Abstract
Peptides, peptidomimetics, and peptide derivatives that self-assemble into fibrillar gels have received increasing interest as synthetic extracellular matrices for applications in 3D cell culture and regenerative medicine. Recently, several of these fibrillizing molecules have been functionalized with bioactive components such as cell-binding ligands, degradable sequences, drug-eluting compounds, and chemical modifications for cross-linking, producing gels that can reliably display multiple factors simultaneously. This capacity for incorporating precise levels of many different biological and chemical factors is advantageous given the natural complexity of cell-matrix interactions that many current biomaterial strategies seek to mimic. In this review, recent efforts in the area of fibril-forming peptide materials are described, and advantages of biomaterials containing multiple modular elements are outlined. In addition, a few hurdles and open questions surrounding fibrillar peptide gels are discussed, including issues of the materials’ structural heterogeneity, challenges in fully characterizing the diversity of their self-assembled structures, and incomplete knowledge of how the materials are processed in vivo.
Introduction
Biomaterial scaffolds in regenerative medicine, tissue engineering, and defined cell culture systems seek to recapitulate the function of natural extracellular matrices (ECMs) by providing supramolecular frameworks capable of bringing about desired cellular or tissue-level responses. However, ECMs are elusive design targets for biomaterial engineers because they are tremendously multifunctional, dynamic, and not fully characterized. ECMs exert their effects on cells and tissues through highly variable cell-matrix binding interactions, mechanical signaling, the controlled diffusion of soluble factors, the spatial and temporal organization of each of these aspects, and interactions with immune and inflammatory processes. It is the summation and integration of all of these signals together that drives cell behavior.1 This presents a great challenge for engineering synthetic scaffolds that likewise can direct specific biological processes.2 Creating synthetic materials that can incorporate many relevant signals and factors in a precise manner is challenging, as is designing systems where each factor can be independently adjusted in order to systematically elucidate complex combinations that promote specific biological responses of interest.
To address this challenge, several strategies for constructing synthetic ECMs from synthetic fibrillizing components have emerged recently.2–8 In general, each of these approaches is characterized by the design of a peptide, peptidomimetic, or peptide derivative that can self-assemble into fibrillar gel networks. Following the design of a basic peptide building block, functional variants of the original are designed, with each variant possessing a unique biological or structural activity. Collectively, these approaches have advantages over polymeric or other covalently built synthetic ECMs in that once a robust self-assembling base material is designed, many different modifications of the base material (e.g. containing ligands, cross-linking domains, degradable sequences, drug-releasing components, etc.) can then be co-assembled into integrated multi-functional scaffolds. Because the materials are constructed non-covalently, the different factors can in principle be explored as combinations much more efficiently than within covalent or polymerized biomaterials. Given the complexity of biological responses to biomaterials and ECMs, this modularity is advantageous. In this review, we will highlight recent advances in the development of fibrillizing peptide gels and discuss the characterization of these materials both in vitro and within physiological environments. We will then summarize encouraging recent successes of this class of materials in vivo and discuss a few challenges that remain for employing them in biomedical applications. Other specific and complementary aspects of these materials are covered in greater detail in other recent reviews, including those focused on stimulus-responsiveness,9 protein and therapeutic delivery from self-assembled scaffolds,3,10 polypeptide-based materials,4–7 peptide-amphiphiles,11 nanofibrous biomaterials,8,12,13 and modular biomaterials.2,6
Expanding the limits of multifunctionality through non-covalent co-assembly
Peptide-based fibrillizing materials (Figure 1) can be exploited for their modular and multifunctional potential. One benefit of modularity is that different molecular features may be varied independently or in conjunction with each other without having to extensively redesign or re-characterize the material. For example, different cell adhesion ligands could be interchanged with other bioactive amino acid sequences or chemical functionalities without significantly altering more global aspects of the material such as fiber size, gelation kinetics, or compliance, allowing one to conclusively relate differences in cell behavior with the presence of a specific ligand.2 Recently, steps have been taken to refine such modularity in self-assembling materials, focusing on different cell-binding ligands, mechanical factors, enzymatically cleavable domains, and controllably released soluble factors. A range of specific behaviors including attachment, migration, proliferation, and more cell type-specific differentiation behaviors have been investigated and are discussed in the sections below.
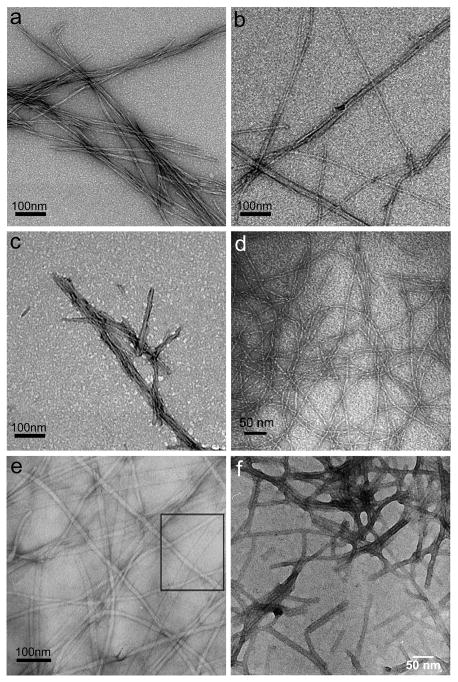
Figure 1.
Negative-stained TEM images of β-rich fibrils from different peptides, PAs, and peptide derivatives. (a) fibrils formed from the peptide Q11; (b) fibrils formed from the peptide RGDS-Q11; (c) fibrils formed from the thiol-presenting peptide Cys-SGSG-Q11; (d) fibrils formed by the β-hairpin peptide MAX1;103 (e) fibrils formed by mixtures of two aromatic short peptides, Fmoc-FF and Fmoc-RGD;60 (f) fibrils formed by a PA terminated with a cyclic RGD sequence.50 (d) is reprinted with permission from Ozbas et al. Macromolecules 2004, 37, 7331–7337; (e) is reprinted with permission from Zhou et al. Biomaterials 2009, 30, 2523–2530; (f) is reprinted with permission from Guler et al. Biomacromolecules 2006, 7, 1855–1863.
Underivatized self-assembling peptides
Synthetic peptides are among the simplest self-assembling molecules employed as biomaterials and are readily produced by standard peptide synthesis protocols, making them particularly accessible. Although fibril-forming peptides have been developed based on several different secondary structures, including α-helices,14 and collagen triple helices,15,16 we will focus here on those capable of forming gels amenable to cell culture or delivery in vivo. In this regard, the bulk of research has focused on β-sheet fibrillizing peptides. To date, several different peptides forming β-sheet fibrils have been studied as foundations for multifunctional biomaterials, including the strictly alternating polar/non-polar peptides first described by Zhang and coworkers,17,18 fibrillizing peptides from laminin,19,20 glutamine-rich sequences such as DN1 and P11 first described by Aggeli and coworkers,21–23 other glutamine-rich peptides such as Q11 that followed these initial designs,24–26 and peptides from amyloidogenic proteins such as transthyretin.27 For an overview of the development of several of these peptides, see the recent review by Semino.4 In strategies aimed at conferring specific biofunctionality to these otherwise purely structural assemblies, functional amino acid sequences have recently been appended to either their N-termini or C-termini. In many cases the functional sequences are presented on the surface of the self-assembled fibrils in configurations that allow them to influence the behaviors of cells in contact with them. For example, Q11 peptides (QQKFQFQFEQQ) with N-terminal IKVAV and RGDS sequences self-assemble into fibrils that functionally display the ligands, which influence endothelial cell growth on the surface of the gels (Figure 2).26 The presence of the ligands on the surface of the fibrils was verified using TEM with gold labeling, and the ligands were surface-displayed both in single-peptide fibrils and in mixtures of the ligand-bearing peptides with unmodified Q11. Moreover, Q11 gels and mixed peptide gels containing 90% unmodified Q11 and 10% ligand-bearing Q11 had similar fibril morphology, stiffness, and secondary structure, indicating modularity in the system and opening the door for exploring these and other ligands systematically in combination.
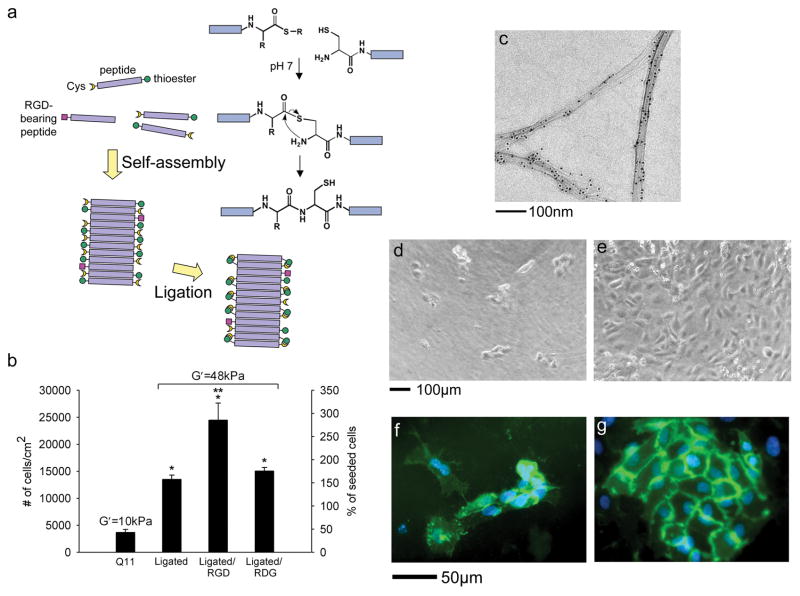
Figure 2.
Co-assembling peptides for controlling matrix mechanics and enhancing cell adhesion. Self-assembling peptides with N-terminal Cys residues and C-terminal thioesters self-assemble into fibrils capable of undergoing native chemical ligation (a). RGD-bearing peptides are co-assembled within these gels to provide for cell attachment. Both ligation and RGD functionalization significantly and additively improved endothelial cell growth (p<0.01 by ANOVA compared to Q11 (*) or all others (**), n=4, means±SD).25 Ligand display was also evidenced by streptavidin-colloidal gold labeling of biotinylated RGDS-Q11 fibrils (c). RGDS-Q11 incorporation significantly improved human umbilical vein endothelial cell (HUVEC) growth (d, Q11 gel; e, Q11 gel containing 10% RGDS-Q11, both at day 7 post-plating).26 Stiffening by native chemical ligation significantly improved CD31 expression in HUVEC cultures (f, Q11 gel; g, ligated CQ11G-thioester; green, CD31; blue, DAPI).25 (d) and (e) were reprinted with permission from Jung et al. Biomaterials 2009, 30, 2400–2410. (b), (f), and (g) were reprinted with permission from Jung et al. Biomaterials 2008, 29, 2143–2151.
Beyond the binding of integrins and other cell attachment receptors, cell behavior can be profoundly influenced by physicochemical factors such as matrix stiffness.28–30 As a strategy for specifically adjusting gel mechanics that is complementary to the ligand display described above, we also designed Q11 peptides containing C-terminal thioesters and N-terminal cysteine residues, enabling matrix stiffening by native chemical ligation (Figure 2).25 Gels with ligated Q11 fibrils showed a 5-fold increase in storage moduli, to nearly 50 kPa, making these among the stiffest self-assembled gels yet reported. Because fibrillar peptide gels tend to be significantly less stiff than this, having a chemoselective means for independently adjusting their cross-linking is useful for tuning their mechanical properties, even in the presence of other unprotected ligands or functional groups. This ability to stiffen the matrices is useful both for specifying the mechanical properties that result in a desired cellular behavior and also for stabilizing the materials for biomedical applications, for example as interfacial coatings on other biomaterials. In mixtures of Q11, RGDS-Q11, and Cys-Q11-Gly-thioester, both ligand presentation and mechanical stiffening were easily combined, and the two factors together increased endothelial cell proliferation and CD31 expression to a much greater degree than either factor alone (Figure 2). In addition, although the strength and elasticity of the stiffened gels was not measured directly in this work, the stiffened gels were significantly easier to handle without fragmentation than unligated gels (JPJ and JHC, unpublished findings). Other groups have also recently developed strategies for modulating stiffness. For example, Schneider, Pochan, and co-workers have reported several approaches for tailoring the stiffness of β-hairpin assemblies, employing borate ion complexation,31 shear thinning,32 strand swapping,33 and control over lateral association.34 Given the highly stable nature of β-rich assemblies, strategies such as these for dynamically tuning their stiffness are significant technological advances. Designed β-hairpins are also similarly non-cytotoxic and biocompatible compared to other fibrillizing peptides and peptide derivatives,32,35,36 and they are highly capable of being tuned to exhibit desirable ranges of other physical parameters such as pH responsiveness and mesh size.32,37
Ligand-bearing versions of β-sheet fibrillar peptides with strictly alternating polar/non-polar amino acid sequences have also been investigated in recent years in a variety of biological contexts. For example the RAD16 family of peptides is well known to form stable β-sheet fibrillar gels in physiologic buffers with peptide concentrations as low as 1–10 mg/mL.38,4 RAD16-I has been commercialized under the trade name PuraMatrix™. Several different pendant peptides have been attached either at the N-terminus, including the laminin-derived YIGSR and RYVVLPR sequences and the type IV collagen TAGSCLRKFSTM sequence;39 or at the C-terminus, including the bone marrow homing SKPPGTSS and PFSSTKT sequences,40 the osteogenic ALKRQGRTLYGF sequence, and the osteopontin-derived DGRGDSVAYG cell adhesion sequence.41 In cultures of human aortic endothelial cells, gelled RAD16-I peptides bearing YIGSR and TAGSCLRKFSTM ligands modulated growth and nitric oxide production, indicating that a sufficient number of ligands were presented by the self-assembled fibrils to induce these changes in cell behavior.39 Gelled RAD16 peptides bearing bone marrow homing sequences at their C-termini were also shown to promote neural stem cell adhesion and differentiation in the absence of soluble neurotrophic factors.40
Beyond non-native designed peptides such as Q11 and RAD16, native amyloid fibrils have also received attention as engineered materials for applications in bionanotechnology.42–45 Some of these are capable of presenting biofunctional amino acid sequences analogously to the engineered peptides described above. In a recent example, amyloid-forming peptides corresponding to residues 105–115 of the protein transthyretin were functionalized with an RGD sequence, and the ligand-bearing peptides formed fibrils exhibiting enhanced fibroblast adhesion.27 The cross-β fibril core was completely preserved, indicating that the conjugation of the ligand did not significantly alter the assembly of the transthyretin peptide. In light of these studies and those discussed above, it is interesting that so many completely unrelated β-sheet fibrillizing peptides, native or de novo, form similar fibrils that have the capacity to display bioavailable ligands on their surfaces (Figure 1). This suggests that there is considerable flexibility in designing fibrillar gels as biomaterials, and that not only the displayed ligands but also the fibrils themselves can be tailored to meet the specific constraints of a particular application. For example, the fibril-forming portions of these peptides may be selected or engineered to address specific biological requirements (e.g. non-toxicity, non-immunogenicity, specific degradation rate), mechanical requirements (e.g. compliance, strength), or economic considerations (e.g. cost of synthesis, ease of sterilization) that may vary from application to application. Examples of such efforts to adjust the properties of the fibrillizing portions of these peptides are discussed below, and it is anticipated that additional strategies will continue to be introduced and refined.
Peptide amphiphiles
Peptide amphiphiles (PAs), like unmodified synthetic peptides, have received significant interest as gel-forming biomaterials owing to their predictable self-assembly, their ease of synthesis, and their capacity for incorporating a wide variety of functional components. Their general construction of an N-terminal alkyl tail, a β-sheet–forming central segment, and a C-terminal functional segment represents a flexible platform for incorporating a variety of different molecular features.46–48 In fact, the presence of the alkyl tail drives self-assembly for peptide sequences, molecular architectures, and peptide concentrations that would be challenging to fibrillize otherwise, making PAs even more widely applicable, in some respects, than the peptides described above. In recent years PAs have been designed with increasingly complex and bulky functional domains, illustrating their ability to assemble a wide variety of cargoes into fibrils. These include PAs with bulky fluorophores, branched PAs that present two RGD ligands or one YIGSR and one IKVAV ligand, cyclic RGD ligands, and others.49–52 Similarly, the N-terminal alkyl tail also provides unique opportunities for modification, as demonstrated by the inclusion of diacetylene groups capable of being polymerized to covalently stabilize the self-assembled fibrils.53 Further flexibility in this system has also been demonstrated by incorporating proteolytically susceptible amino acid sequences in the central portion of the PAs.54,55 In this work, even aggregated fibrils possessed substrate activity, suggesting that other biological functions may also be able to be localized to this region of the molecule as well. In a more practical sense, it was shown that matrix metalloproteinase-2 (MMP-2) substrate-containing PAs could be transformed from fibrils to globular aggregates in the presence of the enzyme, providing a means to design degradable β-sheet fibrillar gels. Ordinarily, β-sheet fibrillar aggregates are highly resistant to proteolysis, so such an approach is an important step towards gels with controllable degradation profiles. Interestingly, in related recent work, MMP substrates were incorporated in the central portion of self-assembling peptides lacking lipid tails, and these assemblies also showed MMP substrate activity.56 However, gel formation appeared to be strongly dependent on the position of the MMP sequence within these peptides, and the gelation of some of the peptides was significantly hindered. This may have been an indirect result of having a proline residue within the MMP substrate portion of the peptide, however, as prolines can disrupt β-sheet fibrillization.
Other Peptide Derivatives
Peptides containing aromatic groups such as di-phenylalanine (FF) or fluorenylmethoxycarbonyl (Fmoc) offer unique advantages and have recently received attention as flexible fibrillizing materials.57–66 In these approaches, the mechanism of self-assembly often includes β-sheet formation, but fibrillization is additionally favored through π-π stacking of the aromatic groups. For example, extremely short Fmoc-FF peptides spontaneously fibrillize in neutral buffers to produce gels with storage moduli between 2–10 kPa owing to the combined effects of π-π stacking by the Fmoc groups and β-sheet formation in the peptide segments.59,60 For biological applications, variations on this overall construction have recently been explored.60,67,68 These include mixtures of Fmoc-FF peptides and Fmoc-RGD peptides, which co-assemble into relatively stiff hydrogels (G′~ 10 kPa) capable of presenting the RGD ligand in a manner similar to the approaches described above for unmodified peptides and PAs.60 In another study, enzyme substrate activity was imparted to self-assembling Fmoc-peptides using the thermolysin substrate Gly-Phe-Cys sequence. In this case, rather than being used as hydrogel materials, the Fmoc-peptides were conjugated to gold nanoparticles to produce highly sensitive optical enzyme sensors.68 One of the significant advantages of this approach using Fmoc and other aromatic groups is that gels can be built using much shorter peptides than with PAs or unmodified peptides. This has an obvious benefit in reducing the cost of peptide synthesis and purification. Moreover, the reduced complexity of these materials may also facilitate a more detailed understanding of their fibrillar structures.59 One aspect that may influence their use as cell culture substrates, however, is their apparent sensitivity to different amino acid sequences.60,67 For example, the stiffness of mixed Fmoc-RGD/Fmoc-FF gels depended significantly and non-linearly on the ratio of the two peptides, which may make independent adjustment of both cell attachment factors and mechanical factors challenging.60 However, if ranges of peptide mixtures can be identified where stiffness is less dependent on ligand amount, this issue can be minimized. Considering the diversity of engineered β-sheet fibrillar structures that have been designed to date, it can be concluded that there are many different molecular strategies that can be employed to produce structurally similar gels of entangled fibrils (Figure 1). In this respect, engineered β-sheet fibrillar materials mirror the diversity of natural amyloids, which can likewise be formed from proteins of widely varying size, sequence, and native function.
Other modular biomaterials approaches beyond fibrillizing peptides
Additional recent work illustrates the breadth of modular approaches currently being developed in the field of biomaterials that extend beyond fibrillizing peptides. We focus on two of them here. The first utilizes supramolecular polymers based on ureido-pyrimidinone (UPy) units. These self-complementary tautomers can dimerize via hydrogen bonding and have been employed to form stable, long polymer chains in a convenient one-step procedure. 69,70 To functionalize these polymers with cell-binding ligands in a modular way, UPy-caprolactones were mixed with UPy-GRGDS and UPy-PHSRN, thus incorporating the ligands via hydrogen bonding in precisely defined amounts. In cultures of fibroblasts, the non-covalently bound ligands led to enhanced attachment.70 Any combination of other UPy-functionalized peptide ligands can also easily be envisioned, illustrating the potential flexibility of this approach. A second class of modular biomaterials is based on recombinant proteins, which provide access to longer, more complexly folded domains than self-assembling peptides. In this regard, Heilshorn and coworkers reported multifunctional protein scaffolds designed to support PC-12 neuronal-like cells. The multi-domain proteins included urokinase plasminogen activator (uPA) cleavage sites for tunable degradation, elastin-like domains to confer elasticity and resilience, and RGD cell-binding sequences.71 With only a 3% change in the total amino acid sequence, the uPA degradation half-life could be varied over two orders of magnitude. In addition, increasing the density of RGD within the matrices led to increased attachment, neuronal differentiation, and neurite extension in cultures of PC-12 cells. It is expected that modular approaches such as these will continue to be developed in the coming years as self-assembling peptides, peptide amphiphiles, peptide derivatives, peptide-polymers, and proteins are pursued in parallel.
Spatial and temporal heterogeneities in self-assembled networks
One of the chief advantages of self-assembling materials, as described in the previous sections of this review, is that once an adaptable base unit is designed, increasingly complex variations on the base material may be produced. Through co-assembly of these variants, multiple specific biofunctionalities can be installed into the matrix. In practice, however, it is not always straightforward to achieve this goal, and in some cases self-assembly can be altered or disrupted by the biofunctional components. In particular, small alterations in folding, β-strand topology, or lateral aggregation imposed by the functional elements may be propagated or magnified by the self-assembly process to significantly alter fibril morphology,72 thus affecting bulk properties such as gel mechanics or overall fibril morphology.60,39 In this way, self-assembling materials are more sensitive to defects than other more top-down approaches such as microfabrication,73–75 where defects tend to be manifested as spatially discrete anomalies rather than global changes in physical or biological properties. To address this issue, it is likely that self-assembling components with even greater tolerances for functionalization will be introduced in the future.
Another consequence of using β-rich self-assembly to construct nanomaterials is that some degree of structural heterogeneity is often present. This heterogeneity can exist on multiple levels. For example, lateral entanglement and aggregation can produce a distribution of lengths between cross-link points, and there can be heterogeneity in the twist, bend, and topology of the β-strands themselves, as has been observed within highly controlled models of β-fibrillization.72 Additionally, lateral aggregation can be significantly influenced by the processing history of the molecule and its storage conditions. Because of the innate fibrillizing behavior of these materials, care must be taken to ensure that parameters such as concentration, pH, temperature, ionic strength, and lyophilization techniques are tightly controlled during all points of synthesis, storage, and use. Imprecision in this regard may result in divergent and unpredictable assembly behavior. For this reason, disaggregation in hexafluoroisopropanol or trifluoroacetic acid has been useful for rendering consistent, repeatable starting materials for gel formation,26,76 and consistent handling practices are essential.77 Further, if ligand-bearing peptides are mixed to form co-assemblies with unmodified peptides, as with several current approaches, there are the attendant heterogeneities associated with the potentially non-uniform display of these ligands that must be considered as well (summarized in Figure 3). One consequence of these structural heterogeneities is that it is difficult to predict or measure the number and spatial distributions of receptor-ligand binding events that occur between cells and fibrillized materials in biological environments. This is a challenging question in any 3D biomaterial. However, recent strategies developed for visualizing ligand-receptor interactions in polymeric scaffolds such as the FRET-based techniques described recently by Mooney and coworkers may be useful for quantifying ligand binding in self-assemblies as well.78,79 Additionally, heterogeneity may be controlled directly by designing fibrils from more predictably folded β-sheets. As an example of work that moves in this direction, Koide and coworkers have designed soluble peptide self-assembly mimics based on the single-layer β-sheet of Borrelia burgdorferi outer surface protein A (OspA) to produce water-soluble, monodisperseβ-sheets.67,80–83 This protein engineering approach has greatly facilitated high-resolution structure determinations of β-sheet assemblies, and an exciting possibility may be to employ these or similar expressed β-rich proteins as nanomaterials themselves through their controlled polymerization.
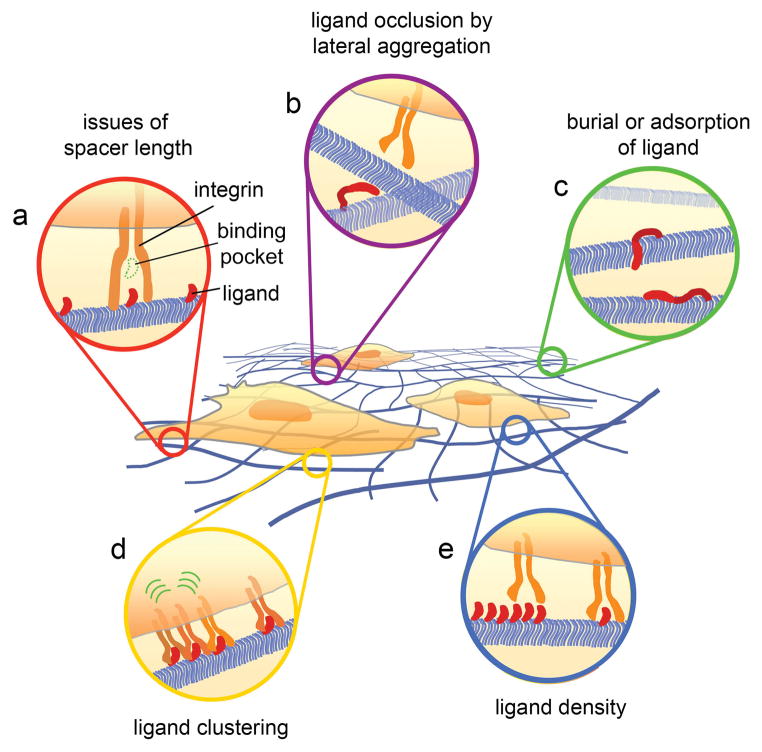
Figure 3.
Potential types of heterogeneity within ligand-bearing self-assemblies. Several structural aspects may influence the number and spatial distributions of ligand-integrin binding events, which collectively determine cell behavior. Peptide ligands must be appropriately spaced from surfaces to efficiently deliver the ligand to the binding pockets of integrins (a). Inter-fibril lateral aggregation (b), ligand burial, or ligand adsorption (c) may also remove a subset of ligands from interacting with integrins. These aspects as well as potential phase separation may influence the clustering of the ligands on the fibril (d), which may in turn influence integrin clustering, focal adhesion assembly, and the resultant intracellular signaling. It is also conceivable that the density of the ligand itself could sterically influence the ability of integrins to bind (e). Strategies to define and control these aspects will contribute significantly to the biological precision possible with these materials.
In addition to the structural heterogeneity that is a natural characteristic of materials produced by bottom-up processes, temporal heterogeneity may also influence the behavior of these materials in many biological contexts. Most fibrillized materials applied within biological environments are presumed to be equilibrated and thermodynamically stable initially, but as they are degraded or remodeled in vitro or in vivo, it is likely that their valency, fibril morphology, and supramolecular organization will undergo transformations. For example, MMP-cleavable peptide-amphiphiles, when degraded, transition from fibrils to globular aggregates.55 Currently, the impact of such morphological changes have not been clearly elucidated in biological contexts, and this is particularly true in vivo. Given the strong dependence of many biological processes such as focal adhesion assembly or immunogenicity on the spatial arrangement of ligands, it is likely that there is a level of detail in the biological interactions of dynamic supramolecular structures that has yet to be fully understood. One way of managing temporal heterogeneity in self-assemblies is by employing degradative processes such as MMP activity so that the action of a predictable enzyme predominantly determines the kinetics of disassembly. This strategy has been applied both to PAs 54 and peptides,56 as described above.
Towards applications in biomedicine and biotechnology
Beta-sheet fibrillar gels are attractive in biological applications because of their modularity and because they form very stable structures that are resistant to proteolysis or denaturation. However, the same properties that make β-sheet fibrillar networks exceptionally useful as engineerable biomaterials also necessitate additional characterization as they move towards applications in vivo. For example, the materials discussed here are all structurally related to natural amyloids, which have been predominantly associated with pathological and neurodegenerative disorders since their discovery over 100 years ago. Interestingly, however, there are numerous recent examples of engineered β-sheet fibrillar materials that have been found to be very well tolerated in vitro25,26,32,35,39–41,47,84 and in vivo26,85–89 and that show no apparent toxicity. In addition, intriguing examples continue to be introduced where β-sheet fibers are not pathological, but where they serve specific and tightly regulated biological functions in organisms from bacteria to humans.90–94 These examples include structural amyloids such as curli fibers of bacteria,94 chorion proteins in the eggshells of insects and fish,95 fungal coat-forming hydrophobins,96 and more complex biofunctional amyloids such as Pmel17 involved in human melanin biosynthesis.92 These and other functional amyloids have been recently reviewed.93 Understanding how certainβ-sheet aggregates are toxic while others are non-toxic or even biologically functional is important for the advancement of self-assembling materials in clinical applications.
Additionally complicating the translation of in vitro work to in vivo settings is the sensitivity of fibrillar peptides to environmental conditions, which is highlighted in recent work by Schneider, Pochan, and co-workers where MAX1 β-hairpin gels were stiffened in the presence of borate but were more compliant in the presence of glucose.31 In vivo, it may be difficult to predict how the presence of small molecules, glycoproteins, polysaccharides, or proteoglycans may similarly influence the lateral aggregation points in fibrillar peptide gels. Further, other work with β-hairpins illustrates how the mechanical properties of fibrillar self-assemblies may be significantly altered by shear forces.32 These studies highlight the importance of evaluating fibrillar peptide assemblies in culture conditions that replicate the in vivo biochemical and mechanical environment as faithfully as possible.
A final critical hurdle for employing fibrillar biomaterials in vivo is understanding the factors that impact their immunogenicity and their interactions with inflammatory cells. Functionalized self-assembled fibrous networks, by virtue of their repeated patterns of epitopes, can in principle induce immune responses, as has been illustrated in other highly oligomerized systems.97–99 However, there have been few studies fully examining the immunogenicity of peptide-based hydrogels. Nitric oxide-releasing PA hydrogels delivered to rat carotid arteries did not elicit any obvious adverse inflammation, which is encouraging.89 Few infiltrating macrophages and neutrophils were observed within RAD16-II assemblies injected within myocardium,85 which is additionally encouraging, but it is indirect evidence that these materials are well tolerated by the immune system. Moreover, not all in vivo studies using self-assembling peptides have resulted in the absence of inflammation.100 To specifically measure the immunogenicity of Q11-based peptides, we recently investigated anti-peptide IgG production against Q11 and RGDS-Q11 in mice, finding immeasurable immunogenicity for Q11 and little to no immunogenicity for RGDS-Q11.26 Low antibody titers have also been reported in rabbits and goats for RAD16 peptides.84 Collectively, these studies represent several examples of minimally immunogenic fibrillizing peptides and PAs, but the highly oligomerized and non-native structure of these materials nevertheless continues to warrant attention with respect to their immunogenicity, especially when functionalized with increasingly complex ligands.
Despite the challenges described above regarding the biological application of fibrillizing materials for clinical use, several recent reports indicate progress in this regard (Figure 4). For example, cardiomyocytes or embryonic stem cells mixed with RAD16 peptides have been delivered to the myocardium (Figure 4a). Co-injection of RAD16 peptides promoted either improved cell transplant survival or the recruitment of progenitor cells around the injected area.86,101 The transplanted embryonic stem cells also showed evidence of cardiomyocyte differentiation after co-delivery with the peptide biomaterial.86 In this way, self-assembling peptides may be a viable option for improving cell-based therapies. Nitric oxide-releasing PAs have also been investigated recently in rats to inhibit neointimal hyperplasia following carotid artery balloon angioplasty (Figure 4c). Nitric oxide-releasing PA gels were applied around the insulted area, resulting in a decrease in vascular smooth muscle cell proliferation and an increase in re-endothelialization.89 Also, similar PA nanofibers have been applied within the interconnected pores of titanium (Ti) foam in order to improve osseointegration into orthopedic implants (Figure 4b).87,102 Combining fibrillar gels with other biomaterials in this way is advantageous for several reasons. First, previously approved materials and devices may facilitate the addition of novel coatings or bioactive components from both a regulatory and a practical standpoint. Having a well-characterized starting material helps enable a more complete understanding of how the novel components behave. In addition, the Ti foam complements the PA by providing mechanical stiffness, which tends to be lacking in self-assembled materials.
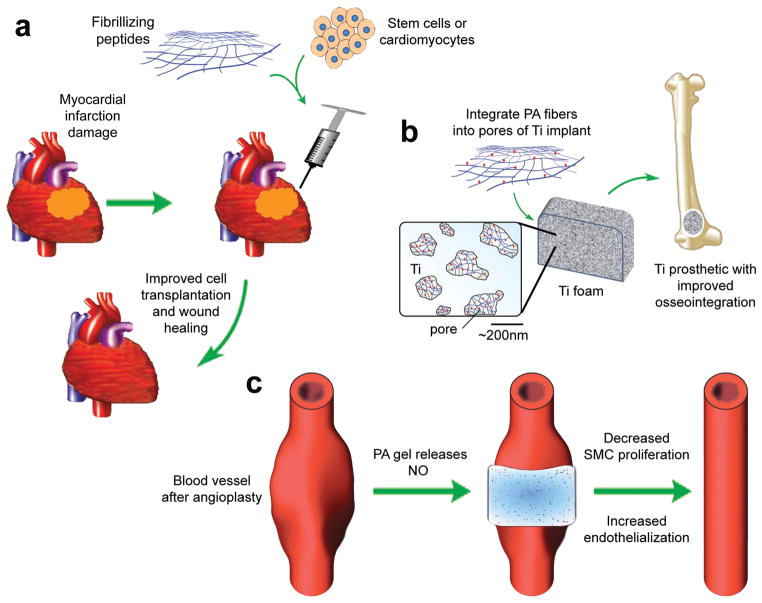
Figure 4.
Potential uses of fibrillar peptide gels for clinical applications. Improvement in cell-based therapies (a). Fibrillizing peptides are mixed with cardiomyocytes or undifferentiated stem cells and injected into damaged myocardium for improved transplanted cell survival and wound healing after myocardial infarction.86,101 Improvement of prosthetics (b). PAs are integrated into the pores of titanium foam to create bioactive composites that induce mineralization and vascularization around and within orthopedic implants.87,102 Direct application of a therapeutic-releasing gel (c). Following angioplasty, a nitric oxide-releasing PA gel is applied directly to the exterior of the vessel at the site of injury, reducing smooth muscle cell proliferation and increasing endothelialization compared to non-hydrogel treated controls.89
The early in vivo successes of fibrillizing peptide biomaterials, along with their expanding capacity for incorporating multiple biological factors, are good evidence that they are advancing towards meaningful biomedical and biotechnological use. Continuing progress will be greatly facilitated by improvements in understanding and controlling these materials’ heterogeneity, their modularity, and the physiological processes that govern their behavior in various biological contexts.
Acknowledgments
The authors thank Kathy Goss and Jai Rudra for helpful comments on the manuscript.
References
- 1.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Collier JH. Soft Matter. 2008;4:2310–2315. doi: 10.1039/b805563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segers VFM, Lee RT. Drug Discov Today. 2007;12:561–568. doi: 10.1016/j.drudis.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Semino CE. J Dent Res. 2008;87:606–616. doi: 10.1177/154405910808700710. [DOI] [PubMed] [Google Scholar]
- 5.Kopecek J, Yang J. Acta Biomater. 2009;5:805–816. doi: 10.1016/j.actbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dankers PYW, Meijer EW. Bull Chem Soc Jpn. 2007;80:2047–2073. [Google Scholar]
- 7.Hamley IW. Angew Chem Int Ed Engl. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 8.Ulijn RV, Smith AM. Chem Soc Rev. 2008;37:664–675. doi: 10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 9.Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Soft Matter. 2006;2:822–835. doi: 10.1039/b607706d. [DOI] [PubMed] [Google Scholar]
- 10.Branco MC, Schneider JP. Acta Biomater. 2009;5:817–831. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer LC, Stupp SI. Acc Chem Res. 2008;41:1674–1684. doi: 10.1021/ar8000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higashi N, Koga T. Self-Assembled Nanomaterials I. Springer-Verlag; Berlin: 2008. pp. 27–68. [Google Scholar]
- 13.Scanlon S, Aggeli A, Boden N, McLeish TCB, Hine P, Koopmans RJ, Crowder C. Soft Matter. 2009;5:1237–1246. [Google Scholar]
- 14.Papapostolou D, Brornley EHC, Bano C, Woolfson DN. J Am Chem Soc. 2008;130:5124–5130. doi: 10.1021/ja0778444. [DOI] [PubMed] [Google Scholar]
- 15.Gauba V, Hartgerink JD. J Am Chem Soc. 2007;129:2683–2690. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 16.Kotch FW, Raines RT. Proc Natl Acad Sci U S A. 2006;103:3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koutsopoulos S, Unsworth LD, Nagai Y, Zhang S. Proc Natl Acad Sci U S A. 2009;106:4623–4628. doi: 10.1073/pnas.0807506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai S, Ohga Y, Mochizuki M, Nishi N, Kadoya Y, Nomizu M. Biopolymers. 2004;76:27–33. doi: 10.1002/bip.10565. [DOI] [PubMed] [Google Scholar]
- 20.Kasai S, Urushibata S, Hozumi K, Yokoyama F, Ichikawa N, Kadoya Y, Nishi N, Watanabe N, Yamada Y, Nomizu M. Biochemistry. 2007;46:3966–3974. doi: 10.1021/bi062097t. [DOI] [PubMed] [Google Scholar]
- 21.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TCB, Pitkeathly M, Radford SE. Nature. 1997;386:259–262. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 22.Carrick LM, Aggeli A, Boden N, Fisher J, Ingham E, Waigh TA. Tetrahedron. 2007;63:7457–7467. [Google Scholar]
- 23.Riley JM, Aggeli A, Koopmans RJ, McPherson MJ. Biotechnol Bioeng. 2009;103:241–251. doi: 10.1002/bit.22274. [DOI] [PubMed] [Google Scholar]
- 24.Collier JH, Messersmith PB. Bioconjug Chem. 2003;14:748–755. doi: 10.1021/bc034017t. [DOI] [PubMed] [Google Scholar]
- 25.Jung JP, Jones JL, Cronier SA, Collier JH. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gras SL, Tickler AK, Squires AM, Devlin GL, Horton MA, Dobson CM, MacPhee CE. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 28.Pelham RJ, Jr, Wang Y. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 30.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozbas B, Rajagopal K, Haines-Butterick L, Schneider JP, Pochan DJ. J Phys Chem B. 2007;111:13901–13908. doi: 10.1021/jp075117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc Natl Acad Sci U S A. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagarkar RP, Hule RA, Pochan DJ, Schneider JP. J Am Chem Soc. 2008;130:4466–4474. doi: 10.1021/ja710295t. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopal K, Ozbas B, Pochan DJ, Schneider JP. Eur Biophys J. 2006;35:162–169. doi: 10.1007/s00249-005-0017-7. [DOI] [PubMed] [Google Scholar]
- 35.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Haines-Butterick LA, Salick DA, Pochan DJ, Schneider JP. Biomaterials. 2008;29:4164–4169. doi: 10.1016/j.biomaterials.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Biomaterials. 2009;30:1339–1347. doi: 10.1016/j.biomaterials.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Gelain F, Zhao X. Semin Cancer Biol. 2005;15:413–420. doi: 10.1016/j.semcancer.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Genove E, Shen C, Zhang S, Semino CE. Biomaterials. 2005;26:3341–3351. doi: 10.1016/j.biomaterials.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Gelain F, Bottai D, Vescovi A, Zhang S. PLoS ONE. 2006;1:e119. doi: 10.1371/journal.pone.0000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horii A, Wang X, Gelain F, Zhang S. PLoS ONE. 2007;2:e190. doi: 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldwin AJ, Bader R, Christodoulou J, MacPhee CE, Dobson CM, Barker PD. J Am Chem Soc. 2006;128:2162–2163. doi: 10.1021/ja0565673. [DOI] [PubMed] [Google Scholar]
- 43.Kodama H, Matsumura S, Yamashita T, Mihara H. Chem Comm. 2004:2876–2877. doi: 10.1039/b409641j. [DOI] [PubMed] [Google Scholar]
- 44.MacPhee CE, Dobson CM. J Am Chem Soc. 2000;122:12707–12713. [Google Scholar]
- 45.Sondheimer N, Lindquist S. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 46.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 47.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 48.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 49.Behanna HA, Rajangam K, Stupp SI. J Am Chem Soc. 2007;129:321–327. doi: 10.1021/ja062415b. [DOI] [PubMed] [Google Scholar]
- 50.Guler MO, Hsu L, Soukasene S, Harrington DA, Hulvat JF, Stupp SI. Biomacromolecules. 2006;7:1855–1863. doi: 10.1021/bm060161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guler MO, Soukasene S, Hulvat JF, Stupp SI. Nano Lett. 2005;5:249–252. doi: 10.1021/nl048238z. [DOI] [PubMed] [Google Scholar]
- 52.Harrington DA, Cheng EY, Guler MO, Lee LK, Donovan JL, Claussen RC, Stupp SI. J Biomed Mater Res A. 2006;78:157–167. doi: 10.1002/jbm.a.30718. [DOI] [PubMed] [Google Scholar]
- 53.Hsu L, Cvetanovich GL, Stupp SI. J Am Chem Soc. 2008;130:3892–3899. doi: 10.1021/ja076553s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galler KM, Cavender A, Yuwono V, Dong H, Shi ST, Schmalz G, Hartgerink JD, D’Souza RN. Tissue Eng Part A. 2008;14:2051–2058. doi: 10.1089/ten.tea.2007.0413. [DOI] [PubMed] [Google Scholar]
- 55.Jun HW, Yuwono V, Paramonov SE, Hartgerink JD. Adv Mater. 2005;17:2612. [Google Scholar]
- 56.Chau Y, Luo Y, Cheung ACY, Nagai Y, Zhang S, Kobler JB, Zeitels SM, Langer R. Biomaterials. 2008;29:1713–1719. doi: 10.1016/j.biomaterials.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 57.Reches M, Gazit E. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 58.Reches M, Gazit E. J Nanosci Nanotechnol. 2007;7:2239–2245. doi: 10.1166/jnn.2007.645. [DOI] [PubMed] [Google Scholar]
- 59.Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Adv Mater. 2008;20:37–41. [Google Scholar]
- 60.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Gu H, Yang Z, Xu B. J Am Chem Soc. 2003;125:13680–13681. doi: 10.1021/ja036817k. [DOI] [PubMed] [Google Scholar]
- 62.Gao J, Wang HM, Wang L, Wang JY, Kong DL, Yang ZM. J Am Chem Soc. 2009;131:11286–7. doi: 10.1021/ja9042142. [DOI] [PubMed] [Google Scholar]
- 63.Yang Z, Liang G, Xu B. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 64.Yang ZM, Gu HW, Fu DG, Gao P, Lam JK, Xu B. Adv Mater. 2004;16:1440–1444. [Google Scholar]
- 65.Yang ZM, Liang GL, Wang L, Xu B. J Am Chem Soc. 2006;128:3038–3043. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 66.Yang ZM, Xu B. Chem Comm. 2004:2424–2425. doi: 10.1039/b408897b. [DOI] [PubMed] [Google Scholar]
- 67.Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough J, Ulijn RV. Adv Mater. 2006;18:611–614. [Google Scholar]
- 68.Laromaine A, Koh L, Murugesan M, Ulijn RV, Stevens MM. J Am Chem Soc. 2007;129:4156–4157. doi: 10.1021/ja0706504. [DOI] [PubMed] [Google Scholar]
- 69.Sontjens SHM, Sijbesma RP, van Genderen MHP, Meijer EW. J Am Chem Soc. 2000;122:7487–7493. [Google Scholar]
- 70.Dankers PYW, Harmsen MC, Brouwer LA, Van Luyn MJA, Meijer EW. Nat Mater. 2005;4:568–574. doi: 10.1038/nmat1418. [DOI] [PubMed] [Google Scholar]
- 71.Straley KS, Heilshorn SC. Soft Matter. 2009;5:114–124. [Google Scholar]
- 72.Makabe K, McElheny D, Tereshko V, Hilyard A, Gawlak G, Yan S, Koide A, Koide S. Proc Natl Acad Sci U S A. 2006;103:17753–17758. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curtis AS, Wilkinson CD. J Biomater Sci Polym Ed. 1998;9:1313–1329. doi: 10.1163/156856298x00415. [DOI] [PubMed] [Google Scholar]
- 74.Russell P, Gasiorowski JZ, Nealy PF, Murphy CJ. Invest Ophthalmol Vis Sci. 2008;49:629–635. doi: 10.1167/iovs.07-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 76.Chen S, Wetzel R. Protein Sci. 2001;10:887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagarkar RP, Schneider JP. Nanostructure Des. 2008:61–77. doi: 10.1007/978-1-59745-480-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kong HJ, Polte TR, Alsberg E, Mooney DJ. Proc Natl Acad Sci U S A. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kong HJ, Boontheekul T, Mooney DJ. Proc Natl Acad Sci U S A. 2006;103:18534–18539. doi: 10.1073/pnas.0605960103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makabe K, Yan S, Tereshko V, Gawlak G, Koide S. J Am Chem Soc. 2007;129:14661–14669. doi: 10.1021/ja074252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makabe K, Koide S. J Am Chem Soc. 2008;130:14370–14371. doi: 10.1021/ja805011h. [DOI] [PubMed] [Google Scholar]
- 82.Makabe K, Biancalana M, Yan S, Tereshko V, Gawlak G, Miller-Auer H, Meredith SC, Koide S. J Mol Biol. 2008;378:459–467. doi: 10.1016/j.jmb.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biancalana M, Makabe K, Koide A, Koide S. J Mol Biol. 2009;385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Proc Natl Acad Sci U S A. 2000;97:6728–6733. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hsieh PCH, Davis ME, Gannon J, MacGillivray C, Lee RT. J Clin Invest. 2006;116:237–248. doi: 10.1172/JCI25878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis ME, Motion JPM, Narmoneva DA, Takahashi T, Hakuno D, Kamm RD, Zhang S, Lee RT. Circulation. 2005;111:442–450. doi: 10.1161/01.CIR.0000153847.47301.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sargeant TD, Guler MO, Oppenheimer SM, Mata A, Satcher RL, Dunand DC, Stupp SI. Biomaterials. 2008;29:161–171. doi: 10.1016/j.biomaterials.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 88.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kapadia MR, Chow LW, Tsihlis ND, Ahanchi SS, Eng JW, Murar J, Martinez J, Popowich DA, Jiang Q, Hrabie JA, Saavedra JE, Keefer LK, Hulvat JF, Stupp SI, Kibbe MR. J Vasc Surg. 2008;47:173–182. doi: 10.1016/j.jvs.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 91.Larsen P, Nielsen JL, Otzen D, Nielsen PH. Appl Environ Microbiol. 2008;74:1517–1526. doi: 10.1128/AEM.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fowler DM, Koulov AV, Balch WE, Kelly JW. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 94.Olsen A, Jonsson A, Normark S. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 95.Iconomidou VA, Vriend G, Hamodrakas SJ. FEBS Lett. 2000;479:141–145. doi: 10.1016/s0014-5793(00)01888-3. [DOI] [PubMed] [Google Scholar]
- 96.Mackay JP, Matthews JM, Winefield RD, Mackay LG, Haverkamp RG, Templeton MD. Structure. 2001;9:83–91. doi: 10.1016/s0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- 97.Bachmann MF, Hengartner H, Zinkernagel RM. Eur J Immunol. 1995;25:3445–3451. doi: 10.1002/eji.1830251236. [DOI] [PubMed] [Google Scholar]
- 98.Boato F, Thomas RM, Ghasparian A, Freund-Renard A, Moehle K, Robinson JA. Angew Chem Int Ed Engl. 2007;46:9015–9018. doi: 10.1002/anie.200702805. [DOI] [PubMed] [Google Scholar]
- 99.Fehr T, Bachmann MF, Bucher E, Kalinke U, Di Padova FE, Lang AB, Hengartner H, Zinkernagel RM. J Exp Med. 1997;185:1785–1792. doi: 10.1084/jem.185.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dubois G, Segers VF, Bellamy V, Sabbah L, Peyrard S, Bruneval P, Hagege AA, Lee RT, Menasche P. J Biomed Mater Res B Appl Biomater. 2008;87:222–228. doi: 10.1002/jbm.b.31099. [DOI] [PubMed] [Google Scholar]
- 101.Davis ME, Hsieh PC, Takahashi T, Song Q, Zhang S, Kamm RD, Grodzinsky AJ, Anversa P, Lee RT. Proc Natl Acad Sci U S A. 2006;103:8155–8160. doi: 10.1073/pnas.0602877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sargeant TD, Oppenheimer SM, Dunand DC, Stupp SI. J Tissue Eng Regen Med. 2008;2:455–462. doi: 10.1002/term.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Macromolecules. 2004;37:7331–7337. [Google Scholar]