Levels of Lethal-7a in KRAS-mutated colorectal carcinomas from patients who received salvage cetuximab plus irinotecan were quantified and studied for association with survival outcomes. In the whole group, higher Lethal-7a levels were significantly associated with better survival outcomes.
Keywords: Colorectal cancer, KRAS, MicroRNA, Let-7, Cetuximab, Prognosis
Abstract
Preclinical and experimental data in vivo indicate that Lethal-7 (Let-7) microRNA downregulates KRAS with antitumor effects in the presence of activating KRAS mutations. We quantified the Let-7a isoform in KRAS-mutated colorectal carcinomas from patients who received salvage cetuximab plus irinotecan. The study population was retrospectively identified among metastatic colorectal cancer patients who underwent third-line therapy with cetuximab plus irinotecan in a period when only epidermal growth factor receptor (EGFR) expression was required for anti-EGFR therapy. In 59 patients harboring KRAS mutations, Let-7a levels were analyzed for association with overall survival (OS) and progression-free survival (PFS) times. An exploratory subgroup analysis was performed using the rs61764370 (LCS6 T>G) polymorphism that experimentally impairs Let-7 binding to KRAS mRNA. In the whole group, higher Let-7a levels were significantly associated with better survival outcomes. For the primary OS endpoint, the multivariate hazard ratio was 0.82 (95% confidence interval, 0.73–0.91; p = .01). The same findings with an accentuated positive effect of high Let-7a levels on both OS and PFS times were observed in an exploratory analysis of the 45 wild-type LCS6 patients (excluding 14 carriers of the LCS6 G allele variant). All survival associations were confirmed after excluding patients with KRAS codon 13 mutations. Among the clinicopathologic features, high Let-7a levels were associated with grade 2–3 skin toxicity (p = .002). In patients with KRAS mutations, Let-7a analysis may serve to identify subgroups of patients who may still benefit from EGFR inhibition and this may open up new perspectives for alternative treatment strategies.
Introduction
Understanding the mechanisms of post-transcriptional regulation of oncogenic pathways [1] may lead to novel and hopefully effective treatment strategies for cancer patients with activated KRAS [2]. In this regard, members of the Lethal-7 (Let-7) family of microRNAs (miRNAs) have been found to display tumor suppressor functions [3, 4] and to possess KRAS downregulating activity [5, 6]. Let-7 induces KRAS downregulation after binding to specific sites in the 3′ untranslated region (3′-UTR) of KRAS mRNA [5]. These findings led to growing interest in Let-7 for its role in cancer development and control [2]. Most relevant, Esquela-Kerscher et al. [3] and Kumar et al. [4] recently showed that exogenous Let-7 reduced tumor formation in vivo in animals expressing the G12D activating KRAS mutation.
In the last few years, anti-epidermal growth factor receptor (EGFR) therapy with the monoclonal antibodies cetuximab and panitumumab has represented a major improvement in the treatment of patients with metastatic colorectal cancer. Activating KRAS mutations (mainly in codon 12 and codon 13) are predictive of disease unresponsive to anti-EGFR therapy [7], and analysis of the KRAS mutational status has become mandatory for their use [8]. However, as recently reported by De Roock et al. [7], tumor shrinkage and intriguing disease control rates (responsive patients and patients with stable disease) may be observed in chemotherapy-refractory patients harboring KRAS mutations treated with salvage cetuximab plus irinotecan.
We hypothesized the existence of a proportion of colorectal cancer patients with KRAS mutations who may still obtain a survival benefit from anti-EGFR therapy when their tumors display upregulated Let-7a levels. With this in mind, we investigated Let-7 miRNA levels in colorectal carcinomas with KRAS mutations in patients treated with salvage cetuximab plus irinotecan. We chose the Let-7a isoform for the assessment. In fact, it has been adequately characterized in tumor models for its KRAS downregulating function [2–5], and Let-7a levels have been assessed in paraffin-embedded tissues from gastrointestinal carcinomas [9].
Evidence of the existence of this effect in vivo may translate into relevant clinical applications. First, whether or not anti-EGFR agents might have some benefit in subgroups of KRAS-mutated patients with a favorable miRNA profile should be reconsidered. Also, this would encourage the clinical development of anti-KRAS, miRNA-based treatment strategies [2].
The overall survival (OS) time was the primary endpoint of the study. Potential nonhomogeneous scheduling of times for radiological assessments prompted us to consider the progression-free survival (PFS) interval as a secondary endpoint. Additional analyses addressed the clinical impact of Let-7a levels in relation to a single nucleotide polymorphism (SNP) in the Let-7a KRAS mRNA binding site and the type of KRAS mutation. In fact, the T>G base change (rs61764370) in a Let-7 complementary site (LCS6) attenuates the binding capability of mature Let-7 to target KRAS mRNA [10], whereas the weight of the detrimental effects of KRAS activation may differ according to mutations in codon 12 or codon 13 [11].
Materials and Methods
Patients
In 2005–2008, 172 patients were treated with cetuximab plus irinotecan as salvage therapy for metastatic colorectal cancer at three medical oncology units in central Italy. During this period, only positive EGFR expression was required for selecting patients to be treated with anti-EGFR therapy. In this group, we retrospectively identified patients who were carriers of KRAS mutations and had a wild-type BRAF status in the primary tumor, and were therefore deemed eligible for the present investigation. They were required to be classified as irinotecan refractory (i.e., progressed ≤3 months after treatment with an irinotecan-based regimen) and were treated with a third-line combination of biweekly irinotecan (180 mg/m2) with weekly cetuximab (400 mg/m2 loading dose followed by 250 mg/m2). In each case, availability of a formalin-fixed paraffin-embedded (FFPE) tumor specimen was mandatory for performing the Let-7a miRNA quantification and LCS6 SNP analysis.
Pretreatment evaluation included a medical history, clinical and physical examinations, Eastern Cooperative Oncology Group (ECOG) performance status evaluation, assessment of metastatic disease based on computed tomography scans, x-rays, or other radiographic means and serum chemistries. The OS time was defined as the time from the beginning of therapy to death or last follow-up. The PFS interval was defined as the time from the beginning of cetuximab–irinotecan therapy to the first appearance of progression or death resulting from any cause. Patient characteristics and their outcomes were unknown to investigators performing genetic analyses. The study was planned according to the Reporting Recommendations for Tumor Marker Prognostic Studies criteria [12] and was approved by local ethics committees.
Samples
Three to five 10-μm sections from FFPE specimens were obtained from the primary tumor. Representative areas from FFPE tumor blocks were evaluated by pathologists. Before cutting sections for miRNA isolation, one slide was prepared for hematoxylin and eosin staining to select only representative samples with almost complete tumor infiltration. Sections were sent to the Laboratory of Molecular Biology, Department of Molecular Sciences, University of Urbino for centralized analyses of the KRAS and BRAF mutational status, Let-7a levels, and KRAS 3′-UTR LCS6 genotypes. All assays were performed by investigators who were blinded to the clinical data of the sample cohort.
RNA Preparation for Let-7a Analysis
Total RNA was isolated from human FFPE tissues using the miRNeasy FFPE Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The extracted RNA was quantified by absorbance at 260 nm and its purity was evaluated by the absorbance ratio at 260 nm/280 nm with a NanoDrop 1000 spectrophotometer (Nanodrop Technologies, Inc., Rockland, DE).
cDNAs were generated using the miScript Reverse Transcription (RT) Kit (Qiagen) according to the manufacturer's instructions. The RT products were diluted 1:50 to obtain a final concentration of 1 ng/μL of reverse-transcribed RNA.
Quantitative Real-Time Polymerase Chain Reaction
Real-time polymerase chain reaction (PCR) was performed using the miScript SYBR Green PCR Kit (Qiagen) on an Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, CA). The 25 μL of PCR mixture included 2 μL diluted RT product, 12.5 μL 2× QuantiTect SYBR Green PCR Master Mix, 2.5 μL 10× miScript Universal Primer, 2 μL 10× mi-Script Primer Assay for Hs_let-7a_1 (Qiagen; catalog number, MS00006482) or miScript Primer Assay for Hs_RNU6B_2 (Qiagen; catalog number, MS000014000), and 5.5 μL distilled water. The reaction mixtures were incubated at 95°C for 15 minutes, followed by 40 amplification cycles at 94°C for 15 seconds, 55°C for 30 seconds, and 70°C for 34 seconds. All reactions were performed in triplicate and each PCR run included a no template control. PCR amplification efficiencies were calculated for Let-7a and RNU6B using the formula E = (10−1/slope − 1) × 100, using the slope of the plot of the cycle threshold (Ct) versus the log input of cDNA (tenfold dilution series). PCR amplification efficiencies were 97.5% and 99.5% for Let-7a and RNU6B, respectively. RNU6B (small nuclear RNA 6B) was used as a reference gene and the relative expression level of Let-7a was expressed as ΔCt = Ct(Let-7a) − Ct(RNU 6B). The Ct was defined as the number of cycles needed for the fluorescence to reach a specific threshold level of detection selected (0.02) and is inversely correlated with the amount of template nucleic acid present in the sample [13]. Therefore, a higher miRNA expression level corresponds to a smaller ΔCt value. All experiments were repeated using a secondary reference gene (RNU48).
Genetic Analyses
DNA was extracted from tumor tissue samples using the Qiamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer's protocol. Methods for assessing the KRAS and BRAF mutational status were described previously [14]. PCR analyses to detect KRAS 3′-UTR LCS6 genotypes were performed as follows. The primers 5′-TTTTAGGAGAGACGGGGTTTCA (forward), 5′-[Btn]-TGAGTTCTGCAAAACAGGTTTATG (reverse), and 5′-TCCTGACCTCAAGTGAT (sequence) were designed using PSQ Assay Design Software (Biotage, Uppsala, Sweden). Each PCR contained 50–150 ng DNA, 0.4 μM of each primer (forward and reverse), 12.5 μL PCR Master Mix (Diatheva, Fano, Italy), and 0.625 U HotStarTaq polymerase (Diatheva) in a total volume of 25 μL. Successful and specific amplification of the region of interest was verified by visualizing 5 μL of the PCR product on a 2% agarose gel. Preparation of the single-stranded DNA template for the pyrosequencing analysis was performed using the PSQ Vacuum Prep Tool (Biotage) according to the manufacturer's instructions. Twenty microliters of biotinylated PCR product was immobilized on streptavidin-coated Sepharose™ High Performance beads (Amersham Biosciences, Piscataway, NJ) and processed to obtain single-stranded DNA using the PSQ 96 Sample Preparation Kit (Biotage) according to the manufacturer's instructions. The template was incubated with 0.4 μmol/L sequencing primer at 80°C for 2 minutes in a PSQ96 plate. Sequencing by synthesis reaction of the complementary strand was automatically performed on a PSQ 96MA instrument (Biotage) using PyroGold reagents (Biotage).
Statistical Analysis
Let-7a miRNA levels were expressed as values obtained from the ΔCt equation with mean and standard deviation (SD) and compared within each analyzed group using a t-test. For prognostic analyses, a recursive descent partition analysis was employed for splitting Let-7a expression into high-level and low-level groups. As previously reported [15] and as used in miRNA investigations [16], this method finds a set of cutpoints of X values (gene expression) that best predict the Y value (survival time). These data splits are done recursively, forming a tree of decision rules until the desired fit is reached; the most significant split is determined by the largest likelihood ratio χ2 statistic. In either case, the split is chosen to maximize the difference in the responses between the two branches of the split. Subsequently, survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. Finally, a Cox proportional hazards model was used to estimate and test Let-7a levels and clinicopathological features for their association with OS and PFS outcomes. Age was included as a continuous variable, whereas Let-7a level (high versus low), KRAS mutation (codon 12 versus codon 13), sex (male versus female), ECOG performance status score (0 versus 1), number of metastatic sites (1 versus ≥2), and skin toxicity (grade 0–1 versus grade 2–3) were included as dichotomous variables. As the decision level for the multivariate analyses, we included variables with a p-value ≤.10 obtained in the univariate analysis. Results are expressed as a hazard ratio (HR) with a 95% confidence interval (95% CI). The assumption of proportional hazard was verified. Statistical significance was set at a two-sided p-value <.05. All statistical analyses were performed using JMP 7.01 software (SAS Institute, Cary, NC).
Results
Characteristics of Patients
Among the 172 patients screened for KRAS and BRAF status, 65 (37%) had KRAS mutations. Six patients were excluded—four were deemed not irinotecan refractory whereas tumor specimens were not sufficient for miRNA analysis in two additional patients. The final study population consisted of 59 patients with a wild-type BRAF status and a KRAS mutation in codon 12 (G12D in 22 patients, G12V in 13 patients, G12C in five patients, G12S in four patients, G12A in four patients, and G12R in one patient) or codon 13 (G13D in nine patients and G13C in one patient).
All patients received third-line therapy with cetuximab (250 mg/m2 i.v.) on day 1 weekly (loading dose, 400 mg/m2, day 1 in the first cycle) and irinotecan (180 mg/m2 i.v.) on day 1 every 2 weeks. The characteristics of the 59 patients are shown in Table 1. Treatment outcome was stable disease in 13 patients (22%) and progression in 46 patients (78%). The median PFS interval was 12 weeks (95% CI, 9–14 weeks) and the median OS duration was 21 weeks (95% CI, 17–26 weeks). All patients died with progression of metastatic disease.
Table 1.
Characteristics of patients and Let-7a levels
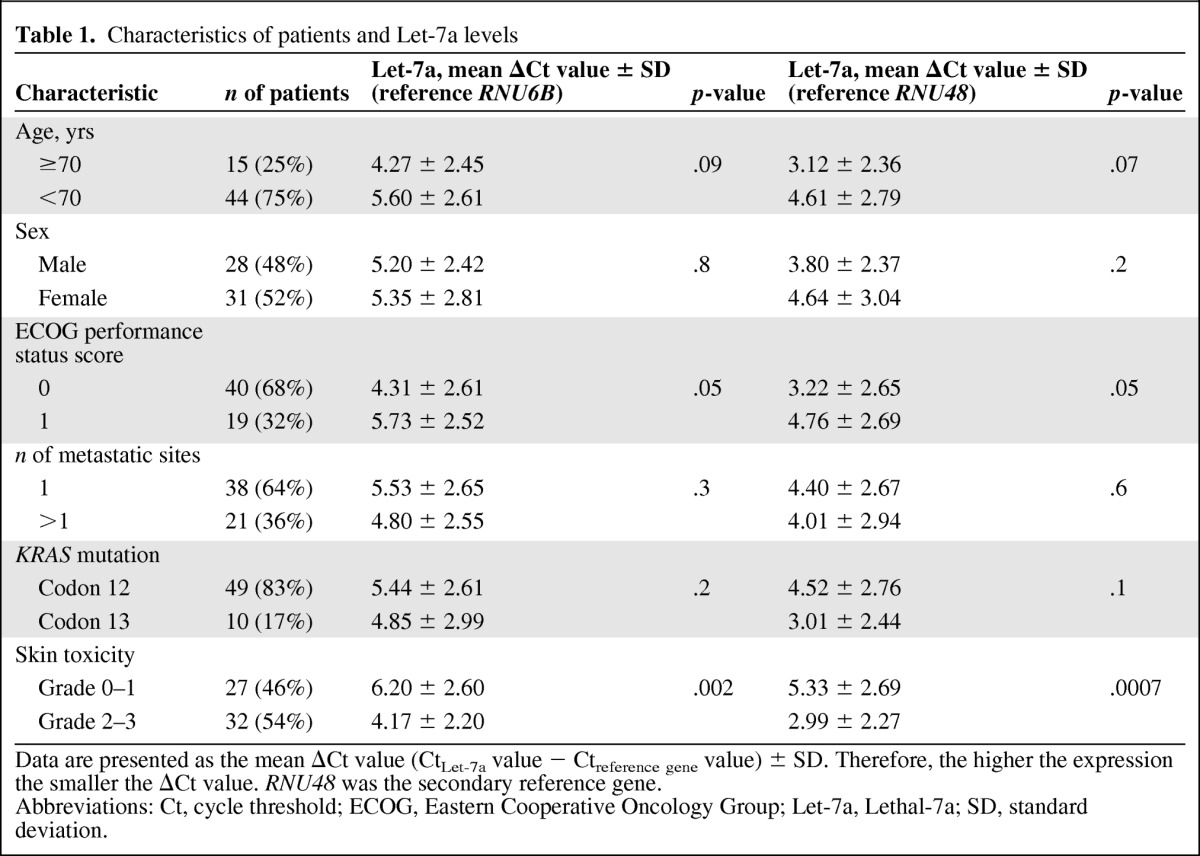
Data are presented as the mean ΔCt value (CtLet-7a value − Ctreference gene value) ± SD. Therefore, the higher the expression the smaller the ΔCt value. RNU48 was the secondary reference gene.
Abbreviations: Ct, cycle threshold; ECOG, Eastern Cooperative Oncology Group; Let-7a, Lethal-7a; SD, standard deviation.
Descriptive Let-7a Analyses
Let-7a levels were detectable in all tumor samples, with lowest and highest ΔCt values of 1.3 and 10.2, respectively. In the overall study population, the mean ΔCt value was 5.27 (±2.6 SD). Association analyses between Let-7a level and clinicopathologic features are shown in Table 1. Data are presented as the mean ΔCt (CtLet-7a value − CtRNU 6B value) ± SD. Therefore, the higher the expression, the smaller the ΔCt value. Among these patients, there was a significant association between Let-7a level and skin toxicity. According to the observed difference in ΔCt value, Let-7a levels were higher in patients with grade 2–3 skin toxicity than in patients with grade 0–1 skin toxicity. All findings were confirmed after repeating experiments adopting the secondary reference gene (Table 1).
There were 45, 13, and one carriers of the KRAS 3′-UTR LCS6 TT, TG, and GG genotypes, respectively. According to the functional data on the G allele and the low frequency of the homozygous variant genotype, it was decided to collapse the GT and the GG genotypes (14 carriers of the G allele genotypes) for subsequent analyses. We found comparable Let-7a levels in carriers of the TT genotype and carriers of the variant G allele. The mean ΔCt value was 5.11 (±2.75 SD) in carriers of the LCS6 TT genotypes and 5.80 (±2.12 SD) in carriers of the LCS6 G allele genotypes (p = .3).
Survival Analyses
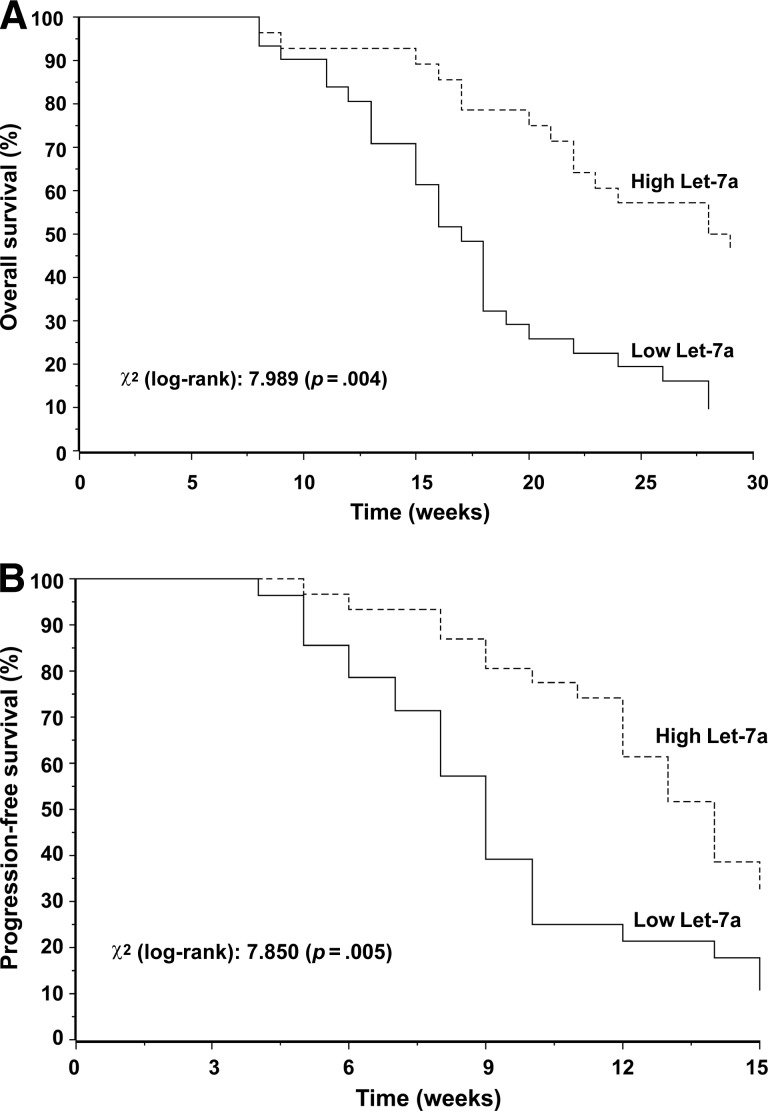
Adopting the primary endpoint of the study (the OS time), recursive partition analysis indicated a 4.2 ΔCt cutoff value for Let-7a level (the 45th percentile). Accordingly, 26 patients were included in the high Let-7a expression group and 33 patients were included in the low Let-7a expression group. There was a trend for the prevalence of stable disease cases in the high Let-7a expression group and disease progression cases in the low Let-7a expression group. Among the 13 stable disease cases, there were nine (69%) and four (31%) patients with high and low Let-7a levels, respectively. Among the 46 patients with progression, there were 17 (37%) and 29 (63%) patients with high and low Let-7a levels, respectively (p = .05). As shown in Figure 1, both the OS and PFS outcomes significantly favored patients with high Let-7a levels. The results of the univariate analysis of clinicopathologic features are reported in Table 2. Accordingly, Let-7a level and skin toxicity were included in the multivariate model. In this analysis, patients with high Let-7a levels had significantly better OS (HR, 0.82; 95% CI, 0.73–0.91; p = .01) and PFS (HR, 0.88; 95% CI, 0.79–0.98; p = .03) outcomes. All survival associations were confirmed using the secondary reference gene RNU48, as well as after excluding the 10 patients harboring KRAS mutations in codon 13 (data not shown).
Figure 1.
Kaplan–Meier curves for overall survival (A) and progression-free survival (B) with results of the log-rank test in the overall population.
Table 2.
Results of the univariate analysis
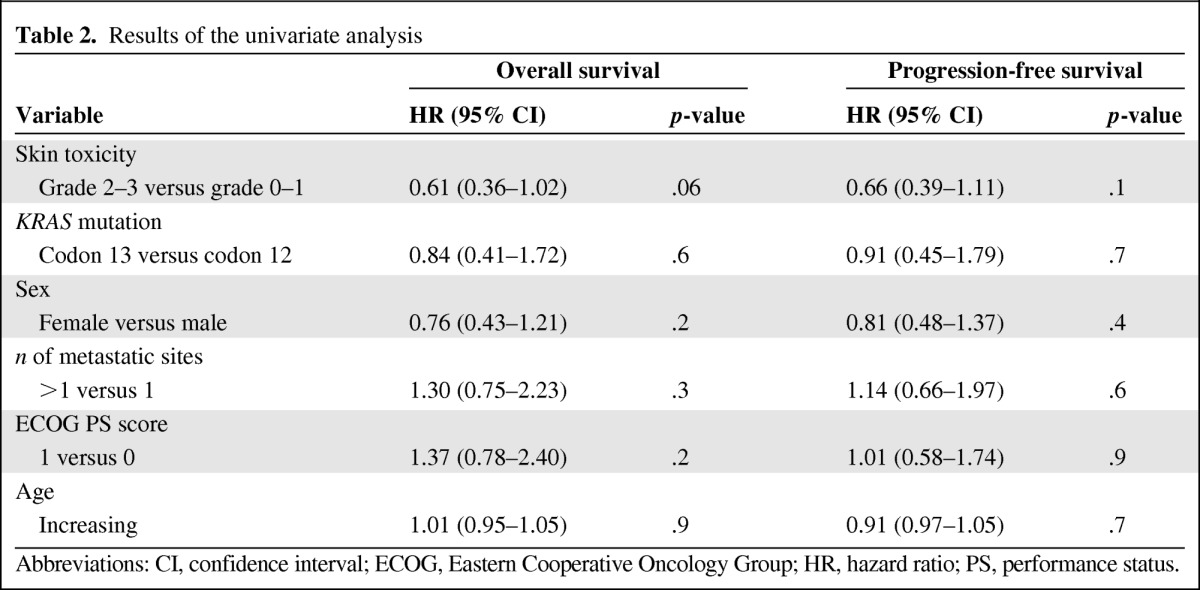
Abbreviations: CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; PS, performance status.
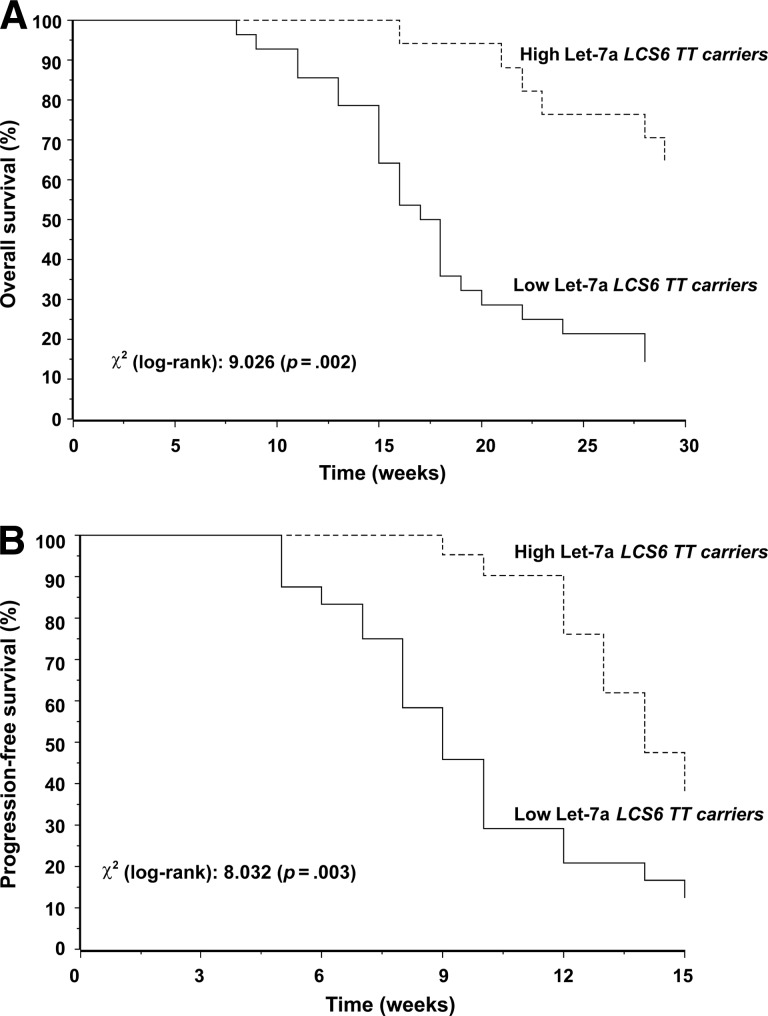
In the exploratory analysis of OS and PFS times in the 45 patients with a wild-type LCS6 status (TT genotype), the survival differences in favor of patients with high Let-7a levels (Fig. 2) seemed more marked than those observed in the overall population (Fig. 1). Only 14 patients were carriers of the LCS6 variant G allele (TG and GG genotypes), and the same analysis was not carried out in this subgroup.
Figure 2.
Kaplan–Meier curves for overall survival (A) and progression-fee survival (B) with results of the log-rank test in the 45 carriers of the wild-type LCS6 TT genotype.
Discussion
The current lack of alternative salvage treatment strategies for patients harboring KRAS mutations makes this setting of particular interest for translational clinical research. In this respect, the existence of mechanisms that post-transcriptionally downregulate mutated KRAS may be clinically relevant to patients with metastatic colorectal cancer [2]. Experimental and early clinical data revealed Let-7 miRNA as a relevant player for obtaining such an effect, with antitumor activity in the presence of KRAS mutations [2, 3]. Therefore, we investigated Let-7a levels in colorectal carcinomas with mutations in codon 12 and codon 13 and in patients with metastatic disease treated with salvage cetuximab plus irinotecan. We hypothesized that EGFR inhibition could retain some favorable clinical effect in the presence of a downstream negative regulator of mutated KRAS. To this end, we designed a retrospective study profiting from the lack of restrictions on anti-EGFR use in terms of KRAS mutational status until 2008.
As recently reported by De Roock et al. [7] using a large cohort of chemotherapy-refractory, metastatic colorectal cancer patients, patients with mutant KRAS tumors treated with salvage cetuximab plus chemotherapy showed rare tumor responses, but disease control in up to 49% of cases. This clinical observation suggests that a proportion of patients with KRAS mutation may obtain a survival gain from anti-EGFR therapy, and according to our findings, this favorable effect may occur when tumors display upregulated Let-7a levels. As pointed out by Kumar et al. [4], Let-7–mediated tumor suppression occurs largely, although not completely, through regulation of the Ras family. Actually, upregulated Let-7 levels may confer a survival advantage not only by turning off mutated KRAS under anti-EGFR therapy. In fact, Let-7 may display additional favorable effects by controlling cell-cycle regulators [17], Myc [18], Bcl-2 [19], signal transducer and activator of transcription 3 [20], and integrins [21]. In our study, a possible indirect indication that the favorable clinical effect of Let-7 works mainly on the EGFR–mutated KRAS axis comes from the exploratory analysis of Let-7a levels in patients with the wild-type LCS6 TT genotype. In that analysis, after excluding carriers of the variant G allele (GT and GG genotypes), we observed a stronger association between Let-7 level and survival outcomes than that observed in the analysis of the overall population. According to the functional role of the LCS6 variant, it is possible that patients with the LCS6 TT genotype fully benefited from the favorable Let-7 effect on KRAS downregulation, whereas this was lower in LCS6 G allele carriers. Combined assessment of miRNA levels (Let-7a) and polymorphisms in the miRNA binding site (LCS6) may be more reliable than clinical analysis of a single marker. This may also explain the conflicting results on the clinical role of the LCS6 polymorphism alone in this setting [22, 23].
Additional endpoints of the study involved Let-7a expression analyses in relation to clinicopathologic features. According to our current knowledge, the association between a high Let-7a level and grade 2–3 skin toxicity is difficult to explain. It has been found that miRNAs are also involved in the regulation of skin development and pathology [24]. However, as recently reported by Hildebrand et al. [25], Let-7 was not identified among miRNAs that participate in the regulation of human keratinocyte differentiation. Nevertheless, it cannot be excluded that, under circumstances that hurt and modify the skin microenvironment at the cellular or molecular level (e.g., exposure to an anti-EGFR agent), dysregulation of specific miRNAs (including those of the Let-7 family) may be critical in derailing the healing sequence in chronic wounds [24].
Limitations of the study are its retrospective nature and the limited sample size. However, consideration should be given to the fact that the study focuses on a homogeneously staged and treated population of patients with KRAS mutations (about 35% of metastatic colorectal cancer patients screened for anti-EGFR therapy). Also, based on the current use of anti-EGFR therapy in wild-type KRAS patients, such an early investigation is only possible in retrospective series. In addition, our findings seem to be confirmed by preliminary results in a similar setting recently reported by other investigators [26]. Notwithstanding this, additional investigations are necessary for confirming our results and excluding false-positive associations. Also, more data will be useful for clarifying the predictive or prognostic role of Let-7. A number of experimental and in vivo studies brought to light the multifaceted functions of this miRNA, which seems to repress several oncogenic pathways [17–21]. Therefore, it is possible that, even after additional effort, the extent of the putative Let-7 effects may render a clearcut distinction of its predictive or prognostic role quite difficult.
In conclusion, further studies are warranted in this field. As a next step, these findings could be verified by analyzing banked tissues from the registrative trials of cetuximab and panitumumab in metastatic colorectal cancer patients. These populations would also offer the opportunity for an appropriate randomized “control arm” of mutated KRAS patients treated using best supportive care. Knowledge on Let-7 and other recently identified KRAS-related miRNAs [2] may open novel perspectives for the treatment of patients with metastatic colorectal cancer with KRAS mutations. EGFR inhibition could be reconsidered in subgroups of patients through the presence of KRAS mutations. Moreover, the development of anti-KRAS therapeutics that mimic miRNA functions could be encouraged in this specific setting [27].
Conclusions
The current lack of alternative salvage treatment strategies in metastatic colorectal cancer patients harboring KRAS mutations makes this setting of particular interest for translational clinical research. Let-7 miRNA post-transcriptionally downregulates KRAS, and Let-7 administration reduced tumor formation in animal cancer models expressing activating KRAS mutations. We investigated Let-7a isoform levels in colorectal carcinomas with KRAS mutations and in patients with metastatic disease treated with salvage cetuximab plus irinotecan. We hypothesized that EGFR inhibition could retain some favorable clinical effect in the presence of such a downstream negative regulator. Our findings support the clinical effects of Let-7 levels in vivo, and the existence of a proportion of patients with a KRAS mutation who may obtain a survival benefit from anti-EGFR therapy if their tumors display upregulated Let-7a. Additional investigations are warranted to confirm these results, because they may lead to novel perspectives in the overall treatment strategy of patients with mutated KRAS and to the development of innovative therapeutics that mimic miRNA functions.
Acknowledgments
This work was partially supported by PROJECT INTEROMICS, Ministero dell'Istruzione, dell'Universití e della Ricerca (MIUR), Italy.
Annamaria Ruzzo and Francesco Graziano contributed equally to this study.
Footnotes
- (C/A)
- Consulting/advisory relationship
- (RF)
- Research funding
- (E)
- Employment
- (H)
- Honoraria received
- (OI)
- Ownership interests
- (IP)
- Intellectual property rights/inventor/patent holder
- (SAB)
- Scientific advisory board
Author Contributions
Conception/Design: Francesco Graziano, Annamaria Ruzzo, Bruno Vincenzi, Giuseppe Perrone, Mauro Magnani
Provision of study material or patients: Francesco Graziano, Annamaria Ruzzo, Bruno Vincenzi, Giuseppe Perrone, Emanuele Canestrari, Vincenzo Catalano, Fotios Loupakis, Carla Rabitti, Daniele Santini, Giuseppe Tonini
Collection and/or assembly of data: Francesco Graziano, Annamaria Ruzzo, Bruno Vincenzi, Giuseppe Perrone, Emanuele Canestrari, Vincenzo Catalano, Fotios Loupakis, Carla Rabitti, Daniele Santini, Giuseppe Tonini, Giammaria Fiorentini, David Rossi, Alfredo Falcone
Data analysis and interpretation: Francesco Graziano, Annamaria Ruzzo, Giuseppe Perrone, Emanuele Canestrari, Giammaria Fiorentini, Alfredo Falcone, Mauro Magnani
Manuscript writing: Francesco Graziano, Annamaria Ruzzo, Mauro Magnani
Final approval of manuscript: Francesco Graziano, Annamaria Ruzzo, Emanuele Canestrari, Giuseppe Perrone, Nadia Galluccio, Vincenzo Catalano, Fotios Loupakis, Carla Rabitti, Daniele Santini, Giuseppe Tonini, Giammaria Fiorentini, David Rossi, Alfredo Falcone, Mauro Magnani
References
- 1.Whelan JT, Hollis SE, Cha DS, et al. Post-transcriptional regulation of the Ras-ERK/MAPK signaling pathway. J Cell Physiol. 2012;227:1235–1241. doi: 10.1002/jcp.22899. [DOI] [PubMed] [Google Scholar]
- 2.Kasinski AL, Slack FJ. Potential microRNA therapies targeting Ras, NFκB and p53 signaling. Curr Opin Mol Ther. 2010;12:147–157. [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 4.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Oh JS, Kim JJ, Byun JY, et al. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76:5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 7.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 8.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HH, Wang XJ, Li GX, et al. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 12.Mallett S, Timmer A, Sauerbrei W, et al. Reporting of prognostic studies of tumour markers: A review of published articles in relation to REMARK guidelines. Br J Cancer. 2010;102:173–180. doi: 10.1038/sj.bjc.6605462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 14.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu KH, Patterson AP, Wang L, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 16.Ohuchida K, Mizumoto K, Kayashima T, et al. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18:2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Q, Meng P, Wang T, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PloS one. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Meng F, Henson R, Wehbe-Janek H, et al. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- 21.Ml̈ler DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 22.Graziano F, Canestrari E, Loupakis F, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10:458–464. doi: 10.1038/tpj.2010.9. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104–109. doi: 10.1093/annonc/mdq315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shilo S, Roy S, Khanna S, et al. MicroRNA in cutaneous wound healing: A new paradigm. DNA Cell Biol. 2007;26:227–237. doi: 10.1089/dna.2006.0568. [DOI] [PubMed] [Google Scholar]
- 25.Hildebrand J, Rẗze M, Walz N, et al. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol. 2011;131:20–29. doi: 10.1038/jid.2010.268. [DOI] [PubMed] [Google Scholar]
- 26.Landi L, Biagioni F, Ludovini V, et al. miR128 and LET-7 microRNAs as potential biomarkers for selection of patients with metastatic colorectal cancer candidate to cetuximab/panitumumab therapy [abstract 3636] Proc Am Soc Clin Oncol. 2011;29:255. [Google Scholar]
- 27.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]