Abstract.
The protein microtubule-associated protein 1, light chain 3 (LC3) functions in autophagosome formation and plays a central role in the autophagy pathway. Previously, we found LC3 diffuses more slowly in cells than is expected for a freely diffusing monomer, suggesting it may constitutively associate with a macromolecular complex containing other protein components of the pathway. In the current study, we used Förster resonance energy transfer (FRET) microscopy and fluorescence recovery after photobleaching (FRAP) to investigate the interactions of LC3 with , a catalytically inactive mutant of the cysteine protease involved in lipidation and de-lipidation of LC3, as a model system to probe protein complex formation in the autophagy pathway. We show is in FRET proximity with LC3 in both the cytoplasm and nucleus of living cells, consistent with previous biochemical evidence that suggests these proteins directly interact. In addition, overexpressed diffuses significantly more slowly than predicted based on its molecular weight, and its translational diffusion coefficient is significantly slowed upon coexpression with LC3 to match that of LC3 itself. Taken together, these results suggest and LC3 are contained within the same multiprotein complex and that this complex exists in both the cytoplasm and nucleoplasm of living cells.
Keywords: Förster resonance energy transfer, fluorescence recovery after photobleaching, diffusion, protein-protein interactions, systems biology, live cells, green fluorescent protein, fluorescence microscopy
1. Introduction
The influx of information about protein-protein interactions from bioinformatics and proteomics analyses has shifted the bottleneck to discovery toward a more thorough characterization of these interactions within the context of living cells.1–4 Fortunately, recent advances in live cell imaging are making it possible to quantitatively characterize the spatial and temporal regulation of green fluorescent protein (GFP) tagged proteins in the complex environment of the living cell. For example, protein-protein interactions are readily monitored through the use of FRET microscopy,5–10 and the dynamics of individual proteins and protein complexes can be quantitatively characterized using techniques sensitive to mobility such as confocal fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS).11–22
One example of an intracellular pathway whose function depends on the coordinated effort of a network of interacting proteins is macroautophagy (referred to here as autophagy).23 The process of autophagy is activated in response to stress signals such as low nutrient availability, and upregulation of the pathway leads to the capture of cytosolic materials in a double membrane structure termed the autophagosome that is subsequently trafficked to the lysosome for degradation and recycling.24–26 LC3 (ATG8 in yeast) plays a central role in the autophagy pathway23,25,26 facilitating the formation of autophagosomes.27–29 Under basal conditions LC3 is known to exist in a soluble form termed LC3-I, and upon upregulation of the autophagy pathway LC3 is converted to a lipidated form termed LC3-II that associates with autophagosomal membranes.30
Recently, a large-scale proteomics screen of the human autophagy pathway revealed LC3 and orthologs interact with a network of 67 proteins.23 In independent studies, our group discovered soluble GFP-LC3 diffuses much more slowly than predicted for a freely diffusing monomer in both the cytoplasm and the nucleus of live cells.13 This raises the possibility that LC3 may constitutively associate with a high molecular weight complex containing multiple proteins, perhaps comprised of one or more LC3-interacting proteins identified in the large-scale proteomics screen.23 Importantly, constitutive association of LC3 with a multiprotein complex could be a potential mechanism for regulating the autophagy pathway. However, it is unclear how many of these proteins come together in live cells to perform the diverse functions associated with autophagy- related proteins, or how these complexes are regulated.
In the current study, we sought to develop tools to further investigate protein complex formation in the autophagy pathway. As a test case, we examined the properties of the complex formed between LC3 and , a catalytically inactive mutant of the cysteine protease known to play important roles in both the lipidation and de-lipidation of LC3.31 Previous studies reported overexpression of wild type Atg4B reduces LC3 lipidation as well as LC3 membrane localization.32 Fujita et al. demonstrated this effect was likely the result of Atg4B nonproductively binding to LC3. In their study, they showed expression of , has similar effects, and proposed the use of this construct as a tool for studying autophagy.33 Given the strong biochemical evidence that shows directly interacts with LC3 to form a stable complex, we used this pair of proteins as a model for developing FRET and diffusion-based methods to characterize protein complexes in living cells.
The basis for using FRET to analyze protein-protein interactions lies in the distance dependence of near field nonradiative energy transfer between two suitable fluorophores known as the donor and acceptor,
| (1) |
where the energy transfer efficiency varies with the inverse sixth power of the separation distance between them,34–36 and the Förster distance is the distance between a given pair of donor and acceptor fluorophores at which . The variable is for commonly used donors and acceptors used in microscopy-based FRET experiments, such as the GFP variants Cerulean and Venus.37 Considering the lengthscale of a typical protein-protein interaction, FRET can be used to determine if two fluorescently labeled proteins are either directly interacting or are close together in a complex. Live cell FRET measurements can be carried out in a number of ways by fluorescence microscopy; for example, by measurements of sensitized acceptor emission, analysis of fluorescence lifetime, or quantification of donor dequenching following acceptor photobleaching.8,34–36
While FRET is clearly a powerful tool for mapping protein-protein interactions in the cell, diffusion-based measurements can provide complementary information about the nature of protein complexes. A commonly used technique to measure the diffusion of fluorescently labeled molecules in single living cells is FRAP. In recent years, FRAP has become a popular technique to study the dynamics of GFP-tagged proteins in intracellular compartments using confocal microscopes.4,12,14,18 In these experiments, cells expressing a fluorescently tagged protein of interest are visualized by fluorescence microscopy. In order to probe the underlying motions of the proteins, an intense pulse of laser light is directed to a defined region of interest in the cell, inducing permanent photobleaching of the fluorophores within that region. Subsequently, unbleached molecules from the surrounding regions exchange with the bleached molecules (termed recovery). The rate at which exchange occurs reveals useful information about the state of the underlying molecules of interest, including the translational diffusion coefficient and the fraction of mobile molecules, commonly referred to as the mobile fraction or Mf.
For the case of simple diffusion of soluble proteins, is related to the size and shape of the diffusing species, the viscosity of the medium, and the absolute temperature . The precise relationship is given by the Stokes-Einstein relation,
| (2) |
for spherical molecules with radius , where is Boltzmann’s constant. Thus, the diffusion coefficients for two soluble proteins within the same multiprotein complex should be identical, and correspond to the size and shape of the complex itself.
To obtain this information from FRAP data, it is necessary to quantitatively analyze the recovery curves to obtain an accurate measurement of . In a classic paper, Axelrod et al. described an analytical solution for FRAP involving a circularly symmetric stationary- focused laser beam that can be used to extract .22 However, several of the assumptions underlying derivation of this equation do not hold when applied to FRAP data obtained using confocal microscopes, such as the assumption that the bleaching event occurs rapidly relative to the characteristic recovery time. Recently, our lab described an approach that generalizes the Axelrod equation to a form that can be applied to data obtained using a laser scanning confocal microscope.22,38 Here, we use FRET microscopy and confocal FRAP as complementary methods to characterize the properties of LC3 and complexes in single living cells.
2. Materials and Methods
2.1. Cell Lines and Constructs
COS-7 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum, 1% PenStrep, and phenol red. EGFP-LC3 and mStrawberry- were the gift of T. Yoshimori (Osaka University).32 Cerulean and Venus were the gift of D. Piston (Vanderbilt University).37 Cerulean and Venus tagged versions of LC3 and were constructed as follows: cDNA for Cerulean and Venus were inserted into Clontech pEGFP-C1 vectors by AgeI and BsrGI double restriction digestion resulting in pCerulean-C1 and pVenus-C1 vectors. Next, we inserted LC3 and cDNA into the pCerulean-C1 and pVenus-C1 multiple cloning sites by BglII and EcoRI double restriction digestion.
2.2. Microscope and Cell Preparation for Live Cell Imaging
All FRET and FRAP microscopy experiments were carried out on a Zeiss LSM 510 confocal microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) using an Argon/2 30 mW laser (458, 488, 514 nm), oil immersion N.A. Zeiss Plan-Neofluar objective, and 1 Airy Unit pinhole.
COS-7 cells were plated on the day before transfection in either MatTek (Ashland, MA) 35 mm No. 1.5 glass bottom culture dishes, or Lab-Tek II 4-well No. 1.5 glass bottom chamber slides (Thermo Fisher Scientific, Rochester, NY). On the following day the cells (50 to 80% confluent monolayer) were transfected with the described mammalian expression constructs using FuGENE 6 (Roche Applied Science, Indianapolis, IN) transfection reagent according to the manufacturer’s recommended protocol.
On the day of the experiment (24 hours after transfection) cell culture medium was rinsed and replaced with phenol red-free DMEM supplemented with 10% fetal calf serum, 1% PenStrep, and 25 mM HEPES. The cells were allowed to come to equilibrium at 37°C before transferring to the temperature- controlled microscope stage set to 37°C.
2.3. Acceptor Photobleaching FRET Data Acquisition
When FRET occurs the intensity of fluorescence emission from the donor is quenched and fluorescence emission from the acceptor is stimulated. The FRET efficiency can be determined by quantifying the relative intensity of fluorescence emission from the donor in the presence and absence of the acceptor. A form of FRET microscopy known as acceptor photobleaching directly measures these quantities on a single sample by irreversibly photobleaching the acceptor. If donor and acceptor are undergoing FRET this procedure results in an increase in the intensity of fluorescence emission from the donor called dequenching.8,35,39 Of the various methods available to experimentally measure FRET in single cells using fluorescence microscopy, the acceptor photobleaching method is a straightforward approach that avoids many of the technical difficulties involved in measuring FRET accurately. A limitation of acceptor photobleaching is that it only provides a single time- point steady-state measurement of FRET.35
To perform acceptor photobleaching FRET experiments the microscope was configured for time-lapse imaging, and several images were collected: first, a 12 bit () image of the donor at digital zoom using 0.375 mW 458 nm excitation followed by acquisition of a second image of the acceptor using 0.075 mW 514 nm excitation (prebleach images). Next, we bleached the acceptor from the entire cell using 30 iterations of 15 mW 514 nm excitation. Finally, we collected another set of images using the same settings as described for the prebleach images (postbleach images). With these settings, were required to acquire a single image of the donor and acceptor, and were required to bleach the acceptor. To separate the excitation and emission wavelengths we used an HFT dichroic, followed by an NFT 515 dichroic. In addition, a 470 to 500 band pass filter was positioned before the donor channel detector, and a LP 530 filter was positioned before the acceptor channel detector.
2.4. Acceptor Photobleaching FRET Data Analysis
To quantify the acceptor photobleaching FRET data, we defined freehand ROIs outlining the cytoplasm and nucleus of each cell using ImageJ (NIH). Next, we measured the mean intensity inside the ROIs for the prebleach images in the donor () and acceptor channels () followed by the mean intensities inside the same ROIs for the postbleach images in the donor () and acceptor () channels.
To accurately determine energy transfer efficiencies from acceptor photobleaching FRET experiments, it is important to subtract the appropriate background from the measured fluorescence intensities, as contributions of detector noise, cell autofluorescence, and the presence of fluorescent material in the media can potentially contribute to the signal. To determine the relative contributions of these parameters, we performed control experiments in which we compared the fluorescence intensity measured in the donor channel in regions of interest centered on unlabeled cells both before and after photobleaching at the acceptor wavelength. We also measured the fluorescence intensity in regions directly adjacent to cells under each of these conditions. We found that the signal was identical for each of these cases, indicating that the major source of background under the conditions of our experiments was from detector noise, and that no bleaching of fluorescent species in the media or in unlabeled cells was occurring. For subsequent analysis we determined the mean intensity inside an ROI placed adjacent to the cells in the prebleach image in the donor channel as a measure of fluorescence background ().
The percent FRET efficiency was then calculated for each cell using
| (3) |
We collected data from multiple cells over several different experiments and report the mean confidence interval for the total number of cells. To ensure significant bleaching of the acceptor we verified,
2.5. Confocal FRAP Data Acquisition
The microscope was configured for time-lapse imaging of a 12-bit () imaging ROI at digital zoom. We defined a circular bleaching ROI (9 pixel radius, 0.99 μm) centered at pixel (, ) within the imaging ROI. Imaging was performed using 0.15 mW 514 nm excitation, and bleaching was performed by scanning 10 iterations of 30 mW 514 nm excitation throughout the bleaching ROI. We utilized bidirectional rastering and maximized the scan speed of our microscope. Under these conditions, 45.1 msec were required to acquire a single image, and 150.1 msec were required to bleach the circular ROI and acquire the next image. We collected 20 prebleach images followed by 280 postbleach images to monitor recovery after the bleach.
2.6. Quantitative FRAP Data Analysis
In this study we analyzed the diffusion of fast- moving proteins such as soluble Venus which have been a challenge to quantitatively measure in cells by confocal FRAP.13,17,19,38 Under our experimental conditions a significant amount of diffusion occurred during the time it took to perform the bleach step (0.1501 sec). In order to quantitatively analyze the FRAP curve to obtain , the initial conditions to solve the diffusion equation in the derivation of the FRAP model must be empirically determined from the intensity profile of the image collected immediately after photobleaching (postbleach profile),11
| (4) |
where for a normalized postbleach profile, is a bleaching parameter, is the radial displacement from the center of the bleaching ROI, and is the effective radius. and reflect the diffusion that occurs in the time it takes to bleach and acquire the postbleach image.
In a previous study, we collected line profiles from the postbleach image and averaged them for multiple cells to obtain the mean .13 This was necessary because line profiles are inherently noisy. Here, we determined on a cell-by-cell basis by increasing the signal to noise of the experimentally determined postbleach profiles as follows. First, we normalized the image acquired immediately after bleaching a circular bleach ROI (postbleach image) to the mean of 10 images acquired immediately prior to the postbleach image (pre-bleach images). Next, we calculated the radial displacement for each pixel in the image from the center of the circular bleach ROI (, ). The symmetry of a circular bleach ROI allows us to reduce the dimensions of the postbleach profile by plotting the intensity of a pixel in the normalized postbleach image vs. its radial displacement from the center of the circular bleaching ROI, .
To calculate , we utilized a series representation of a closed-form analytical FRAP equation describing free diffusion of unbleached molecules into a circular bleach ROI which is applicable to data obtained on a laser scanning confocal microscope,38
| (5) |
where for a normalized FRAP curve, is time, is the diffusion coefficient, and Mf is the mobile fraction. This equation is a modified form of the Axelrod equation22 where a Gaussian laser profile, and an approximation for the diffusion of molecules that occurred before acquisition of the postbleach image, are taken into account by incorporation of and parameters from the postbleach profile.38 To obtain a good match between from the postbleach profile and from the FRAP data, we numerically evaluated using Eq. (5) at .
To obtain an experimental FRAP curve we measured the mean intensity inside the circular bleaching ROI () at the location defined during data acquisition for each image in the time-lapse data set . Next, we normalized by the mean of for ten prebleach images.
Under our experimental conditions, there was a small amount of unintentional photobleaching that occurred throughout FRAP data acquisition. We verified this decay could be approximated by a first order exponential rate equation,
| (6) |
where is the first order rate constant for the decay. To correct the experimental curves, we acquired time-lapse data significantly longer than required for complete recovery after photobleaching and used at these later time points to estimate .17 The corrected FRAP curve is then,
| (7) |
By fitting the corrected data to Eq. (5) we obtain the parameters and Mf. However, because a significant fraction of fluorophores are irreversibly lost as the result of the photobleaching event, Mf is underestimated by this approach. To determine the true fraction of immobile fluorophores on the time scale of our FRAP experiments, we calculated the difference between and the mean intensity inside an adjacent circular ROI, , at time points after the recovery was complete.40
2.7. Other Data Analysis
Image analysis was performed using ImageJ (NIH), and nonlinear least squares analysis was performed using custom routines written in MatLab (MathWorks; Natick, MA). All scatter plots and bar graphs were created using MatLab and Adobe Illustrator (Adobe Systems Incorporated; San Jose, CA). All reported p values were calculated using an unpaired two sample t-test assuming unequal variances. In the figures, we summarized the results of this analysis with the following notation: NS signifies p > 0.05, * signifies p < 0.05, and ** signifies p < 0.001.
3. Results
3.1. Subcellular Localization of LC3 is Altered upon Coexpression of
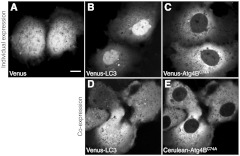
The interaction of with LC3 has been reported to cause a shift in the subcellular localization of LC3, leading to its sequestration in the cytoplasm.13,32 To confirm this shift in localization occurs under the conditions of our experiments, we compared the subcellular distribution of Venus- and Cerulean-tagged forms of LC3 and expressed individually versus under conditions where they were coexpressed using confocal microscopy (Fig. 1). COS-7 cells were used as a model system for these studies, and, as a control, we confirmed Venus itself is evenly distributed between the nucleus and cytoplasm [Fig. 1(a)].
Fig. 1.
Subcellular localization of Venus- and Cerulean-tagged versions of and LC3 when expressed individually or in combination. COS-7 cells were transiently transfected with the indicated constructs and imaged live. (a) Venus is evenly distributed between the cytoplasm and nucleus. (b) Venus-LC3 is enriched in the nucleus relative to the cytoplasm. (c) Venus- is enriched in the cytoplasm relative to the nucleus. (d) In cells coexpressing Venus-LC3 and Cerulean-, LC3 is pulled out of the nucleus. The relative distribution of LC3 between the nucleus and cytoplasm varies between cells depending on levels of present (not shown). (e) In cells coexpressing Venus- and Cerulean-LC3, is further enriched in the cytoplasm compared to its distribution in cells expressing Venus- alone. Bar, 10 μm.
When expressed on its own, Venus-LC3 was found localized in both the cytoplasm and the nucleoplasm but was notably enriched in the nucleoplasm [Fig. 1(b)]. Venus- expressed individually was also localized in both the cytoplasm and nucleus; however, unlike Venus-LC3, Venus- was enriched in the cytoplasm over the nucleus [Fig. 1(c)]. We next compared the distribution of Venus-LC3 and Cerulean- when the two proteins were coexpressed in the same cells. Under these conditions, Cerulean- was further concentrated in the cytoplasm and almost completely absent from the nucleus [Fig. 1(e)]. Similarly, a significant shift in the distribution of Venus-LC3 out of the nucleus was observed in cells coexpressing Cerulean- [Fig. 1(d)].
These findings show Venus- and Cerulean-tagged versions of these proteins behave as expected, consistent with a previous study where we showed by quantitative image analysis that the levels of EGFP-LC3 are significantly higher in the nucleus relative to the cytoplasm, and that coexpression of Strawberry- with EGFP-LC3 results in a redistribution of EGFP-LC3 out of the nucleus.13,32
3.2. and LC3 Are within FRET Proximity in Living Cells
Catalytically inactive mutants were previously shown to constitutively bind to LC3.32,41 In addition, a recent crystal structure shows two molecules of LC3 bind to catalytic and regulatory domains on a single molecule of .41 These findings suggest LC3 and should be within FRET proximity in living cells. To test this prediction, we performed acceptor photobleaching FRET microscopy measurements using Cerulean and Venus as the FRET donor and acceptor, respectively.
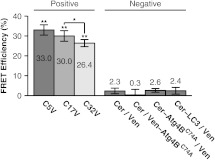
To establish conditions for the FRET assay, we measured a series of FRET standards consisting of Cerulean and Venus separated by 5, 17, or 32 amino acids as positive controls.42 The measured energy transfer efficiencies () under our conditions (Fig. 2) were slightly less than the values previously reported, possibly due to a small population () of acceptor remaining after bleaching.42 However, the relative trend between samples was identical. As negative controls, we measured FRET in cells coexpressing Cerulean and Venus-LC3, Cerulean and Venus-, Cerulean-LC3 and Venus, and Cerulean- and Venus. For these constructs, the mean ranged from to 2.6% (Fig. 2).
Fig. 2.
Controls for FRET microscopy. Mean percent energy transfer efficiency for positive FRET controls consisting of Cerulean and Venus linked by 5, 17, or 32 amino acids, and negative FRET controls (cells coexpressing Cerulean and Venus, Cerulean and Venus- or Cerulean- and Venus). Error bars represent 95% CI with cells from at least two independent experiments.
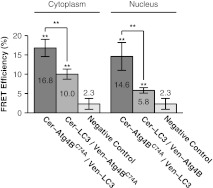
We measured significant FRET in the cytoplasm and nucleus of cells coexpressing either Cerulean-LC3 and Venus- or Cerulean- and Venus-LC3 (Fig. 3). Additionally, the measured FRET efficiencies were times larger when the FRET acceptor, Venus, was attached to LC3 compared to when Venus was attached to in the cytoplasm and nucleus (Fig. 3). These results show and LC3 are within FRET proximity in the cytoplasm of living cells, in agreement with previous biochemical evidence showing the two proteins constitutively interact in solution.32 In addition, our results suggest LC3 and also interact in the nuclear compartment.
Fig. 3.
FRET is detected between LC3 and in both the cytoplasm and nucleus of living cells. FRET efficiency between Cerulean- LC3 and Venus-, or Cerulean- and Venus-LC3 as measured using acceptor photobleaching FRET. Data are shown for measurements in both the cytoplasm and the nucleus. FRET efficiencies between the negative control Cerulean and Venus are shown for comparison. Error bars represent 95% CI with COS-7 cells from at least two independent experiments.
3.3. Both and LC3 Diffuse More Slowly than Freely Diffusing Monomers as Measured by Confocal FRAP
In a previous study, we found EGFP-LC3 diffuses more slowly than expected for a freely diffusing monomer, suggesting it may be part of a multiprotein complex13 This raises the possibility that other proteins known to interact with LC3 may also be part of this complex and diffuse similarly to LC3. Since is predicted to interact with endogenous LC3, we hypothesized may also diffuse as if it is part of a much larger complex, rather than a freely diffusing monomer.
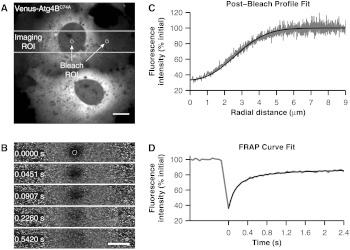
To test this, we performed measurements of Venus- using confocal FRAP to obtain a translational diffusion coefficient, which is related to its size assuming a particular geometry, such as a sphere or rod. Although monomers and dimers are difficult to discern from one another using this approach, molecules that exhibit large differences in molecular weight or shape from one another can be distinguished. As internal controls, we performed parallel FRAP studies for Venus and Venus-LC3, where Venus serves as an inert reporter of free diffusion.19 Soluble proteins like Venus diffuse rapidly, making it technically challenging to analyze their diffusional mobility because they recover completely within seconds following a photobleaching event. Because a laser scanning confocal microscope takes to acquire a full frame () image, it is not possible to resolve the early time points of the recovery curve while collecting images of this size. Therefore, to analyze the diffusional mobility of these proteins, we bleached a small circular region of interest located either in the nucleoplasm or in the cytoplasm, and imaged only the area immediately surrounding the bleach ROI during the recovery [Figs. 4(a) and 4(b)].11 Under these conditions, we were able to acquire images every 0.0451 s.
Fig. 4.
Confocal FRAP assay. (a) Example of the imaging ROI used for confocal FRAP experiments, and the position and size of bleach ROIs used to perform FRAP in the nucleus and cytoplasm. The image is representative of COS-7 cells expressing Venus-. Bar, 10 μm. (b) Normalized images of selected time points from a representative FRAP experiment on in the nucleus. The white circle shows the nominal bleach ROI. Bar, 10 μm. For visualization purposes a 1 pixel median filter was applied to the images. (c) Representative example of a fit [black line, Eq. (4)] to the mean postbleach profile of Venus- in the nucleus (gray line, cells) obtained from images such as in B at . The bleach depth () and effective radius () parameters from the postbleach profile fit provide an empirical estimate of the initial conditions for the diffusion equation. The normalized postbleach fluorescence intensity was obtained as described in Sec. 2. (d) Representative example of the mean confocal FRAP data from Venus- in the nucleus (gray line, ) fit with a theoretical FRAP curve assuming a single diffusing species [Eq. (5)].
We bleached a 0.99 μm circular bleach ROI positioned in either the cytoplasm or the nucleus by scanning 10 iterations of a higher intensity laser light, requiring approximately 0.15 s [Figs. 4(a) and 4(b)]. Because the bleaching event is not instantaneous and soluble proteins diffuse rapidly, some diffusion of the protein from the bleached region to the surrounding area occurs in the time it takes to acquire the postbleach image. This is evidenced by the presence of bleached molecules outside of the user-defined bleach ROI [Fig. 4(b)]. It is important to take this into account, because the intensity profile of the bleach spot defines the initial conditions to solve Fick’s second law in the derivation of the FRAP equation [Eq. (5)].11,38 This is accomplished by fitting the postbleach profile to obtain parameters and [Fig. 4(c)] [Eq. (4)].11 With the parameters and in hand, the corrected FRAP data is fit with Eq. (5) to obtain parameters and Mf [Fig. 4(d)].38 The value of the diffusion coefficient obtained for this example is , and the mobile fraction Mf is 0.82. However, because a significant fraction of fluorophores is lost as the result of the photobleaching event, this does not reflect the true fraction of mobile molecules. To obtain a more accurate estimate of (, 95%, CI ) we compared the fluorescence intensity inside an ROI positioned adjacent to the bleach regions after the fluorescence had completely recovered.40
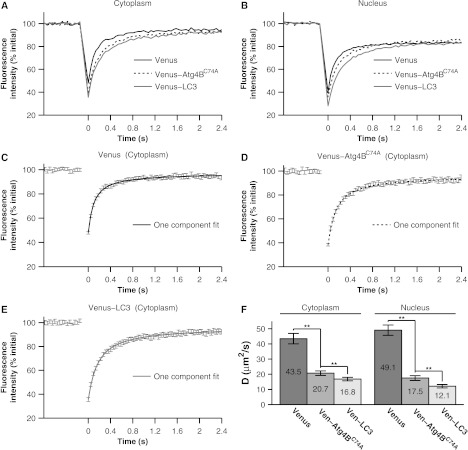
By comparison of the mean FRAP curves we found Venus-LC3 and Venus- both diffuse more slowly than Venus in the cytoplasm [Fig. 5(a)] as well as in the nucleus [Fig. 5(b)]. We measured mobile fractions close to 100% for all of these proteins on the timescale of our experiments (Table 1). To obtain values, the FRAP curves for Venus, Venus-LC3, and Venus- were fit with a one- component diffusion model [Eq. (5)]. This model fit the recovery curves effectively in both the cytoplasm [Figs. 5(c)–5(e)] and nucleus (data not shown). The mean from fits to Venus-LC3 and Venus- FRAP curves were and times smaller respectively than the mean for Venus in both the cytoplasm and the nucleus (95% CI, ) [Fig. 5(f)].
Fig. 5.
The diffusional mobilities of Venus-LC3 and Venus- in both the cytoplasm and the nucleus are significantly slower than those of Venus as assessed by confocal FRAP. (a) Comparison of mean FRAP curves for Venus, Venus-LC3, and Venus- in the cytoplasm ( cells from 3 independent experiments). (b) Comparison of mean FRAP curves for Venus, Venus-LC3, and Venus- in the nucleus ( cells from 3 independent experiments). (c)–(e) Representative examples of fits using a one-component diffusion model to the experimentally determined recovery curves for Venus (c), Venus- (d), and Venus-LC3 (e) in the cytoplasm (error bars represent 95% CI cells from 3 independent experiments). (f) Diffusion coefficients for Venus, Venus-LC3, and Venus- in the cytoplasm and nucleus. Error bars represent 95% CI cells from 3 independent experiments.
Table 1.
Mobile fractions () for Venus-LC3 and Venus-.
| Protein |
CI () |
|
|---|---|---|
| Cytoplasm | Nucleus | |
| Venus | (30) | (30) |
| Venus-LC3 | (30) | (30) |
| Venus- | (30) | (30) |
| Venus-LC3 () | (30) | (30) |
| Venus- () | (20) | |
Using the Stokes-Einstein relation (, with Venus as a standard for MW),19 we determined Venus-LC3 diffuses with an apparent molecular weight of , and Venus- diffuses slightly faster with an apparent molecular weight of in the cytoplasm assuming a spherical geometry (95% CI, ). In the nucleus we see a similar trend; Venus-LC3 diffuses with an apparent molecular weight of , and Venus- diffuses with an apparent molecular weight of (95% CI, ). Importantly, these molecular weights are much larger than the expected molecular weights for monomeric Venus-LC3 (45 kDa) and Venus- (72 kDa) (Table 2).
Table 2.
Comparison of the experimentally determined apparent molecular weights to the expected molecular weights for Venus- and Venus-LC3.
| Protein | Expected MW (kDa) | Apparent MW (kDa)a |
|
|---|---|---|---|
| Cytoplasm | Nucleus | ||
| Venus- | 72 | (30) | (30) |
| Venus-LC3 | 45 | (30) | (30) |
| Venus-LC3 () | 117 | (30) | (30) |
| Venus- () | 117 | (20) | N/A |
Values represent the mean CI. The number of observations is in parentheses.
Calculated assuming a spherical shape.
The above estimates of the size of the putative complex containing and LC3 assume a spherical geometry. Given the crystal structure of the LC3-Atg4B complex appears roughly rod shaped,41 if we estimate a fluorescently labeled complex is long, and wide, the expected diffusion coefficient is ,43 consistent with our diffusion measurements in the cytoplasm. However, in the nucleus the measured diffusion coefficient of LC3 is significantly smaller, suggesting the formation of a higher molecular weight complex or a much more anisotropic complex in this compartment.
3.4. and LC3 Coexpression Slows Diffusion as Measured by Confocal FRAP
If and LC3 are bound to the same complex, we expect the diffusion coefficients for these proteins should be identical and correspond to the size and shape of the complex. However, we determined for Venus- is while for Venus-LC3 is slightly slower at in the cytoplasm (95% CI ) [Fig. 5(f)]. A simple explanation for this finding is that there may be different fractions of bound and unbound forms of LC3 and . For example, 100% of Venus- may not be bound to LC3 if endogenous LC3 levels are limiting. Thus, the recoveries for Venus- may contain contributions from both rapidly diffusing unbound protein and slowly diffusing bound protein. If this is the case, we reasoned it may be possible to drive additional complex formation by transiently expressing additional LC3. Therefore, we next performed confocal FRAP measurements of Venus- in cells coexpressing additional Cerulean-LC3 (Fig. 6). As a control, we also measured the diffusional mobility of Venus-LC3 in cells coexpressing additional Cerulean-.
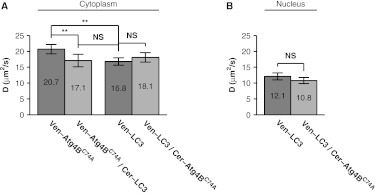
Fig. 6.
The diffusional mobility of Venus- is significantly slower upon coexpression with Cerulean-LC3, whereas no significant change was observed for the mobility of Venus-LC3 upon coexpression with Cerulean-. (a) Mean diffusion coefficients for Venus- Venus- coexpressed with Cerulean-LC3, Venus-LC3, and Venus-LC3 coexpressed with Cerulean in the cytoplasm ( COS-7 cells). (b) Mean diffusion coefficients for Venus-LC3 and Venus-LC3 coexpressed with Cerulean in the nucleus. Error bars represent 95% CI with COS-7 cells.
The FRAP curves for Venus- and Venus-LC3 in cells coexpressing either Cerulean-LC3 or Cerulean- respectively were, again, effectively fit by a pure diffusion model in both the cytoplasm and the nucleus (data not shown). Under these conditions, essentially 100% of the molecules were mobile on the timescale of our experiments (Table 1). The mean for Venus-LC3 coexpressed with Cerulean- was identical within error to that obtained for Venus-LC3 expressed alone (Fig. 6). This suggests excess Cerulean- binding to Venus-LC3 does not significantly influence the size or dynamics of the putative macromolecular complex containing Venus-LC3. In contrast, the mean for Venus- in the cytoplasm of cells coexpressing Cerulean-LC3 was significantly slower at compared to cells expressing Venus- on its own, and is identical to that of LC3 itself (95% CI, ).
Using the Stokes-Einstein relation (, with Venus as a standard for MW),19 we determined the apparent molecular weight of Venus-LC3 diffusing in cells coexpressing Cerulean- is in the cytoplasm and in the nucleus, which is statistically identical to Venus-LC3 expressed on its own (95% CI, ). This is also identical to the apparent molecular weight of for Venus- diffusing in the cytoplasm of cells coexpressing Cerulean-LC3 (95% CI, ) (Table 2). These results indicate the level of LC3 in the cytoplasm of cells is an important determinant of the diffusional mobility of when LC3 concentrations are limiting, and suggests not only are LC3 and likely directly interacting in live cells, but they also are part of the same multiprotein complex. As discussed above, if we assume the complex has a rod shape instead of spherical, the actual MW may be smaller than estimated here.
3.5. LC3 and LC3 are within FRET Proximity in the Nucleus, but Not the Cytoplasm of Living Cells
The results of the above experiments suggest both LC3 and are part of the same multiprotein complex in cells. However, it is unclear whether these complexes contain multiple copies of LC3 or . Furthermore, it was reported that an LC3 homolog, GABARAP, forms homo-oligomers.44,45 Therefore, the formation of large homo-oligomers of Venus-LC3 could possibly explain LC3’s slower than expected diffusional mobility. To test for the presence of homo-oligomers, as well as to test if multiple copies of LC3 may be present within the same high molecular weight complex, we performed acceptor photobleaching FRET experiments on cells coexpressing Venus-LC3 and Cerulean-LC3.
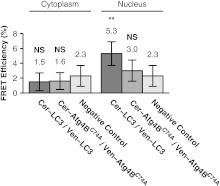
We detected no significant difference between for cells coexpressing Venus-LC3 and Cerulean-LC3 in the cytoplasm and for negative controls (, ) (Fig. 7). However, we observed a small but significant amount of energy transfer () in the nucleus of cells expressing Venus-LC3 and Cerulean-LC3 compared to the negative control () (95% CI, ) (Fig. 7). These results suggest LC3 either homo-oligomerizes, or more than one LC3 is in close proximity within a multiprotein complex in the nucleus but not the cytoplasm of live cells.
Fig. 7.
FRET occurs between donor and acceptor-labeled LC3 in the nucleus but not the cytoplasm of living cells, whereas no FRET is detected between donor and acceptor-labeled in either the cytoplasm or the nucleus. FRET efficiency between Cerulean-LC3 and Venus-LC3, or Cerulean- and Venus- as measured using acceptor photobleaching FRET. Data are shown for measurements in both the cytoplasm and the nucleus. FRET efficiencies between the negative control Cerulean and Venus are shown for comparison. Error bars represent 95% CI with COS-7 cells from at least two independent experiments.
3.6. and are not in FRET Proximity in Either the Cytoplasm or Nucleus of Living Cells
We also considered the possibility that forms homo-oligomers or that multiple copies of may be present within the same multiprotein complex. To test this, we performed acceptor photobleaching FRET experiments on cells coexpressing Venus- and Cerulean-. The energy transfer efficiencies between Venus- and Cerulean- were identical within error to values obtained for the negative controls (Fig. 7). These results demonstrate if multiple copies of are present in the same multiprotein complex, they are not close enough to yield detectable FRET.
4. Discussion
In the current study, we used FRET and FRAP to characterize the properties of a protein complex formed by the interaction of LC3 with a catalytically inert mutant form of Atg4B, the protease required for both lipidation and delipidation of LC3. Our findings reveal several novel properties of the complex formed between and LC3, as well as each of these molecules individually.
As a starting point for our study, we confirmed coexpression of Cerulean- with Venus-LC3 results in a detectable shift in the subcellular localization of LC3 as reported previously.13,32 In particular, upon coexpression with , LC3 is de-enriched from the nucleus and becomes sequestered in the cytoplasm. This suggests the localization of is dominant over that of LC3. Why this is the case has not been established. However, a predictive sequence analysis for nuclear export signals indicates contains a putative consensus leucine rich NES at residues 225 to 230.46 Thus, it is possible is actively exported from the nucleus, leading to the accumulation of both LC3 and in the cytoplasm. LC3 was also predicted to have an NES; however, this NES does not appear to be functional under steady-state conditions.13 Therefore, a functional NES on may control the nucleo-cytoplasmic distribution of both and LC3.
Our FRET measurements provide supporting evidence that LC3 and likely directly interact in the cytoplasm of living cells. The observation that these two proteins are in FRET proximity is in good agreement with previous biochemical evidence showing these two proteins form a stable complex.32,41 We also show here the interaction of LC3 and is not exclusively confined to the cytoplasm, where the lipidation and delipidation of LC3 and autophagosome formation are known to occur. Instead, they interact within the nucleus as well, indicating either LC3 or Atg4B or both may have currently unidentified functions in the nucleus, consistent with our previous findings showing soluble LC3 itself is enriched in the nucleus relative to the cytoplasm.13
In addition, we observed a significant increase in energy transfer efficiency when the acceptor LC3 is labeled with the acceptor as opposed to when is labeled with the acceptor. In our experiments the manner in which Cerulean or Venus is attached to LC3 and Atg4B is identical. Assuming the average relative orientation of transition dipole moments in a complex between LC3 and is the same regardless of which protein is labeled with Cerulean or Venus, the only variable in a comparison of FRET experiments where the donor and acceptors are switched is the relative amount of donor to acceptor. Based on the recent crystal structure for the interaction, it is plausible two molecules of acceptor labeled LC3 are within close proximity of a single donor labeled , providing significantly more opportunity for energy transfer to occur.6,41 This suggests the -LC3 complex may not only occur under crystallography conditions, but also occur in the native environment of live cells, and may represent an important mechanism for regulating LC3 post-translational modification. However, slight structural differences in the two different fluorescent protein labels could lead to differences in the average relative orientation of transition dipole moments when bound in the complex.47 For this reason, brightness analysis or time-resolved fluorescence anisotropy could be used in the future to further validate a 2:1 stoichiometry for the LC3- complex in live cells.34,48
In a previous study, we found the mobility of LC3 is much slower than expected for a free monomer in the cytoplasm and nucleus consistent with that of a high molecular weight complex.13 The actual molecular weight of this complex is currently unknown, as the relationship between and MW is dependent on the geometry of the complex, and for anisotropic molecules will be dominated by the longest dimension of the molecule. Since should bind to endogenous LC3, we speculated it may also become incorporated into a slowly diffusing complex as the result of this interaction. Here, we tested this possibility by performing quantitative FRAP experiments using a laser scanning confocal microscope. If the transport process leading to recovery of fluorescence after photobleaching is simple diffusion, quantitative analysis of a FRAP curve yields the diffusion coefficient of the diffusing species, which is related to the temperature and viscosity of the medium as well as to the size and shape of the diffusing species. To obtain quantitative estimates of the diffusion coefficients from FRAP data we utilized a recently developed analytical FRAP model applicable to laser scanning confocal microscopes [Eq. (5)].38 The model is a generalized form of the classical Axelrod equation which assumed a stationary Gaussian laser profile, but takes into account diffusion which occurs before acquisition of the first image in the time series by determining the initial conditions from the postbleach image intensity profile [Figs. 4(b) and 4(c)].11
Our FRAP measurements revealed the diffusion coefficient of expressed in live cells was much slower than expected for a freely diffusing monomer according to the Stokes-Einstein relation. Although it is difficult to distinguish a monomer from a dimer using this method, a monomer is readily discernible from a much larger or more anisotropic complex. Thus, our finding strongly suggests does not exist as a monomer, but rather is incorporated into a larger complex in live cells. However, we noted for was slightly faster than for LC3. We hypothesized the difference in between these proteins expressed individually could be due to limiting concentrations of endogenous LC3 resulting in a larger fraction of unbound from the complex. To test this, we coexpressed LC3 in an attempt to drive additional complex formation and reduce the fraction of unbound . Interestingly, the for Venus- in cells expressing additional LC3 became significantly slower to match that of LC3 itself. These results complement our conclusions based on the FRET data and further suggest LC3 and are likely directly interacting to form a complex in live cells.
As a control, we also expressed additional and looked for changes in for Venus-LC3. Interestingly, for Venus-LC3 was unchanged in cells coexpressing additional . These results suggest there was little LC3 unbound from the complex, and that expression and binding of to LC3 does not interfere with the size or dynamics of the LC3 interacting complex normally slowing its diffusion. Fluorescence correlation spectroscopy (FCS) could be used in future experiments to further validate these findings based on its ability to readily resolve multiple diffusing components.
Given the approximate rod shape of the complex between LC3 and as observed from a recent crystal structure41, we examined the possibility that LC3 and may be diffusing as a rod shaped complex in live cells. We found our diffusion measurements for LC3 and were consistent with their incorporation into a complex of this size and shape in the cytoplasm, but fluorescence anisotropy measurements could potentially be used to further validate this finding. Interestingly, the size and shape of the LC3-Atg4B crystallography complex cannot account for the very slow diffusion of LC3 observed in the nucleus. This implies LC3 may constitutively associate with a larger or more anisotropic complex with currently unknown composition in this compartment.
Our FRAP measurements from cells expressing LC3 alone, alone, as well as cells coexpressing the two proteins, all displayed mobile fractions in the regions of the cells where we performed our measurements. This suggests the majority of LC3 and does not stably associate with cellular superstructures, e.g., microtubules or DNA which are essentially immobile on the time scale of our FRAP experiments. We deliberately avoided bright punctate () structures, which presumably represent autophagosome vesicles. The large vesicles are also effectively stationary on the time scale of our FRAP experiments. If these structures were selected for FRAP experiments, the mobile fractions would likely be less than 100%.
Since we found both LC3 and appear to diffuse much slower than soluble Venus, this raised the possibility that the slower than expected diffusion may be the result of homo-oligomerization of LC3 or , or that multiple copies of LC3 and may be contained within the same multiprotein complex. To test this possibility we performed acceptor photobleaching FRET measurements on cells coexpressing Venus- and Cerulean-tagged versions of LC3 and . In the cytoplasm we found no evidence for the presence of LC3 or homo-oligomers. The absence of detectable FRET between LC3 and LC3, or between and , implies that if more than one copy of LC3 or is present in the same slowly diffusing complex, these molecules are positioned at distances greater than 10 nm from one another in the cytoplasm. However, due to the orientation requirements for FRET, the absence of energy transfer efficiency cannot rule out the possibility of homo-oligomerization, or more than one LC3 in a complex. For example, if two LC3’s are bound to as seen in the crystal structure, it is possible their N-terminal fluorescent protein labels are positioned on opposite sides of (), preventing a detectable amount of energy transfer from occurring.
Conversely, in the nucleus we observed a detectable level of energy transfer between Cerulean-LC3 and Venus-LC3 compared to negative controls. Therefore, in the nucleus, LC3 may homo-oligomerize or more than one LC3 molecule is present in close proximity within the same multiprotein complex. Further, this suggests the organization of LC3 within complexes in the nucleus may differ from that in the cytoplasm, bringing the proteins into sufficiently close proximity to give rise to FRET.
In summary, our data are consistent with a model in which and LC3 not only directly interact but also associate with a slowly diffusing complex in both the cytoplasm and the nucleus of living cells. This complex could either have a relatively high molecular weight or be elongated in shape. The identities of the components of the putative large complexes in the nucleus are not yet known, but are likely to involve other components of the conjugation machinery in the autophagy pathway. In addition, the organization of LC3 within complexes in the nucleus may be different from that in the cytoplasm, or alternatively LC3 may homo-oligomerize in the nucleus. These data also strongly suggest LC3 and may both have currently undefined functions in the nucleus. Ultimately, these types of fundamental measurements of live- cell protein diffusion and complex formation described here can be used to extend biochemical studies to the single living cell in order to better understand intracellular pathways such as autophagy.
Acknowledgments
We sincerely thank M. Kang for assistance with FRAP data analysis, and J. Greer for technical assistance with cloning. We also thank the other members of the Kenworthy lab for helpful discussions about the project. We thank S. Vogel for providing the FRET standards, T. Yoshimori for providing the EGFP-LC3 and mStrawberry- constructs, and D. Piston for providing the Cerulean and Venus constructs. Support from R01 GM073846, 3R01 GM73846-4S1, and NSF/DMS 0970008 is gratefully acknowledged. The funding sources had no role in the study design, collection, analysis or interpretation of data, writing the report, or the decision to submit the paper for publication.
References
- 1.Verveer P. J., Bastiaens P. I., “Quantitative microscopy and systems biology: seeing the whole picture,” Histochem. Cell Biol. 130(5), 833–843 (2008). 10.1007/s00418-008-0517-5 [DOI] [PubMed] [Google Scholar]
- 2.Dehmelt L., Bastiaens P. I., “Spatial organization of intracellular communication: insights from imaging,” Nat. Rev. Mol. Cell Biol. 11(6), 440–452 (2010). 10.1038/nrm2903 [DOI] [PubMed] [Google Scholar]
- 3.Sung M. H., McNally J. G., “Live cell imaging and systems biology,” Wiley Interdiscip. Rev. Syst. Biol. Med. 3(2), 167–182 (2011). 10.1002/wsbm.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J., Snapp E., Kenworthy A. K., “Studying protein dynamics in living cells,” Nat. Rev. Mol. Cell Biol. 2, 444–456 (2001). 10.1038/35073068 [DOI] [PubMed] [Google Scholar]
- 5.Periasamy A., Day R. N., “Visualizing protein interactions in living cells using digitized GFP imaging and FRET microscopy,” Methods Cell Biol. 58, 293–314 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Vogel S. S., Thaler C., Koushik S. V., “Fanciful FRET,” Sci. STKE 2006(331), re2- (2006). [DOI] [PubMed] [Google Scholar]
- 7.Demarco I. A., Periasamy A., Booker C. F., Day R. N., “Monitoring dynamic protein interactions with photoquenching FRET,” Nat. Methods 3(7), 519–524 (2006). 10.1038/nmeth889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jares-Erijman E. A., Jovin T. M., “FRET imaging,” Nat. Biotechnol. 21(11), 1387–1395 (2003). 10.1038/nbt896 [DOI] [PubMed] [Google Scholar]
- 9.Wouters F. S., et al. , “FRET microscopy demonstrates molecular assocation of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes,” EMBO J. 17, 7179–7189 (1998). 10.1093/emboj/17.24.7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon G. W., et al. , “Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy,” Biophys. J. 74(5), 2702–2713 (1998). 10.1016/S0006-3495(98)77976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braga J., Desterro J. M., Carmo-Fonseca M., “Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes,” Mol. Biol. Cell 15(10), 4749–4760 (2004). 10.1091/mbc.E04-06-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., et al. , “Methods to measure the lateral diffusion of membrane lipids and proteins,” Methods 39(2), 147–153 (2006). 10.1016/j.ymeth.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 13.Drake K. R., Kang M., Kenworthy A. K., “Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3,” PloS One 5(3), e9806 (2010). 10.1371/journal.pone.0009806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin J. S., Kenworthy A. K., “Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells,” Methods 37(2), 154–164 (2005). 10.1016/j.ymeth.2005.05.013 [DOI] [PubMed] [Google Scholar]
- 15.Kang M., Day C. A., DiBenedetto E., Kenworthy A. K., “A quantitative approach to analyze binding diffusion kinetics by confocal FRAP,” Biophys. J. 99(9), 2737–2747 (2010). 10.1016/j.bpj.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michelman-Ribeiro A., et al. , “Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy,” Biophys. J. 97(1), 337–346 (2009). 10.1016/j.bpj.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller F., Wach P., McNally J. G., “Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching,” Biophys. J. 94(8), 3323–3339 (2008). 10.1529/biophysj.107.123182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reits E. A. J., Neefjes J. J., “From fixed to FRAP: measuring protein mobility and activity in living cells,” Nat. Cell Biol. 3, E145–E147 (2001). 10.1038/35078615 [DOI] [PubMed] [Google Scholar]
- 19.Sprague B. L., et al. , “Analysis of binding reactions by fluorescence recovery after photobleaching,” Biophys. J. 86(6), 3473–3495 (2004). 10.1529/biophysj.103.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S. A., Heinze K. G., Schwille P., “Fluorescence correlation spectroscopy in living cells,” Nat. Methods 4(11), 963–973 (2007). 10.1038/nmeth1104 [DOI] [PubMed] [Google Scholar]
- 21.Slaughter B. D., Schwartz J. W., Li R., “Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging,” Proc. Natl. Acad. Sci. U.S.A. 104(51), 20320–20325 (2007). 10.1073/pnas.0710336105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axelrod D., et al. , “Mobility measurement by analysis of fluorescence photobleaching recovery kinetics,” Biophys. J. 16(9), 1055–1069 (1976). 10.1016/S0006-3495(76)85755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behrends C., et al. , “Network organization of the human autophagy system,” Nature 466(7302), 68–76 (2010). 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z., Klionsky D. J., “Eaten alive: a history of macroautophagy,” Nat. Cell Biol. 12(9), 814–822 (2010). 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine B., Mizushima N., Virgin H. W., “Autophagy in immunity and inflammation,” Nature 469(7330), 323–335 (2011). 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine B., Kroemer G., “Autophagy in the pathogenesis of disease,” Cell 132(1), 27–42 (2008). 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geng J., Klionsky D. J., “The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series,” EMBO Rep. 9(9), 859–864 (2008). 10.1038/embor.2008.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichimura Y., et al. , “A ubiquitin-like system mediates protein lipidation,” Nature 408(6811), 488–492 (2000). 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 29.Ohsumi Y., “Molecular dissection of autophagy: two ubiquitin-like systems,” Nat. Rev. Mol. Cell Biol. 2(3), 211–216 (2001). 10.1038/35056522 [DOI] [PubMed] [Google Scholar]
- 30.Kabeya Y., et al. , “LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing,” EMBO J. 19(21), 5720–5728 (2000). 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirisako T., et al. , “The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway,” J. Cell Biol. 151(2), 263–276 (2000). 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita N., et al. , “An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure,” Mol. Biol. Cell 19(11), 4651–4659 (2008). 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N., Noda T., Yoshimori T., “Atg4B(C74A) hampers autophagosome closure: a useful protein for inhibiting autophagy,” Autophagy 5(1), 88–89 (2009). 10.4161/auto [DOI] [PubMed] [Google Scholar]
- 34.Periasamy A., Clegg R. M., FLIM Microscopy in Biology and Medicine, Taylor & Francis, Boca Raton: (2010). [Google Scholar]
- 35.Periasamy A., Day R. N., Molecular Imaging: FRET Microscopy and Spectroscopy, Oxford University Press Inc., New York, New York: (2005). [Google Scholar]
- 36.Lakowicz J. R., Principles of Fluorescence Spectroscopy, Springer, New York: (2006). [Google Scholar]
- 37.Rizzo M. A., et al. , “An improved cyan fluorescent protein variant useful for FRET,” Nat. Biotechnol. 22(4), 445–449 (2004). 10.1038/nbt945 [DOI] [PubMed] [Google Scholar]
- 38.Kang M., et al. , “A generalization of theory for two-dimensional fluorescence recovery after photobleaching applicable to confocal laser scanning microscopes,” Biophys. J. 97(5), 1501–1511 (2009). 10.1016/j.bpj.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenworthy A. K., “Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy,” Methods 24(3), 289–296 (2001). 10.1006/meth.2001.1189 [DOI] [PubMed] [Google Scholar]
- 40.Hinow P., et al. , “The DNA binding activity of p53 displays reaction-diffusion kinetics,” Biophys. J. 91(1), 330–342 (2006). 10.1529/biophysj.105.078303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satoo K., et al. , “The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy,” EMBO J. 28(9), 1341–1350 (2009). 10.1038/emboj.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koushik S. V., et al. , “Cerulean, Venus, and VenusY67C FRET reference standards,” Biophys. J. 91(12), L99–L101 (2006). 10.1529/biophysj.106.096206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia de la Torre J., Lopez Martinez M. C., Tirado M. M., “Dimensions of short, rodlike macromolecules from translational and rotational diffusion-coefficients—study of the gramicidin dimer,” Biopolymers 23(4), 611–615 (1984). 10.1002/bip.360230402 [DOI] [Google Scholar]
- 44.Nymann-Andersen J., Wang H., Olsen R. W., “Biochemical identification of the binding domain in the GABA(A) receptor-associated protein (GABARAP) mediating dimer formation,” Neuropharmacology 43(4), 476–481 (2002). 10.1016/S0028-3908(02)00165-X [DOI] [PubMed] [Google Scholar]
- 45.Pacheco V., et al. , “Assessment of GABARAP self-association by its diffusion properties,” J. Biomol. NMR 48(1), 49–58 (2010). 10.1007/s10858-010-9437-5 [DOI] [PubMed] [Google Scholar]
- 46.la Cour T., et al. , “Analysis and prediction of leucine-rich nuclear export signals,” Protein Eng. Des. Sel. 17(6), 527–536 (2004). 10.1093/protein/gzh062 [DOI] [PubMed] [Google Scholar]
- 47.Piston D. W., Kremers G. J., “Fluorescent protein FRET: the good, the bad and the ugly,” Trends Biochem. Sci. 32(9), 407–414 (2007). 10.1016/j.tibs.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 48.Chen Y., et al. , “The photon counting histogram in fluorescence fluctuation spectroscopy,” Biophys. J. 77(1), 553–567 (1999). 10.1016/S0006-3495(99)76912-2 [DOI] [PMC free article] [PubMed] [Google Scholar]