (triangle and rhombus, respectively). An engineered linker between A- and B-chains (electric blue) impedes formation of this partial fold, which may otherwise either (i) unfold completely as an off-pathway event (top) or (ii) aggregate to form a nucleus en route to a protofilament (far right). Sufficient play must be present in the linker to permit induced fit on receptor binding (bottom). Figure adapted from Nielsen and colleagues.28
Use of insulin analogs in pumps (such as Humalog or Novolog) was based on the principle that mutational destabilization of subunit interfaces within the hexamer would facilitate absorption.7,16–20 Such analogs have indeed enabled better glycemic control with reduced risk of hypoglycemia.21–23 Although advantageous for pump delivery,21 this strategy enhances rates of misfolding and fibrillation.24,25 Case reports have documented occasional pump occlusion due to fibrillation,25,26 posing a risk of metabolic decompensation. Although ultrarapid formula-tions would enhance the safety and efficacy of closed-loop systems, extension of this mutational strategy encounters a molecular trade-off: accelerated disassembly ordinarily promotes degradation.
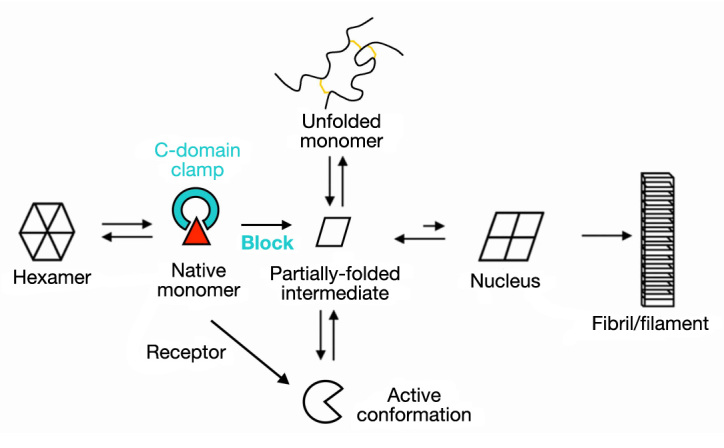
We sought to circumvent the trade-off of conventional prandial insulin design through the use of single-chain insulin (SCI) analogs, also designated miniproinsulins.27 Although SCI analogs are refractory to fibrillation,9,28 their investigation has been discouraged by low activities.29 An extreme combination of these properties is exemplified by SCI-50, a single-chain analog whose crystal structure (Figure 3B) is essentially identical to that of wild-type insulin (Figure 3A).29 Whereas SCI-50 contains a single peptide bond between LysB29 and GlyA1, in wild-type insulin, the C-terminus of the B-chain adjoins (and is often salt-bridged to) the N-terminus of the A-chain. Uniquely among insulin analogs, SCI-50 is both refractory to fibrillation30 and devoid of biological activity.29 The tether between chains presumably constrains conformational changes required in concert for nonnative cross-b assembly and receptor binding.31,32 This approach thus seemed only to replace the trade-off of prandial analog design with another and more severe limitation.
Figure 3.
Molecular structures of insulin and insulin analogs. (A)Ribbon model of an isolated protomer (T-state conformation; PDB entry 4INS). The A-chain is shown in red, and B-chain in blue; disulfide brides are not depicted. (B) Ribbon model of des-ThrB30-SCI-50, an inactive 50-residue single-chain analog in which a peptide bond links LysB29 to GlyA1.29 The structure represents the T-state protomer of a T3Rf3 zinc hexamer (PDB entry 1PID). (C) NMR-derived ensemble of SCI-57 as an engineered monomer in solution.34 The A-chains are shown in red, B-chains in blue, and linkers (sequence GGGPRR) in gray. Coordinates were aligned according to the main-chain atoms of residues B4–B28 and A2–A20. (D) A rotated view of SCI-57 highlights the linker (sticks; electric blue) as a tether between A- and B-domains (space-filling representation). The surface is color-coded according to electrostatic potential (red, electronegative; purple, electropositive; and white, neutral). Images in panels C and D were obtained from PDB entry 2JZQ.
Our interest in the potential therapeutic use of SCI analogs was motivated by two findings. On the one hand, molecular models of insulin fibrils in general posit parallel extension of A- and B-chains with retention of native disulfide pairing;12 in particular, cross-b assembly requires splaying of the C-terminus of the B-chain from the N-terminus of the A-chain (despite their proximity in native structures) by distances greater than 30 Å.30 On the other hand, systematic studies of cross-linked insulin analogs (i.e., containing chemical tethers) have suggested that these termini are between 16 and 24 Å apart in a hormone-receptor complex.33 Together, these findings suggested a reciprocal modulation of fibrillation rate and biological activity as a function of connecting-peptide length: whereas tight tethers (as in SCI-50) would preclude fibrillation at the expense of activity, loose tethers (as in proinsulin) would, at least in part, restore activity at the cost of fibrillation.30 Might there be an optimal intermediate length that is both protective and compatible with native activity? We have described the chemical synthesis and nuclear magnetic resonance (NMR) structure of an SCI of 57 residues (SCI-57) whose connecting peptide lies within the receptor-active distance range (six residues; ∼16–24 Å in length) and whose insulin moiety has been optimized with respect to stability and receptor binding.34 The structure of SCI-57 (Figure 3C) is remarkable for the damping of conformational fluctuations by its tether, packed as a clamp within an interchain crevice (Figure 3D). Here, we demonstrate that SCI-57 retains native potency in a diabetic rat model, exhibits native mitogenicity in a human cancer cell line, and displays extraordinary resistance to chemical and physical degradation as a monomer at an elevated temperature. Together, our findings highlight the promise of intermediate-length SCI analogs as ideal pump insulin analogs.
Methods
Elements of Protein Design
SCI-57 was designed to achieve the following objectives:34
Optimization of α-Helical Stability
Because physical degradation involves a change in conformation from α-helix to β-sheet, we sought to optimize intrinsic helical propensity. Similarly, because rates of chemical degradation are inversely correlated with their thermodynamic stabilities,7 we sought to maximize stability.34
Engineered Monomer
Because insulin absorption is slowed by native assembly, we sought an analog lacking functional dimer-, trimer-, or hexamer-forming surfaces.35
Electrostatic Balance
Because an optimal analog must be stored at high concentration, we sought to maintain a native-like isoelectric point.
Potency
To control glycemia, we sought an analog with potency similar to that of human insulin.
Resistance to Fibrillation
Because cross-b assembly requires splaying of A- and B-chains,12,30,36 we sought an SCI framework as a topo-logical barrier to fibrillation.
Mitogenicity
To reduce the risk of tumorigenesis,37 we sought an analog whose mitogenicity was similar to that of human insulin.
Progress toward the first three objectives was documented in our initial study;34 see also Appendix.
Materials
Insulin and proinsulin were provided by Eli Lilly and Co. (Indianapolis, IN). Insulin-like growth factor-I (IGF-I) was obtained from GroPep, Ltd. (Adelaide, Australia) and thioflavin T (ThT) from Sigma (St. Louis, MO).
Chemical Protein Synthesis
SCI analogs were prepared by native fragment ligation as described.34 [HisA8, AspB10, AspB28, ProB29]-insulin was prepared by chain combination following solid-phase synthesis of the A- and B-chain analogs.32 Predicted molecular masses were verified by mass spectrometry (MS). Purity was >98% as assessed by reverse-phase high-performance liquid chromatography (rp-HPLC).
Mass Spectrometry
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) employed a pulsed nitrogen laser source (l = 337 nm) and acceleration voltage of 28 kV operated in the linear mode (Voyager Biospectro-metry Workstation, PerSeptive Biosystems, Framingham, MA); 100–200 spectra were obtained per sample.
Thermodynamic Modeling
Guanidine denaturation was monitored by circular dichroism (CD) at 222 nm; data were fitted by nonlinear least squares to a two-state model as described.30,38
Preparation of Fibrils
Proteins were made 60 mM in degassed phosphate-buffered saline [PBS; 10 mM phosphate, 140 mM sodium chloride (NaCl), pH 7.4], incubated in presterilized glass vials with airtight sealed caps (Allergy Labs, Oklahoma City, OK), and rocked on a Becton-Dickinson nutator (BD Diagnostic Systems, Franklin Lakes, NJ) at ∼60 rpm. Aliquots, with-drawn with a sterilized single-use syringe, were added to ThT solution for fluorescence assays of fibrillation.
Degradation Assays
Aliquots obtained at indicated time points (10 ml) were mixed with 3 ml ThT assay buffer [5 mM ThT in 50 mM tris(hydroxymethyl)aminomethane, hydrochloride (pH 7.5) and 100 mM NaCl]. Fluorescence emission spectra were collected from 470–500 nm following excitation at 450 nm; ThT in buffer without protein was used as baseline. Changes in covalent structure were evaluated by rp-HPLC and MS.
Modeling of Protofilament
A model of a two-layered insulin protofilament was constructed using InsightII (Accelrys, Inc., San Diego, CA) as described30 based on earlier cryo-electron microscopy studies.12
Receptor Binding
Assays employed lectin-purified and detergent-solubilized insulin receptor [insulin receptor isoform B (IR-B)] or insulin-like growth factor-1 receptor (IGF-1R) as described.39,40 Respective dissociation constants were determined by competitive displacement of 125I-[TyrA14]-insulin or 125I- [Tyr31]-IGF-I (kindly provided by Novo-Nordisk, A/S, Bagsværd, Denmark). Each determination was performed with at least three replicates. The percentage of tracer bound in the absence of competing ligand was <15% to avoid ligand-depletion artifacts.
Mitogenicity
The human breast cancer cell line MCF-7 was maintained in culture essentially as described.37 Medium was changed every 2 days and cells were subcultured when they reached 90% of confluence. Cell proliferation was determined by the incorporation of bromodeoxyuridine (BrdU) using a kit (Roche Diagnostic, Indianapolis, IN). Approximately 3.5 × 103 cells were seeded into each well of a 24-well plate and cultured for 24 h; medium was then replaced with fresh serum-free medium containing 0.1% bovine serum albumin. After 24 h, insulin analogs or IGF-I were added at various concentrations in fresh medium; media were changed each day for 5 days. BrdU incorporation was then determined according to the manufacturer’s directions.
Biological Potency
Male Lewis rats (∼250 g body weight) were rendered diabetic with streptozotocin as described.41 Human insulin and analogs (SCI-57 and a two-chain version lacking the six-residue linker) were purified by HPLC, lyophilized, and dissolved in insulin-related sterile diluent (Eli Lilly and Co., Indianapolis, IN). Rats were injected subcutaneously at time 0 with 1.5 U/kg body weight in 100 ml of diluent. Blood was obtained from clipped tip of the tail at time 0 and every 10 min up to 90 min. Blood glucose was measured using a Hypoguard Advance Micro-Draw (Hypoguard USA, Inc., Minneapolis, MN) meter.
Results
Our study focused on properties pertinent to implantable pumps and closed-loop systems in real-world environ-ments. The thermodynamic stability of SCI-57 at 37 °C (ΔGu 4.3 kcal/mol), as probed by chemical denaturation studies (Figure 4A), is significantly higher than the stabilities of wild-type insulin, KP-insulin, or AspB28-insulin (ΔGu 2.4 kcal/mol; Figure 4B); such stabilities (column two of Table 1) were obtained under zinc-free conditions predictive of relative susceptibility to chemical degradation.6 The net gain in stability (ΔΔGu 1.9 ± 0.2 kcal/mol) presumably reflects the combined effects of stabilizing (HisA8, AspB10, and linker contacts) and destabilizing modifications (AspB28 and ProB29). Loose tethering, as probed by comparison of insulin and proinsulin, resulted in itself in only a small gain in stability (ΔΔGu 0.4 ± 0.2 kcal/mol). In accordance with inferred free-energy values, 50% unfolding of SCI-57 required a higher concentration of guanidine (5.5 ± 0.1 M; column three of Table 1) than did 50% unfolding of proinsulin (4.0 ± 0.2 M), wild-type insulin or prandial insulin analogs (4.1 M).
Figure 4.
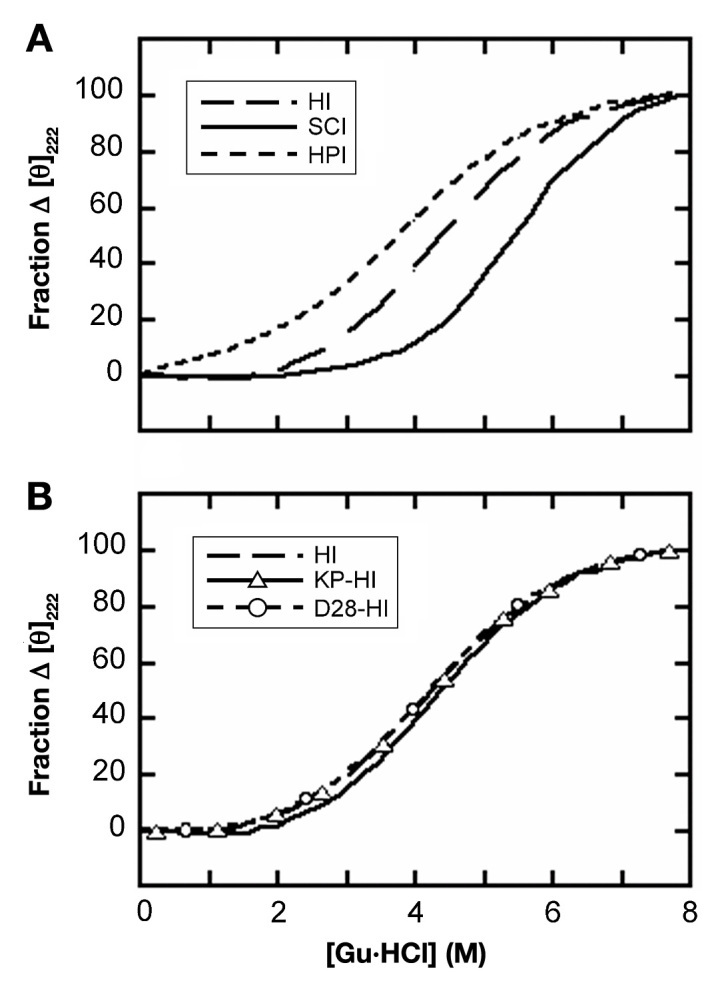
Guanidine (Gu) denaturation studies. (A) Comparison of human insulin (HI), human proinsulin (HPI), and 57-residue single-chain insulin (SCI). (B) Comparison of HI, [LysB28-ProB29]-insulin (KP-HI; the active component of Humalog), and AspB28-insulin (D28-HI; the active component of Novolog). Studies were conducted at 37°C. Thermodynamic parameters are given in Table 1; unfolding was monitored by CD at helix-sensitive wavelength 222 nm.
Table 1.
| Analog | ΔGu (kcal/mol) | Cmid(M) | m(kcal/mol/M) | Fibrillation lag time (days) |
|---|---|---|---|---|
| Human insulin | 2.4 ± 0.1 | 4.1 ± 0.2 | 0.57 ± 0.02 | 3.5 ± 0.6 |
| AspB28-insulinc | 2.4 ± 0.1 | 4.1 ± 0.1 | 0.60 ± 0.01 | 1.7 ± 0.3 |
| KP-insulind | 2.4 ± 0.1 | 4.1 ± 0.1 | 0.59 ± 0.01 | 2.6 ± 0.3 |
| Human proinsulin | 2.8 ± 0.1 | 4.0 ± 0.2 | 0.71 ± 0.03 | 15 ± 2.5 |
| SCI-57 | 4.3 ± 0.1 | 5.5 ± 0.1 | 0.79 ± 0.02 | >180 |
Thermodynamic stabilities were inferred from CD-detected guanidine denaturation studies by application of a two-state model.34 Data are obtained in part from Table 1 of Hua and colleagues.34 Stability (kcal/mol) is defined as the free energy of unfolding (ΔGu) at 37 °C as extrapolated to zero denaturant concentration;38 uncertainties are ±0.1 kcal/mol. Cmid is defined as that molar concentration of guanidine-HCl associated with 50% protein unfolding. The m value (kcal/mol/M) is defined as the slope in plotting the unfolding free energy versus molar concentration of denaturant; this slope is often found to be proportional to the protein surface area exposed on unfolding.
Fibrillation lag times were determined by ThT fluorescence following gentle agitation in the presence of an air-liquid interface as described.30 Proteins were made in 60 mM PBS (pH 7.4) in the absence of zinc ions and gently rocked at 37 °C in a glass vial in the presence of an air-water interface.42
This analog, the active component of Novolog, contains the amino-acid substitution ProB28→Asp.
This analog, the active component of Humalog, contains amino-acid substitutions ProB28→Lys and LysB29→Pro.
Degradation was monitored on gentle agitation in glass vials at 37 °C (Figure 5 and column five of Table 1). Whereas wild-type insulin or prandial analogs were inactivated by fibrillation within 5 days and proinsulin within 15 ± 2.5 days, SCI-57 exhibited no signs of fibrillation up to 180 days (at which point the study was terminated). Analysis of chemical-degradation products by rp-HPLC and MS revealed only a des-amido species, consistent with conversion of Asn→Asp. Despite the harsh conditions employed (elevated temperature and gentle agitation against a nonsilanized glass surface in the presence of an air-liquid interface), no chain cleavage or formation of covalent polymers was detected. Further, no reductions were observed in receptor-binding activity as evaluated at successive time points during the test; representative data are shown in Figure 6A. By contrast, residual binding of the degraded solutions of insulin or prandial analogs was reduced by more than 100-fold.
Figure 5.
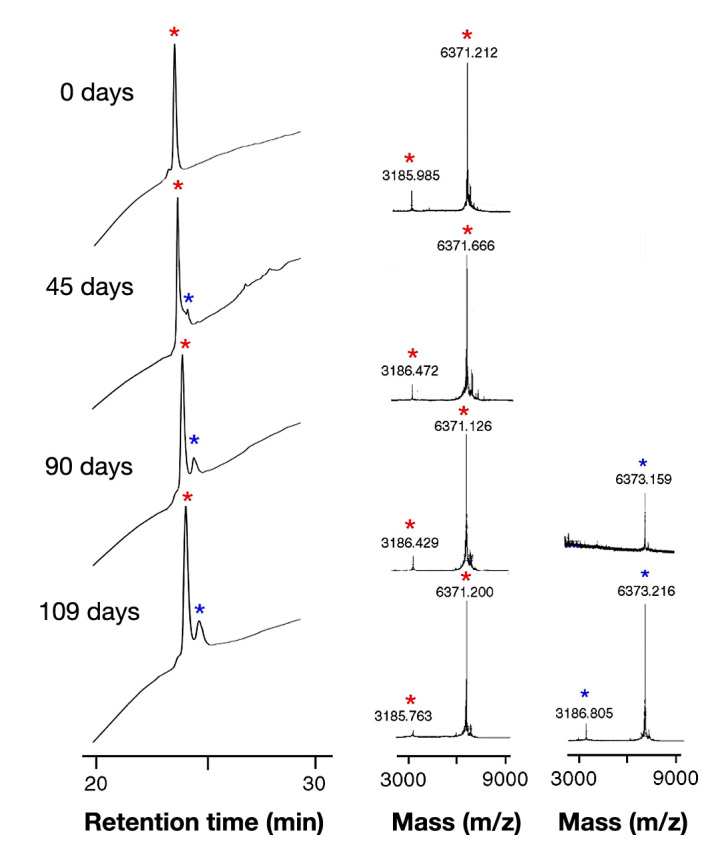
Chemical and physical stability of SCI-57: time course of rp-HPLC chromatogram (left) and MALDI-TOF mass spectrum (right) from 0–109 days. The SCI-57 solution remained clear and without visible precipitation during incubation (see main text). At indicated times (0–109 days), aliquots were applied onto a reverse-phase C8 HPLC column; components were eluted with a methanol gradient. Molecular masses were determined by MS using vendor-provided software (Perkin-Elmer, Wellesley, MA). The difference of 1–2 mass units between the native analog and degradation products represents deamidation (likely to be conversion of AsnA21 to Asp). Respective red and blue asterisks represent major peak (native analog) and minor degradation product. Mass 3186 Da represents the SCI with two charges.
Figure 6.
Receptor-binding studies and biological activity in rats. (A) Stability of the receptor-binding activity of SCI-57 and wild-type insulin on prolonged gentle agitation at 37 °C. The following representative competitive-displacement curves are shown: assays conducted at the start of the incubation (SCI-57 ▪; insulin •), assay performed after 42 days of agitation (HI ∘), or assay performed after 90 days (SCI-57 ▫). Results are expressed as the ratio (B/B0) of 125I-[TyrA14]-insulin specifically bound at a given concentration of cold insulin relative to bound 125I-[TyrA14]-insulin specifically bound in absence of competing unlabeled insulin. (B)Competitive binding of insulin analogs to IGF-1R: SCI-57 (▪), wild-type insulin (●), [HisA8, AspB10, AspB28, ProB29]-insulin (▲), AspB10-insulin (▼), and human IGF-I (♦). (C) Decrease in blood glucose concentration with time (in minutes): regular human insulin (◊), the 57-residue single-chain analog (SCI; ▪), and its two-chain derivative lacking the linker (▲). Initial rates and maximal extents of glucose-lowering activity were indistinguishable. (D) A log plot of concentration-dependent efficacy in the rat shows no difference relative to wild-type insulin.
Because of the presence of AspB10 in SCI-57 and the association of this substitution with increased mitogenicity and carcinogenicity in rats,37 cross-binding to IGF-1R was evaluated in relation to IGF-1, wild-type insulin, AspB10-insulin, and a corresponding two-chain insulin analog lacking a connecting domain ([HisA8, AspB10, AspB28, ProB29]-insulin) (Figure 6B). In accordance with earlier studies,40 baseline affinities (dissociation constants Kd) of IGF-1 and wild-type insulin were 0.041 ± 0.005 and 9.57 ± 0.31 nM, respectively. As expected, AspB10-insulin and [HisA8, AspB10, AspB28, ProB29]-insulin exhibited augmented binding to IGF-1R (Kd 2.7 ± 0.003 and 4.31 ± 0.006 nM, respectively). Remarkably, interposition of the six-residue linker reduced IGF-1R binding to a level lower than that of wild-type insulin (Kd 9.89 ± 0.035 nM). Mitogenicity was directly evaluated in cell culture employing human breast-cancer cell line MCF-7. Whereas mitogenicities of AspB10-insulin and [HisA8, AspB10, AspB28, ProB29]-insulin were intermediate between those of wild-type insulin and IGF-1, as probed by both cell number and DNA content, no significant differences were observed between SCI-57, wild-type insulin, and KP-insulin (data not shown). Thus, the linker may be regarded as a tuning element that downregulates potential mitogenicity.
The biological potency of SCI-57 was assessed in a streptozotocin-induced rat model of diabetes mellitus. No significance differences were observed between SCI-57 and human insulin with respect to initial rate of fall in blood glucose concentration (Figure 6C) and dose-response relationship (Figure 6D). In this model, Humulin®-R U-100 (Eli Lilly and Co.) insulin and Humalog exhibit indistinguishable pharmacodynamics (presumably due to differences between rat skin and human skin and to differences in subcutaneous depot sizes) but each is markedly more rapid than Lantus® (Sanofi, Paris, France) or neutral protamine Hagedorn (NPH) insulin (Humulin N; Eli Lilly and Co.).
Discussion
Insulin functions in the bloodstream as a monomer, and yet it is the monomer that is susceptible to fibrillation and most forms of chemical degradation.9 Degradation in vials and delivery devices has long been delayed through protective self-assembly. Use of Zn2+-coordinated insulin hexamers as a formulation vehicle recapitulates the logic of the pancreatic β-cell whose secretory granules contain microcrystalline arrays of zinc insulin hexamers.43 X-ray structures of such hexamers are of exquisite beauty (Figure 1). Multiple crystal forms have been described in vitro, defining three structural families (T6, T3Rf3, and R6). The T-state (Figure 3A) resembles the structure of an insulin monomer in solution.31,44,45
Absorption of regular insulin is limited by the kinetic lifetime of the Zn-insulin hexamer as disassembly to smaller dimers and monomers enables capillary absorption.35 The essential idea underlying design of Humalog and Novolog was thus to accelerate disassembly. This was accomplished by destabilization of the classical dimer-forming surface (the C-terminal antiparallel β-sheet). Humalog contains substitutions ProB28→Lys and LysB29→Pro, an inversion that mimics the sequence of IGF-I; Novolog contains substitution ProB28→Asp.46,47 Although the substitutions impair dimerization, the analogs are competent for assembly of a phenol-stabilized zinc hexamer.47 This assembly protects the analog from fibrillation in the vial but following injection, the hexamer rapidly dissociates. The instability of these analogs on dilution underlies their reduced shelf life after opening, especially on dilution. In addition to effects of reduced self-assembly, the present studies have shown that zinc-free monomers of AspB28-insulin and KP-insulin at 37 °C are intrinsically more susceptible to fibrillation than is wild-type insulin. Because of the importance of formulation stability, a molecular trade-off is thus posed by the use of destabilizing mutations to enhance rates of absorption.
A NMR study of an engineered insulin monomer demonstrated that the N-terminal A-chain segment (residues A1–A8), although a canonical α-helix in crystal structures (red in Figure 1), exhibits anomalous 13C chemical shifts in solution.41 These findings provided evidence that, in the absence of self-assembly, this segment undergoes significant conformational fluctuations. This interpretation was supported by analysis of amide-proton exchange rates in D2O: protection factors in this segment were observed to be negligible.48 Such anomalous NMR features suggest that the distorted monomeric conformation mediating formation of an amyloidogenic nucleus (trapezoid in Figure 2) contains a partial fold in which the N-terminal segment of the A-chain is poorly ordered (central panel of Figure 7A). Solid-state NMR studies have shown that in a fibril, this segment (like most of the A- and B-chains) forms a β-sheet (Figure 7B). Because retention of native disulfide pairing imposes a parallel orientation between chains,12 this class of models requires splaying of the two chains with distances between the C-terminus of the B-chain and N-terminus of the A-chain greater than 30 Å (Figure 7C).30 It may therefore be of critical importance that the length of the six-residue linker of SCI-57, even at maximal extension, is constrained by covalent structure to be less than 24 Å.
Figure 7.
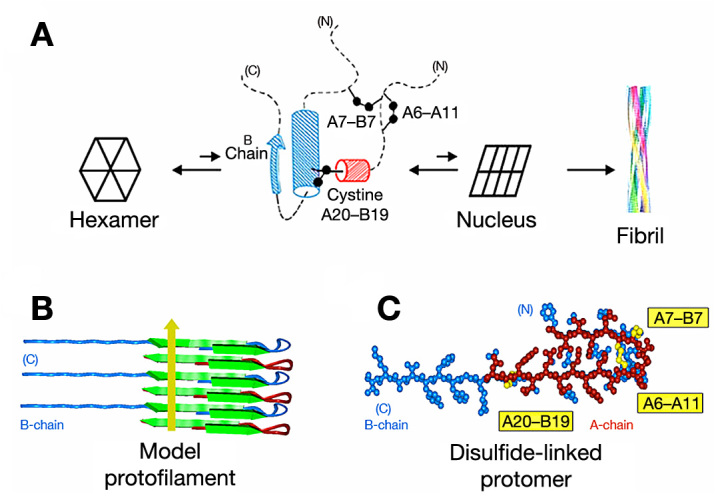
Structural model of insulin protofibrils. (A) Structures at successive stages of insulin fibrillation. Hexameric insulin is shown in cartoon form at far left. Putative amyloidogenic intermediate (cylinder model)30 and unknown nucleus (cartoon) are shown at center. Model of an insulin fibril12 is shown at far right. In the partial fold, cylinders and the arrow indicate molten supersecondary structure (residues B9–B26 and A16–A20). Dashed lines indicate disordered regions; disulfide bridges are indicated by black balls (sulfur atoms). (B) Schematic representation of stacked insulin main chains in a protofilament as proposed by Jimenez and colleagues.12 The cross-b unit consists of alternatively stacked A- and B-chains (red and blue, respectively) to form in-register parallel β-sheets (segments A1–A6, A12–A21, B1–B6, and B12–B30). The yellow arrow indicates direction of long axis of fibril. (C) An insulin protomer in a protofilament (as viewed down its long axis) is predicted to exhibit an extended polypeptide conformation with central bend; disulfide bridges are in yellow. Models were constructed wherein two β-strand regions of the A-chain (A1–A5 and A12–A21) were built with dihedral angles f and y set at -140 and 140 °, respectively; corresponding B-chain segments (B1–B5 and B12–B30) were likewise constructed, and the two chains connected by disulfide bridges (A7–B7 and A20–B19). Interchain β-sheet-related hydrogen bonds were then imposed by time-averaged simulated-annealing.49
The present study suggests that monomeric SCI analogs may be engineered to exhibit a combination of favorable features and thereby circumvent the trade-off of conventional prandial insulin analog design. We envisage that the foreshortened connecting domain acts as a clamp to stabilize the α-helical conformation of the N-terminal A-chain segment and to disallow splaying of the chains as required for cross-b assembly. Potential advantageous features promise to combine native potency and mitogenicity with marked resistance to chemical and physical degradation as encountered in the reservoir of an insulin pump under real-world conditions. Although such extensive modification of the insulin molecule may in principle be precluded by immunogenicity in patients on long-term use (an open question), we envisage that an engineered ultrastable monomeric SCI analog may yield ultrafast rates of absorption, enhancing the efficacy and safety of closed-loop systems.50 Further studies of the pharmacokinetic and immunogenic properties of SCI analogs in large animal models and healthy human volunteers will be required to test these ideas in an effort to reduce this molecular technology to practice.
Acknowledgments
We thank Ms. Martha Raymond and Ms. Crista Moeller for assistance in preparation of the manuscript. This work was supported by grants from the National Institutes of Health (NIH DK079233 and DK040949) and the American Diabetes Association to two of the authors (Michael Weiss and Nelson Phillips; ADA 1-08-RA-218 and 1-08-RA-149). We thank Drs. K. Huang and Q.X. Hua for participation in early stages of this work and assistance with preparation of figures; S. Nakagawa for chemical protein synthesis; B. Li for MCF-7 cellular assays; W. Jia and L. Whittaker for technical assistance; Y.S. Chen and N. Stokes for assistance with figures; Dr. Y. Yang for management of the NMR facilities; and Prof. G. G. Dodson (University of York, UK), Prof. S. B. Kent (University of Chicago, USA) and Dr. R. Tycko (NIH, USA) for helpful discussion. This article is a contribution from the Cleveland Center for Membrane and Structural Biology and Case Western Reserve University Comprehensive Cancer Center.
Appendix
Summary of SCI-57 Design and Prior Characterization
SCI-57 contains two known helix-stabilizing substitutions. One (ThrA8→His)1 enhances the C-terminal residue (C-cap)2 of the N-terminal A-chain α-helix;3,4 the other (HisB10→Asp)5 enhances the N-terminal residue (N-cap)2 of the central B-chain α-helix.3,6 HisA8 and AspB10 singly and in combination augment the affinity of two-chain insulin analogs for the insulin receptor.3–5 In the context of SCI-57, interposition of the six-residue linker offsets such augmentation, returning receptor-binding affinity (as measured using human placental membranes) to a near-native value.7 Whereas AspB10 blocks the trimer-forming surface of insulin and prevents HisB10-mediated binding of zinc ions, the dimer-forming surface is destabilized by the additional substitutions ProB28→Asp (as in Novolog) and LysB29→Pro (as in Humalog). 1H-NMR studies demonstrated a pattern of chemical shifts characteristic of an insulin monomer; dimer- or trimer-related Nuclear Overhauser Effects8 were not observed.7 An accounting of formal charges predicted a shift of -1 in net charge at neutral pH. This calculation highlights the loss of one basic residue (LysB29) in the context of otherwise offsetting modifications, i.e., a gain of two basic residues (in linker sequence GGGPRR) offset by two acidic residues (AspB10 and AspB28), gain of a titratable imidazolic group (HisA8) offset by loss of HisB10, and the balanced elimination of opposing chain termini (A1 a-amino group and B30 a-carboxylate). Isoelectric focusing studies revealed an isoelectric point of 5.3, indeed lower than that of wild-type insulin (pI 5.8).7
Table A1.
Mitogenicity Assaya
| A. Relative to IGF-I | |
|---|---|
| IGF-I:AspB10-insulin | p = .040 |
| IGF-I:KP-insulin | p = .021 |
| IGF-I:SCI-57 | p = .028 |
| IGF-I:two-chain controlb | p = .041 |
| IGF-I:insulin | p = .031 |
| B. Relative to wild-type insulin | |
|---|---|
| Insulin:AspB10-insulin | p = .051 |
| Insulin:KP-insulin | p = .068 |
| Insulin:SCI-57 | p = .091 |
| Insulin:two-chain controlb | p = .049 |
Statistical analysis of BrdU studies as depicted in Figure A1.
Two-chain control designates [HisA8, AspB10, AspB28, ProB29]-insulin, corresponding to the insulin core of SCI-57.
Figure A1.
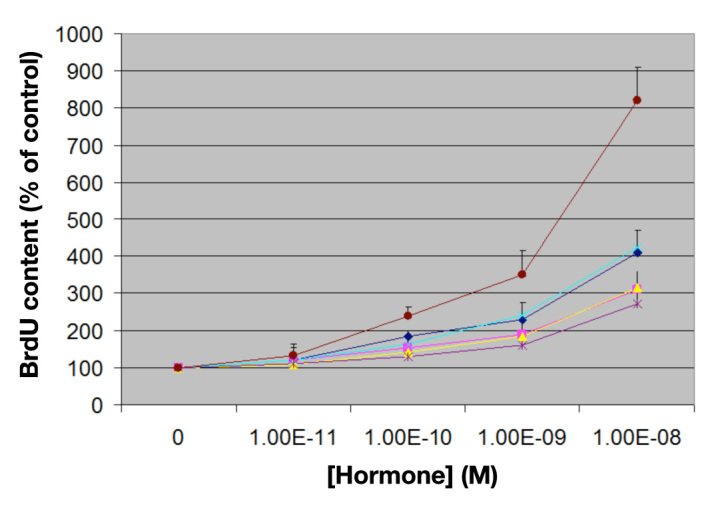
Mitogenicity studies of MCF-7 human breast-cancer-derived cell line. Relative changes in BrdU content after 5 days in culture (vertical axis) are shown as a function of hormone concentration (horizontal axis). From top to bottom: (maroon) human IGF-I; (light blue) AspB10-insulin; (dark blue) [HisA8, AspB10, AspB28, ProB29]-insulin; (yellow) SCI-57; (pink) KP-insulin; and (purple) wild-type insulin. Error bars indicate standard deviation of triplicate determinations. Statistical p values are provided in Table A1. [HisA8, AspB10, AspB28, ProB29]-insulin corresponds to the insulin core of SCI-57 and so provides a two-chain control. KP-insulin designates [LysB28, ProB29]-insulin, the active component of Humalog.
Appendix References:
- 1.Chu YC, Burke GT, Gammeltoft S, Chan SJ, Steiner DF, Katsoyannis PG. High-potency hybrid compounds related to insulin and amphioxus insulin-like peptide. Biochemistry. 1994;33(44):13087–13092. doi: 10.1021/bi00248a018. [DOI] [PubMed] [Google Scholar]
- 2.Doig AJ, Baldwin RL. N- and C-capping preferences for all 20 amino acids in alpha-helical peptides. Protein Sci. 1995;4(7):1325–1336. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaarsholm NC, Norris K, Jorgensen RJ, Mikkelsen J, Ludvigsen S, Olsen OH, Sørensen AR, Havelund S. Engineering stability of the insulin monomer fold with application to structure-activity relationships. Biochemistry. 1993;32(40):10773–10778. doi: 10.1021/bi00091a031. [DOI] [PubMed] [Google Scholar]
- 4.Wan Z, Xu B, Huang K, Chu YC, Li B, Nakagawa SH, Qu Y, Hu SQ, Katsoyannis PG, Weiss MA. Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues. Biochemistry. 2004;43(51):16119–16133. doi: 10.1021/bi048223f. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz GP, Burke GT, Katsoyannis PG. A superactive insulin: [B10-aspartic acid]insulin(human) Proc Natl Acad Sci USA. 1987;84(18):6408–6411. doi: 10.1073/pnas.84.18.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss MA, Hua QX, Jia W, Nakagawa SH, Chu YC, Katsoyannis PG. Activities of insulin analogues at position A8 are uncorrelated with thermodynamic stability. In: Dieken ML, Federwisch M, De Meyts P, Wollmer A, editors. Insulin and related proteins: structure to function and pharmacology. Dordrecht (The Netherlands): Kluwar Academic Publishers; 2002. pp. 103–119. [Google Scholar]
- 7.Hua QX, Nakagawa SH, Jia W, Huang K, Phillips NB, Hu SQ, Weiss MA. Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications. J Biol Chem. 2008;283(21):14703–14716. doi: 10.1074/jbc.M800313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby E, Hua QX, Stern AS, Frank BH, Weiss MA. Structure and dynamics of a protein assembly. 1H-NMR studies of the 36 kDa R6 insulin hexamer. J Mol Biol. 1996;258(1):136–157. doi: 10.1006/jmbi.1996.0239. [DOI] [PubMed] [Google Scholar]
Abbreviations
- (BrdU)
bromodeoxyuridine
- (CD)
circular dichroism
- (Gu)
guanidine
- (HI)
human insulin
- (HPI)
human proinsulin
- (HPLC)
high-performance liquid chromatography
- (IGF-1R)
insulin-like growth factor-1 receptor
- (IGF-I)
insulin-like growth factor-I
- (IR-B)
insulin receptor isoform B
- (MALDI-TOF MS)
matrix-assisted laser desorption/ionization-time of flight mass spectrometry
- (MS)
mass spectrometry
- (NaCl)
sodium chloride
- (NMR)
nuclear magnetic resonance
- (NPH)
neutral protamine Hagedorn
- (PBS)
phosphate-buffered saline
- (PDB)
Protein Data Bank
- (rp-HPLC)
reverse-phase high-performance liquid chromatography
- (SCI)
single-chain insulin
- (ThT)
thioflavin T
Disclosures
The intellectual property pertaining to the present single-chain insulin analogs is owned by Case Western Reserve University and licensed to Thermalin Diabetes, LLC. Michael Weiss owns stock in this company, serves as Chief Scientific Officer, and is a member of its Board of Directors. Michael Weiss has also served as a consultant to Merck and the DEKA Research and Development Corporation. Nelson Phillips and Jonathan Whittaker are consultants to Thermalin Diabetes, LLC; Faramarz Ismail-Beigi owns stock in Thermalin Diabetes, LLC. Jonathan Whittaker owns stock in Novo-Nordisk A/S.
References
- 1.Dobson CM, Karplus M. The fundamentals of protein folding: bringing together theory and experiment. Curr Opin Struct Biol. 1999;9(1):92–101. doi: 10.1016/s0959-440x(99)80012-8. [DOI] [PubMed] [Google Scholar]
- 2.Brunetti P, Orsini Federici M, Massi Benedetti M. The artificial pancreas. Artif Cells Blood Substit Immobil Biotechnol. 2003;31(2):127–138. doi: 10.1081/bio-120020169. [DOI] [PubMed] [Google Scholar]
- 3.Hovorka R. Continuous glucose monitoring and closed-loop systems. Diabet Med. 2006;23(1):1–12. doi: 10.1111/j.1464-5491.2005.01672.x. [DOI] [PubMed] [Google Scholar]
- 4.Brange J, Langkjaer L, Havelund S, Vølund A. Chemical stability of insulin. 1. Hydrolytic degradation during storage of pharmaceutical preparations. Pharm Res. 1992;9(6):715–726. doi: 10.1023/a:1015835017916. [DOI] [PubMed] [Google Scholar]
- 5.Brange J, Havelund S, Hougaard P. Chemical stability of insulin. 2. Formation of higher molecular weight transformation products during storage of pharmaceutical preparations. Pharm Res. 1992;9(6):727–734. doi: 10.1023/a:1015887001987. [DOI] [PubMed] [Google Scholar]
- 6.Brange J, Langkjaer L. Chemical stability of insulin. 3. Influence of excipients, formulation, and pH. Acta Pharm Nord. 1992;4(3):149–158. [PubMed] [Google Scholar]
- 7.Brange J. Chemical stability of insulin. 4. Mechanisms and kinetics of chemical transformations in pharmaceutical formulation. Acta Pharm Nord. 1992;4(4):209–222. [PubMed] [Google Scholar]
- 8.Brange J, Hallund O, Sorensen E. Chemical stability of insulin. 5. Isolation, characterization and identification of insulin transforma-tion products. Acta Pharm Nord. 1992;4(4):223–232. [PubMed] [Google Scholar]
- 9.Brange J, Andersen L, Laursen ED, Meyn G, Rasmussen E. Toward understanding insulin fibrillation. J Pharm Sci. 1997;86(5):517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 10.Brange J, Langkjaer L. Insulin formulation and delivery. Pharm Biotechnol. 1997;10:343–409. doi: 10.1007/0-306-46803-4_13. [DOI] [PubMed] [Google Scholar]
- 11.Murphy RM. Peptide aggregation in neurodegenerative disease. Annu Rev Biomed Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99(14):9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brange J, Dodson GG, Edwards DJ, Holden PH, Whittingham JL. A model of insulin fibrils derived from the x-ray crystal structure of a monomeric insulin (despentapeptide insulin) Proteins. 1997;27(4):507–516. [PubMed] [Google Scholar]
- 15.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, Uversky VN, Fink AL. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40(20):6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 16.DiMarchi RD, Mayer JP, Fan L, Brems DN, Frank BH, Green JK, Hoffman JA, Howey DC, Long HB, Shaw WN, Shields JE, Slieker LJ, Su KSE, Sundell KL, Chance RE. Synthesis of a fast-acting insulin based on structural homology with insulin-like growth factor I. In: Smith JA, Rivier JE, editors. Peptides: Proceedings of the Twelfth American Peptide Symposium. Leiden (The Netherlands): ESCOM Science Publishers; 1992. pp. 26–28. [Google Scholar]
- 17.Howey DC, Bowsher RR, Brunelle RL, Woodworth JR. [Lys(B28), Pro(B29)]-human insulin. A rapidly absorbed analogue of human insulin. Diabetes. 1994;43(3):396–402. doi: 10.2337/diab.43.3.396. [DOI] [PubMed] [Google Scholar]
- 18.Brange J. The new era of biotech insulin analogues. Diabetologia. 1997;40(Suppl 2):S48–S53. doi: 10.1007/s001250051400. [DOI] [PubMed] [Google Scholar]
- 19.Brange J, Vølund A. Insulin analogs with improved pharmaco-kinetic profiles. Adv Drug Deliv Rev. 1999;35(2–3):307–335. doi: 10.1016/s0169-409x(98)00079-9. [DOI] [PubMed] [Google Scholar]
- 20.DeFelippis MR, Chance RE, Frank BH. Insulin self-association and the relationship to pharmacokinetics and pharmacodynamics. Crit Rev Ther Drug Carrier Syst. 2001;18(2):201–264. [PubMed] [Google Scholar]
- 21.Raskin P, Holcombe JH, Tamborlane WV, Malone JI, Strowig S, Ahern JA, Lavent F. A comparison of insulin lispro and buffered regular human insulin administered via continuous subcutaneous insulin infusion pump. J Diabetes Complications. 2001;15(6):295–300. doi: 10.1016/s1056-8727(01)00168-4. [DOI] [PubMed] [Google Scholar]
- 22.Vajo Z, Fawcett J, Duckworth WC. Recombinant DNA technology in the treatment of diabetes: insulin analogs. Endocr Rev. 2001;22(5):706–717. doi: 10.1210/edrv.22.5.0442. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch IB. Insulin analogues. N Engl J Med. 2005;352(2):174–183. doi: 10.1056/NEJMra040832. [DOI] [PubMed] [Google Scholar]
- 24.Lougheed WD, Zinman B, Strack TR, Janis LJ, Weymouth AB, Bernstein EA, Korbas AM, Frank BH. Stability of insulin lispro in insulin infusion systems. Diabetes Care. 1997;20(7):1061–1065. doi: 10.2337/diacare.20.7.1061. [DOI] [PubMed] [Google Scholar]
- 25.Wright AW, Little JA. Cannula occlusion with use of insulin lispro and insulin infusion system. Diabetes Care. 1998;21(5):874–875. doi: 10.2337/diacare.21.5.874a. [DOI] [PubMed] [Google Scholar]
- 26.Wolpert HA, Faradji RN, Bonner-Weir S, Lipes MA. Metabolic decompensation in pump users due to lispro insulin precipitation. BMJ. 2002;324(7348):1253. doi: 10.1136/bmj.324.7348.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steiner DF, Clark JL. The spontaneous reoxidation of reduced beef and rat proinsulins. Proc Natl Acad Sci USA. 1968;60(2):622–629. doi: 10.1073/pnas.60.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen L, Frokjaer S, Brange J, Uversky VN, Fink AL. Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry. 2001;40(28):8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
- 29.Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Bing X, Markussen J. X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J Mol Biol. 1991;220(2):425–433. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang K, Maiti NC, Phillips NB, Carey PR, Weiss MA. Structure-specific effects of protein topology on cross-beta assembly: studies of insulin fibrillation. Biochemistry. 2006;45(34):10278–10293. doi: 10.1021/bi060879g. [DOI] [PubMed] [Google Scholar]
- 31.Hua QX, Shoelson SE, Kochoyan M, Weiss MA. Receptor binding redefined by a structural switch in a mutant human insulin. Nature. 1991;354(6350):238–241. doi: 10.1038/354238a0. [DOI] [PubMed] [Google Scholar]
- 32.Hua QX, Jia WH, Bullock BP, Habener JF, Weiss MA. Transcriptional activator-coactivator recognition: nascent folding of a kinase-inducible transactivation domain predicts its structure on coactivator binding. Biochemistry. 1998;37(17):5858–5866. doi: 10.1021/bi9800808. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa SH, Tager HS. Perturbation of insulin-receptor interactions by intramolecular hormone cross-linking. Analysis of relative movement among residues A1, B1, and B29. J Biol Chem. 1989;264(1):272–279. [PubMed] [Google Scholar]
- 34.Hua QX, Nakagawa SH, Jia W, Huang K, Phillips NB, Hu SQ, Weiss MA. Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications. J Biol Chem. 2008;283(21):14703–14716. doi: 10.1074/jbc.M800313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brange J, Ribel U, Hansen JF, Dodson G, Hansen MT, Havelund S, Melberg SG, Norris F, Norris K, Snel L, Sørensen AR, Voigt HO. Monomeric insulins obtained by protein engineering and their medical implications. Nature. 1988;333(6174):679–682. doi: 10.1038/333679a0. [DOI] [PubMed] [Google Scholar]
- 36.Choi JH, May BC, Wille H, Cohen FE. Molecular modeling of the misfolded insulin subunit and amyloid fibril. Biophys J. 2009;97(12):3187–3195. doi: 10.1016/j.bpj.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milazzo G, Sciacca L, Papa V, Goldfine ID, Vigneri R. ASPB10 insulin induction of increased mitogenic responses and phenotypic changes in human breast epithelial cells: evidence for enhanced interactions with the insulin-like growth factor-I receptor. Mol Carcinog. 1997;18(1):19–25. doi: 10.1002/(sici)1098-2744(199701)18:1<19::aid-mc3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 38.Sosnick TR, Fang X, Shelton VM. Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 39.Whittaker J, Whittaker L. Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J Biol Chem. 2005;280(22):20932–20936. doi: 10.1074/jbc.M411320200. [DOI] [PubMed] [Google Scholar]
- 40.Zhao M, Wan ZL, Whittaker L, Xu B, Phillips NB, Katsoyannis PG, Ismail-Beigi F, Whittaker J, Weiss MA. Design of an insulin analog with enhanced receptor binding selectivity: rationale, structure, and therapeutic implications. J Biol Chem. 2009;284(46):32178–32187. doi: 10.1074/jbc.M109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Petkova A, Huang K, Xu B, Hua QX, Ye IJ, Chu YC, Hu SQ, Phillips NB, Whittaker J, Ismail-Beigi F, Mackin RB, Katsoyannis PG, Tycko R, Weiss MA. An Achilles’ heel in an amyloidogenic protein and its repair: insulin fibrillation and therapeutic design. J Biol Chem. 2010;285(14):10806–10821. doi: 10.1074/jbc.M109.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang K, Dong J, Phillips NB, Carey PR, Weiss MA. Proinsulin is refractory to protein fibrillation. Topological protection of a precursor protein from cross-beta assembly. J Biol Chem. 2005;280:42345–42355. doi: 10.1074/jbc.M507110200. [DOI] [PubMed] [Google Scholar]
- 43.Michael J, Carroll R, Swift HH, Steiner DF. Studies on the molecular organization of rat insulin secretory granules. J Biol Chem. 1987;262(34):16531–16535. [PubMed] [Google Scholar]
- 44.Hua QX, Hu SQ, Frank BH, Jia W, Chu YC, Wang SH, Burke GT, Katsoyannis PG, Weiss MA. Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue. J Mol Biol. 1996;264(2):390–403. doi: 10.1006/jmbi.1996.0648. [DOI] [PubMed] [Google Scholar]
- 45.Olsen HB, Ludvigsen S, Kaarsholm NC. Solution structure of an engineered insulin monomer at neutral pH. Biochemistry. 1996;35(27):8836–8845. doi: 10.1021/bi960292+. [DOI] [PubMed] [Google Scholar]
- 46.Dodson GG, Dodson EJ, Turkenburg JP, Bing X. Molecular recognition in insulin assembly. Biochem Soc Trans. 1993;21(3):609–614. doi: 10.1042/bst0210609. [DOI] [PubMed] [Google Scholar]
- 47.Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin. Structure. 1995;3(6):615–622. doi: 10.1016/s0969-2126(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 48.Hua QX, Jia W, Weiss MA. Conformational dynamics of insulin. Front Endocrin. 2011;2:48. doi: 10.3389/fendo.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Renard E. Implantable closed loop glucose sensing and insulin delivery: The future for insulin pump therapy. Curr Opin Pharmacol. 2002;2:708–716. doi: 10.1016/s1471-4892(02)00216-3. [DOI] [PubMed] [Google Scholar]