Abstract.
Ovarian carcinoma has the highest lethality rate of gynecologic tumors, largely attributed to the late-stage diagnosis of the disease. Reliable tools for both accurate diagnosis and early detection of disease onset are lacking, and presently less than 20% of ovarian cancers are detected at an early stage. Protein biomarkers that allow the discrimination of early and late stages of ovarian serous carcinomas are urgently needed as they would enable monitoring pre-symptomatic aspects of the disease, disease progression, and the efficacy of intervention therapies. We compare the absolute and relative protein levels of six protein biomarkers for ovarian cancer in five different established ovarian cancer cell lines, utilizing both quantitative immunoblot analysis and a guided-mode resonance (GMR) bioassay detection system that utilizes a label-free optical biosensor readout. The GMR sensor approach provided highly accurate, consistent, and reproducible quantification of protein biomarkers as validated by quantitative immunoblotting, as well as enhanced sensitivity, and is therefore suitable for quantification and detection of novel biomarkers for ovarian cancer. We identified fibronectin, apolipoprotein A1, and TIMP3 as potential protein biomarkers for the differential diagnosis of primary versus metastatic ovarian carcinoma. Future studies are needed to confirm the suitability of protein biomarkers tested herein in patient samples.
Keywords: biomedical optics, biophotonics, medicine, nanophotonics, photonics, resonators
1. Introduction
Ovarian carcinoma has the highest lethality rate of gynecologic tumors, with over 16,000 cases reported in the United States in 2005.1 The current five-year survival rate is only 50%, largely attributed to the late-stage diagnosis of the disease.2 Ovarian serous papillary carcinoma is the most prevalent among ovarian carcinomas,1 yet reliable tools for accurate diagnosis and early detection of disease onset are lacking, and presently less than 20% of ovarian cancers are detected at an early stage.1–3 Protein biomarkers that allow the discrimination of early and late stages of ovarian serous carcinomas, such as metastatic versus primary ovarian serous carcinomas,4,5 could provide important insights by allowing monitoring pre-symptomatic aspects of the disease, disease progression, and the efficacy of intervention therapies.
Several studies have identified potential indicators and screening targets for the early detection and diagnosis of ovarian serous papillary carcinoma to monitor pre-symptomatic aspects of the disease as well as disease progression.4–6 Some of these biomarker proteins are differentially upregulated in metastatic or primary ovarian serous papillary carcinoma. While each of those protein biomarkers may prove insufficient to serve as an accurate predictor of pathology,2 combining multiple biomarkers into a diagnostic panel may provide great diagnostic benefit. We investigated a panel of six biomarkers, each of which was previously reported to have at least a twofold up-regulation in either metastatic or primary ovarian carcinoma. Specifically, we chose collagen type I,7–11 tissue inhibitor of metalloproteinases 3 (TIMP-3),12–15 fibronectin,16 and calreticulin,17–21 which in previous studies showed twofold or higher up-regulation in metastatic over primary ovarian serous papillary carcinoma, and apolipoprotein A-I22–24 and mitogen-activated protein kinase 13 (MAPK13),25–30 which have previously been reported as differentially up-regulated in primary over metastatic ovarian serous papillary carcinoma.
Current technologies for biomarker array diagnostics from blood samples are associated with significant cost and time and often are not suitable for high-throughput screening.2,6,31 In the diagnostic laboratory setting, serum and plasma biomarker proteins are typically quantified using enzyme-linked immunosorbant assay (ELISA) technology;32 however, given the lack of established biomarkers for ovarian cancer, no validated ELISA tests are currently available. Alternatives for protein detection include quantitative immunoblotting, which is laborious and associated with a large margin of experimental error and variability given the complex protocol involved. Novel technologies that can provide an easy-to-use, rapid, and accurate differential analysis of protein biomarkers are urgently needed.2,6,31,32
In this study, we utilize a novel optical biosensor technology based on the guided-mode resonance (GMR) effect that occurs in waveguide gratings to perform the label-free analysis. We compare the quantification of protein biomarkers from established ovarian cancer models with a standard, quantitative immunoblotting assay and found good correlation.
We conclude that this novel optical biosensor technology is a high-accuracy, portable sensor system that yields rapid detection of biomarker proteins. Application of this device to diagnostic screening will allow healthcare providers to monitor pre-symptomatic aspects of the disease, disease progression, and the efficacy of intervention therapies with improved reliability and efficiency.
2. Materials and Methods
2.1. Cell Lines and Sample Preparation
Five different established cell lines were used as in vitro model systems for ovarian carcinoma.33–37 All cell lines were obtained from the American Tissue Type Collection (ATTC) and cultured and maintained according to the supplier’s recommendations. TOV-112D (Cat. #CRL-11731), TOV-21G (Cat. #CRL-11730), NIH:OVCAR-3 (Cat. #HTB-161), Caov-3 (Cat. #HTB-75) and SK-OV-3 (Cat. #HTB-77) cells were grown in T75 tissue culture flasks (TPP; Midsci, St. Louis, MO). At passage 4, cells were seeded at a density of 1 million cells per flask and maintained for 48 hours, upon which the supernatant was aspirated, centrifuged for 5 min at to remove any residual cellular debris, aliquoted, flash frozen in liquid nitrogen, and stored at . Aliquots of the same flask were used for protein determination utilizing the RSI bioassay detection system and quantitative immunoblotting.
2.2. Antibodies
All antibodies were obtained from commercial sources, and antibodies of the same lot number were used in both experiments, i.e., the RSI detection system and the validation experiments using quantitative immunoblotting. The following antibodies were used in the present study: mouse anti-fibronectin (MAB1918; RnD Systems, Minneapolis, MN), mouse anti-apolipoprotein A1 (Cat. # 20-783-73037; GenWay Biotech Inc., San Diego, CA); mouse anti-calreticulin (Cat. # SPA-601F; Assay Designs, Enzo Life Sciences Inc., Farmingdale, NY), rabbit anti-collagen type 1 (Ab34710; AbCam, Cambridge, MA), mouse anti-mitogen-activated protein kinase 13 (Cat. # H00005603-M01; Assay Designs, Enzo Life Sciences Inc., Farmingdale, NY) and mouse anti-tissue inhibitor of metalloproteinases 3 (MAB973; RnD Systems, Minneapolis, MN).
2.3. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
For sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), samples were denatured in in sample buffer (final concentrations: SDS 10%, glycerol 10%, -mercaptoethanol 1%, bromophenol blue 0.004%, Tris-HCl 0.5 M, pH 6.8) and boiled for 5 min in a heating block. Equal volumes of supernatants and growth media were loaded on midi-size gradient gels (4% to 12% bis-tris gels, 4% tris-glycine, or 3% to 8% tris acetate; Invitrogen, Carlsbad, CA). Proteins were separated electrophoretically in running buffer containing 3-(N-morpholino) propanesulfonic acid (MOPS) or tris acetate, respectively (both from Invitrogen, Carlsbad, CA), for 55 min at 200 V. The total protein concentration in cell supernatants was determined using the bicinchoninic acid (BCA) assay (Pierce #23225, Thermo Scientific, Rockford, IL), according to the manufacturer’s instructions, and ranged from 1 to 80 μg per lane, depending on the concentration in the supernatant sample and the sensitivity of the antibody. Recombinant proteins were used to establish a standard curve. Proteins were transferred onto a 0.2 μM nitrocellulose membrane (Pall Life Sciences, Ann Arbor, MI) in transfer buffer containing 25 mM Tris pH 8.6, 192 mM glycine, 0.1% SDS, and 20% methanol for 1 hr at 100 V. Membranes were blocked with either 5% milk, 0.2% Tween-20 in PBS, or 0.5%casein/0.05% Tween-20 in PBS for 1 hr, incubated with primary antibody overnight at 4°C, washed three times with either 2.5% milk/0.2% Tween-20 in PBS or 0.25%casein/0.025% Tween-20 in PBS and probed with horseradish peroxidase-conjugated secondary antibody ( dilution; donkey anti-rabbit or donkey anti-mouse obtained from GE Healthcare [Piscataway, NJ] or donkey anti-sheep [Sigma Aldrich, St. Louis, MO]) for 1 h at ambient temperature. Immunoblots were developed using the Luminata Forte Western HRP substrate (Millipore, Billerica, MA), or for enhanced sensitivity, the Western Lightning Ultra Chemoluminescence substrate (Perkin Elmer, Waltham, MA). Membranes were imaged using film (Thermo Scientific, Rockford, IL) and processed on a Minolta film processor (Konica Minolta Medical Imaging USA, Inc., Wayne, NL).
2.4. Determination of Standard Curves for Quantification of Immunoblots
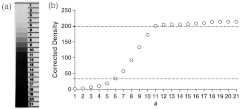
Films were digitalized using a commercial desktop scanner under standardized conditions at a resolution of 2,400 dpi to uncompressed TIFF format, and densitometry was performed using ImageJ software (National Institute of Health, Bethesda, MD). In order to assess the linear range of the film, a densitometer standard was applied to every sheet of film using a Model 303 Sensitometer (X-Rite Company, Grand Rapids, MI). Film exposure to the membranes was adjusted such that all standards and samples were within the linear range of the film (Fig. 1). By measuring the density, corrected for background, of known amounts of recombinant proteins standards for each antibody, a standard curve was generated, which allowed the calculation of the specific protein concentration in ovarian cancer cell supernatants and media only controls.
Fig. 1.
Quantitative analysis of immunoblots using light-sensitive film. (a) A densitometry standard was applied to every sheet of film processed, and exposure of the film adjusted such that bands to be quantified were within the linear range of the film. (b) Densitometry analysis of the densitometry standard range. Dotted lines mark the linear range of the film, encompassing approximately 1.5 orders of magnitude of change that can accurately be detected using this method.
2.5. Guided-Mode Resonance Detection System
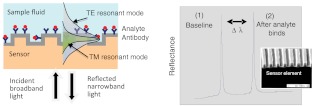
The detection system (provided by Resonant Sensors Incorporated, Arlington, TX) used in this work is based on the GMR effect that occurs in sub-wavelength dielectric waveguide gratings. As shown in Fig. 2, when these diffractive elements are illuminated with a broadband light source, a specific wavelength of light is reflected (or transmitted) at a specific angle. The binding interaction between an immobilized receptor and its analyte can be monitored in real time without the use of reporter labels (such as fluorescent or radioactive tags) by following the corresponding resonance wavelength shift with an optical spectrum analyzer. Test time is limited solely by the chemical binding dynamics between the receptor and its target. Specificity is imparted on the sensor surface by covalently attaching a selective layer (such as antibodies or DNA). It is multifunctional as only the sensitizing surface layer needs to be chemically altered to detect different targets. Repeatable fabrication processes are in place to produce the resonant grating sensor element in low-cost polymer and other dielectric materials.
Fig. 2.
Schematic of a proposed label-free GMR sensor system (single channel illustrated) operating in reflection mode. The collimated beam from a broadband source is incident on the sensor at normal incidence. The reflected spectral response is monitored in real time with an optical spectrum analyzer. As binding events occur at the sensor surface, resonance peak changes (only one polarization depicted in plot) can be tracked as a function of wavelength (). Modified from Ref. 38.
Since the resonance layer is polarization-sensitive, separate resonance peaks occur for incident TE (electric vector normal to the plane of incidence) and TM (magnetic vector normal to the plane of incidence) polarization states. This dual-peak feature provides cross-referenced data useful for increasing detection accuracy. These distinct resonant modes interact differently with the surrounding media, enabling the polarization-based differentiation.38 This sensor technology is broadly applicable to medical diagnostics, drug discovery and development, industrial process control, and environmental monitoring.
In 1992, Magnusson and Wang39 suggested employing the GMR effect for sensor applications and disclosed GMR filters that were tunable on variation in resonance structure parameters including thickness and refractive index. Wawro et al. presented GMR biosensor embodiments and system architectures.40 Following this, others have also discussed the use of these resonant elements as biosensors.41,42 Most commonly, the input light is efficiently reflected in a narrow spectral band whose central wavelength is highly sensitive to chemical reactions occurring at the surface of the sensor element. The sensor’s operating spectral region is determined by the physical waveguide-grating parameters, such as the grating period and chosen dielectric materials. Sensor designs responsive to thickness changes from the nanoscale () to several μm have been analyzed. These studies indicate that the proposed sensor technology can be used to detect binding events at the molecular level as well as bacterial analytes with micron-scale dimensions.
2.6. Protein Determination using GMR Bioassay Detection Sensor
The GMR sensor plates (Resonant Sensors Incorporated, Arlington, TX) are coated with a commercially available silane, which provides a means to covalently bond the antibody to the sensor surface. The specific antibody for each biomarker protein is immobilized on the sensor using a crosslinking agent. To minimize nonspecific binding, the plate is blocked using a bovine serum albumin solution (BSA).To generate a standard curve, dilutions of standard protein are prepared using reagent diluent (3% BSA in PBS). Neat reagent diluent is used as a baseline measurement and blank reference. Spike and recovery samples are run for each assay performed, with each cell line’s media having a known spiked protein and compared with the standard value in reagent diluent.43 This ensures the supernatant/media sample’s matrix is not interfering with the detection of the protein. All ovarian carcinoma cell media and supernatant samples are testing unprocessed, with no sample preparation, unless stated otherwise. All samples are incubated on the prepared sensor surfaces for 60 min (unless otherwise stated) at 37°C, then washed with PBS/Tween to remove unbound material and subsequently measured on the RSI detection system. Results on all protein detection data are based on difference of initial and final baseline readings and are repeated in quadruplicate and averaged, with major outliers removed.
3. Results and Discussion
3.1. Label-Free Optical Biosensor Can Accurately Determine Protein Biomarkers
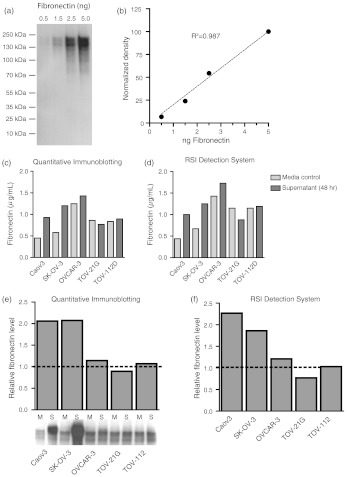
Aliquots of identical samples were processed using either quantitative immunoblotting or the novel label-free GMR-based RSI detection system. For fibronectin, we established a standard curve ranging from 0.5 to 5 ng recombinant protein that could be detected by Western blot [Fig. 3(a)]. Densitometry and subsequent analysis yielded a highly reproducible standard curve across this protein range [Fig. 3(b)]. Absolute fibronectin levels were between 0.45 and in the control media and the supernatant at 48 hr, respectively [Fig. 3(c)]. Nearly identical values were obtained when aliquots of the same sample were analyzed using the label-free RSI detection system [Fig. 3(d)]. Given the distinct growth media for the five different ovarian cancer cell lines used (i.e., containing different amounts of serum), baseline levels were different between cell lines. We therefore calculated the relative fibronectin level in the supernatant versus the control. Thus a relative level of higher than 1 is suggestive of release into the supernatant, whereas a level lower than 1 suggests uptake or degradation. The relative fibronectin level in supernatants of Caov3 and SK-OV-3 cells was 2.06 and 2.07, respectively, whereas OVCAR-3, TOV-21G, and TOV-112D had levels similar to media control (1.14, 0.89, and 1.07, respectively), as determined by quantitative immunoblotting [Fig. 3(e)]. Similar relative levels were detected utilizing the RSI detection system. Using the RSI detection system, we measured similarly high levels in supernatants of Caov3 and SK-OV-3 cells (2.27 and 1.86, respectively) and levels similar to media control in OVCAR-3, TOV-21G, and TOV-112D supernatants [1.21, 0.77, 1.03, respectively; Fig. 3(f)].
Fig. 3.
Fibronectin protein levels are increased in primary ovarian carcinoma and can accurately be detected using the novel label-free optical biosensor RSI detection system. (a) The monoclonal antibody MAB1918 yields a single specific band when probing against human recombinant fibronectin. Representative immunoblot shown. (b) Standard curve derived from immunoblot highlighting that protein concentrations can accurately be determined. The line of best fit following linear regression is shown (). (c) Histogram showing the quantification of cell supernatant levels of fibronectin using quantitative immunoblotting. One representative experiment is shown. (d) Histogram showing the quantification of cell supernatant levels of fibronectin using the novel RSI detection system. One representative experiment is shown. (e) Relative fibronectin protein levels, expressed as the ratio of protein in the supernatant after 48 h of cell growth and maintenance versus media control. Fibronectin levels have increased approximately twofold over a period of 48 hr in Caov3 and SK-OV-3 lines, whereas no change was detected in the other three cell lines. The same representative experiments as in (c) is shown. indicates media control, indicates supernatant. (f) Similar data are obtained when calculating the relative protein level measured with the RSI detection system.
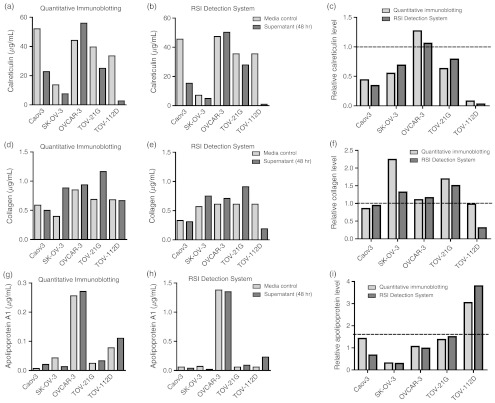
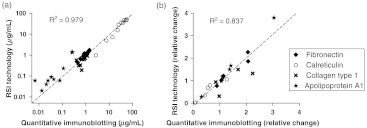
We performed a similar analysis for calreticulin, collagen type 1, and apolipoprotein A1 (Fig. 4). For all these proteins, the RSI detection system yielded similar concentrations as quantitative immunoblotting. In order to mathematically validate the accuracy of the label-free GMR approach, we plotted the absolute concentrations of all biomarkers and cell lines obtained from the RSI detection system against the values obtained by quantitative immunoblotting [Fig. 5(a)]. Linear regression analysis, taking into account all protein biomarkers jointly, resulted in an value of 0.979, indicative of an exact correlation between both measurements. Furthermore, the values obtained for individual biomarkers were overall similar: fibronectin, ; calreticulin, ; collagen type 1, ; apolipoprotein A1, . Our correlational analysis for the relative protein biomarker correlations resulted in similarly linear relationship [Fig. 5(b)] with an for the linear regression of 0.837. The individual values obtained again were overall similar: fibronectin, ; calreticulin, ; collagen type 1, ; apolipoprotein A1, .
Fig. 4.
Assessment of potential biomarkers for differential diagnosis of primary versus metastatic ovarian cancer carcinoma. (a) through (c) Calreticulin protein levels were detected in cell supernatants and media controls using quantitative immunoblotting (a) and the RSI detection system. (b) Relative protein levels were calculated for measurements obtained from both systems, (c) showing high concordance between the two systems. (d) through (f) The same analyses were performed for collagen type 1. Collagen showed larger variation than the other antibodies used in the present study, which may be the result of nonspecific antibody binding to other targets as evident by nonspecific bands (data not shown). (g) through (i) Apolipoprotein A1 protein levels were quantified using both approaches. As corroborated by both approaches, apolipoprotein A1 levels were much higher than in media control in TOV-112D cells, yet much lower in SK-OV-3 cells, making apolipoprotein A1 a prime candidate for future protein biomarker panels for the differential diagnosis of primary versus metastatic ovarian carcinoma.
Fig. 5.
Validation of the label-free, optical RSI protein detection system can accurately measure protein levels in physiological samples. (a) We validate the accuracy of the RSI detection system by linear regression analysis. Taking into account all protein biomarkers jointly, the line of best fit had an value of 0.979, indicative of high correlation between both measurements. (b) Similarly, correlational analysis of the relative protein biomarker concentrations resulted in a similarly linear relationship with an value of the line of best fit of 0.837.
The antibody for collagen type 1 exhibited the highest degree of deviation between the measurements obtained using the two experimental approaches [Fig. 4(d)–4(f)], as evident by the lowest values derived from linear regression (Fig. 5) and the highest coefficient of variance calculated (). This level of variation may be the results of nonspecific antibody-epitope interactions, which are amplified by the use of labeled secondary antibodies and evident on the nitrocellulose membrane as non-specific bands (data not shown). Given the results from the other protein biomarkers shown herein, it may be speculated that the measurements obtained from the RSI detection system represent the more accurate quantification of collagen type 1 in the ovarian cancer cell samples tested here.
Overall, our data shows a high degree of accordance between the two technologies, i.e., quantitative immunoblotting and the RSI bioassay detection system, confirming that the label-free GMR system can provide an accurate quantification of biomarker protein levels in cellular supernatant.
3.2. Label-Free Optical Biosensor Detection System Shows Enhanced Sensitivity
In addition to the four protein biomarkers described above, we tested MAPK13 and TIMP3. MAPK13 could not be detected in supernatant samples, neither by quantitative immunoblotting nor by using the RSI detection system (data not shown). The mathematically derived theoretical threshold for detection, based on the detection limit of recombinant MAPK13 protein, is . In order to exclude technical problems associated with nonspecific antibody-epitope interactions in our biological samples, we tested our antibody against supernatant concentrated using centrifugation columns and total protein extracts from cell pellets. In both instances, MAPK13 could be detected (data not shown), excluding this possibility.
TIMP3 has been implicated in ovarian carcinoma as a target of preferential methylation in ovarian cancer lines as well as implicated with tumor invasionx.44–48 Using quantitative immunoblotting, we could accurately detect 5 ng TIMP3 recombinant protein, translating into a detection limit of approximate when taking into account the total protein concentration and maximal loading volume for quantitative immunoblotting. Under these conditions, we were unable to detect any quantifiable amounts of TIMP3 in the supernatant samples (data not shown). In contrast, reproducible measurements for TIMP3 in cell supernatant samples could be obtained using the RSI detection system (Table 1). The maximum concentration of TIMP3 detected using this method was and thus around the calculated theoretical detection threshold for quantitative immunoblotting. Furthermore, concentrations as low as could successfully and routinely be measured, highlighting the enhanced sensitivity over traditional immunoblotting techniques.
Table 1.
TIMP3 levels are selectively elevated in TOV-112D cells. The RSI detection system showed enhanced sensitivity over traditional quantitative immunoblotting approaches and reproducibly and accurately detected TIMP3 levels in supernatants. Of particular clinical relevance, TIMP3 levels were higher only on TOV-112D cell supernatant after 48 hr in culture. This is the first report of elevated TIMP3 protein levels in a relevant model for metastatic ovarian cancer, making it a prime candidate for future experiments in human serous samples assessing its potential as biomarker for the differential diagnosis of ovarian cancer.
| Caov-3 | SK-OV-3 | OVCAR-3 | TOV-21G | TOV-112D | |
|---|---|---|---|---|---|
| Media control () | 0.05 | 0.43 | 1.70 | 0.05 | 0.05 |
| Supernatant () | 0.05 | 0.47 | 2.61 | 0.29 | 0.25 |
| Relative protein level | 1.00 | 1.09 | 1.54 | 5.80 | 5.00 |
3.3. Feasibility of Development of Protein Biomarker Assay Panels for Ovarian Cancer Diagnosis Utilizing Label-Free Optical Biosensor Detection
In order to assess their initial usefulness as components of a protein biomarker assay panel for ovarian cancer diagnosis and detection, we studied six potential protein biomarkers for ovarian cancer in five established ovarian cancer cell lines. Four of these proteins could reliably and routinely be detected in both quantitative immunoblotting and the RSI detection system.
Fibronectin is a matrix adhesion protein of known involvement in ovarian cancer, and fibronectin synthesis has been hypothesized to be up-regulated in response to oxidative stress occurring during early malignant progression of the disease.49 One pathological study measuring immunoreactivity of fibronectin in a cohort of 211 German ovarian cancer patients found a significant association with tumor stage and growth fraction,50 while another recent study found increased fibronectin plasma levels in a number of gynecological cancers, but not ovarian cancer.51 These conflicting data further highlight the need for novel protein biomarker assay panels that can quantify multiple relevant biomarkers both accurately and cost-effectively. Herein, we showed a relative fibronectin level of in Caov-3 and SK-OV-3 cell supernatants, whereas no evidence of fibronectin secretion was detected in the other cell line systems tested [Fig. 2(c) through 2(f)]. Our data is in line with the hypothesis that fibronectin is upregulated and secreted during early malignancy49 and provides further support for pursuing fibronectin as a potential protein biomarker for the diagnosis and assessment of progression of ovarian carcinomas.
In TOV-112D cells, we detected a significant level secretion of apolipoprotein A1 into the supernatant [Fig. 4(g) through 4(i)], while SK-OV-3 cell supernatants had dramatically lower levels than media control. The other cell lines tested had similar or slightly higher levels than the media only control [Fig. 4(g) through 4(i)]. Apolipoprotein A1 has previously been suggested as a potential protein biomarker for the differential diagnosis of benign pelvic mass versus ovarian cancer and has most recently been incorporated into the DK-index as a correlative proteomic marker in a cohort of Danish patients.52,53 Our data not only substantiates the possible involvement of apolipoprotein A1 in ovarian cancer, but is suggestive that apolipoprotein A1 may serve as a powerful predictive protein biomarker for the differential diagnosis of ovarian cancer.
TIMP3 has been implicated in ovarian carcinoma as a target of preferential methylation in ovarian cancer lines as well as implicated with tumor invasion.44–48 While we were unable to detect TIMP3 by means of quantitative immunoblotting, results using the RSI detection system show an approximate fivefold-higher level in the supernatants of TOV-21G and TOV-112D cells compared with control (Table 1). Both TOV cell lines were originally obtained from grade 3, stage IIIc tumors and represent preclinical models for advanced, metastatic ovarian cancer.35 To our knowledge, this is the first report clearly implicating elevated TIMP3 levels at the protein rather than merely genetic level in advanced, metastatic ovarian cancer.
3.4. Advantages of Label-Free Optical Biosensor Assays
Quantification using Western blot analysis is associated with very large intrinsic variation, which is a result of the multitude of experimental steps and readouts required, including the initial assessment of protein quantification, loading of the SDS-PAGE gel, transfer efficiency, specificity of the antibodies, amplification of the signal using secondary antibodies, the linearity of the detection reagent as well as the limited linear range of film or the low sensitivity of CCD cameras. Similar considerations are necessary for ELISA assays, which are currently the most frequently employed diagnostic assays for protein biomarkers.32 In contrast, label-free optical biosensor detectors such as the RSI detection system yielded reproducible datasets and exhibited greater sensitivity than that of traditional Western blotting. In the present study, this is highlighted by our results for TIMP3 levels (Table 1). While we were not able to detect TIMP3 by classical immunoblotting approaches, the RSI detection system yielded highly consistent, accurate, and reproducible measurements that for the first time implicate elevated TIMP3 protein levels in metastatic ovarian cancer. These results demonstrate the RSI bioassay system is a prime candidate for future experiments in human serous samples assessing its potential as biomarker for the differential diagnosis of this devastating gynecologic carcinoma.
4. Conclusions
We compared the absolute and relative protein levels of protein biomarkers for ovarian cancer in the supernatants of ovarian cancer cell lines of various disease stages utilizing traditional quantitative immunoblot analysis and the novel bioassay system, which utilized a label-free optical biosensor readout. Quantification of biomarker proteins was consistent between Western blot and the GMR biosensor approach. We conclude that the RSI detection system is suitable for quantification and detection of novel biomarkers of primary and metastatic ovarian cancer. Furthermore, we identified fibronectin, apolipoprotein A1, and TIMP3 as potential protein biomarkers for the differential diagnosis of primary versus metastatic ovarian carcinoma. Future studies are needed to confirm the suitability of protein biomarkers tested herein in patient plasma and serum samples.
Acknowledgments
This study was supported in part by NIH SBIR grant R43CA135960 (DW and PK), the Felix and Carmen Sabates Missouri Endowed Chair in Vision Research and the Vision Research Foundation of Kansas City, NSF SBIR grant 0724407 (DW), and the State of Texas Emerging Technology Fund (DW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. Part of the work described herein was presented in abstract form at the 2011 SPIE/OSA European Conference on Biomedical Optics 8090-25: Novel Biophotonic Techniques and Applications. We thank Margaret, Richard, and Sara Koulen for generous support and encouragement.
References
- 1.Jemal A., et al. , “Cancer statistics,” CA Cancer J. Clin. 55, 10–30 (2005). 10.3322/canjclin.55.1.10 [DOI] [PubMed] [Google Scholar]
- 2.Lutz A. M., et al. , “Early diagnosis of ovarian carcinoma: is a solution in sight?,” Radiology 259, 329–345 (2011). 10.1148/radiol.11090563 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R., et al. , “Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths,” CA Cancer J. Clin. 61, 212–236 (2011). 10.3322/caac.v61:4 [DOI] [PubMed] [Google Scholar]
- 4.Liotta L. A., et al. , “Importance of communication between producers and consumers of publicly available experimental data,” J. Natl. Cancer Inst. 97, 310–314 (2005). 10.1093/jnci/dji053 [DOI] [PubMed] [Google Scholar]
- 5.Bignotti E., et al. , “Gene expression profile of ovarian serous papillary carcinomas: identification of metastasis-associated genes,” Am. J. Obstet. Gynecol. 196, 245.e1–245.e11 (2007). 10.1016/j.ajog.2006.10.874 [DOI] [PubMed] [Google Scholar]
- 6.Kulasingam V., Pavlou M. P., Diamandis E. P., “Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer,” Nat. Rev. Cancer 10, 371–378 (2010). 10.1038/nrc2831 [DOI] [PubMed] [Google Scholar]
- 7.Werkmeister J. A., Peters D. E., Ramshaw J. A., “Development of monoclonal antibodies to collagens for assessing host-implant interactions,” J. Biomed. Mater. Res. 23, 273–283 (1989). 10.1002/(ISSN)1097-4636 [DOI] [PubMed] [Google Scholar]
- 8.Werkmeister J. A., Ramshaw J. A., “Monoclonal antibodies to collagens for immunofluorescent examination of human skin,” Acta Derm Venereol 69, 399–402 (1989). [PubMed] [Google Scholar]
- 9.Werkmeister J. A., Ramshaw J. A., Ellender G., “Characterisation of a monoclonal antibody against native human type I collagen,” Eur. J. Biochem. 187, 439–443 (1990). 10.1111/ejb.1990.187.issue-2 [DOI] [PubMed] [Google Scholar]
- 10.Chesney J., et al. , “Regulated production of type I collagen and inflammatory cytokines by peripheral blood fibrocytes,” J. Immunol. 160, 419–425 (1998). [PubMed] [Google Scholar]
- 11.Harle J., et al. , “Effects of ultrasound on the growth and function of bone and periodontal ligament cells in vitro,” Ultrasound Med. Biol. 27, 579–586 (2001). 10.1016/S0301-5629(00)00326-4 [DOI] [PubMed] [Google Scholar]
- 12.Wilde C. G., et al. , “Cloning and characterization of human tissue inhibitor of metalloproteinases-3,” DNA Cell Biol 13, 711–718 (1994). 10.1089/dna.1994.13.711 [DOI] [PubMed] [Google Scholar]
- 13.Apte S. S., Mattei M. G., Olsen B. R., “Cloning of the cDNA encoding human tissue inhibitor of metalloproteinases-3 (TIMP-3) and mapping of the TIMP3 gene to chromosome 22,” Genomics 19, 86–90 (1994). 10.1006/geno.1994.1016 [DOI] [PubMed] [Google Scholar]
- 14.Apte S. S., Olsen B. R., Murphy G., “The gene structure of tissue inhibitor of metalloproteinases (TIMP)-3 and its inhibitory activities define the distinct TIMP gene family,” J. Biol. Chem. 270, 14313–14318 (1995). 10.1074/jbc.270.24.14313 [DOI] [PubMed] [Google Scholar]
- 15.Yang P., Baker K. A., Hagg T., “A disintegrin and metalloprotease 21 (ADAM21) is associated with neurogenesis and axonal growth in developing and adult rodent CNS,” J. Comp. Neurol. 490, 163–179 (2005). 10.1002/(ISSN)1096-9861 [DOI] [PubMed] [Google Scholar]
- 16.Epstein A. L., et al. , “Identification of a monoclonal antibody, TV-1, directed against the basement membrane of tumor vessels, and its use to enhance the delivery of macromolecules to tumors after conjugation with interleukin,” Cancer Res. 55, 2673–2680 (1995). [PubMed] [Google Scholar]
- 17.Michalak M., et al. , “Endoplasmic reticulum form of calreticulin modulates glucocorticoid-sensitive gene expression,” J. Biol. Chem. 271, 29436–29445 (1996). 10.1074/jbc.271.25.15153 [DOI] [PubMed] [Google Scholar]
- 18.Waser M., et al. , “Regulation of calreticulin gene expression by calcium,” J. Cell. Biol. 138, 547–557 (1997). 10.1083/jcb.138.3.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burns K., Opas M., Michalak M., “Calreticulin inhibits glucocorticoid—but not cAMP-sensitive expression of tyrosine aminotransferase gene in cultured McA-RH7777 hepatocytes,” Mol. Cell. Biochem. 171, 37–43 (1997). 10.1023/A:1006865108833 [DOI] [PubMed] [Google Scholar]
- 20.Coppolino M. G., et al. , “Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion,” Nature 386, 843–847 (1997). 10.1038/386843a0 [DOI] [PubMed] [Google Scholar]
- 21.Krause K. H., Michalak M., “Calreticulin,” Cell 88, 439–443 (1997). 10.1016/S0092-8674(00)81884-X [DOI] [PubMed] [Google Scholar]
- 22.Liao W., Yeung S. C., Chan L., “Proteasome-mediated degradation of apolipoprotein B targets both nascent peptides cotranslationally before translocation and full-length apolipoprotein B after translocation into the endoplasmic reticulum,” J. Biol. Chem. 273, 27225–27230 (1998). 10.1074/jbc.273.42.27225 [DOI] [PubMed] [Google Scholar]
- 23.Liao W., Kobayashi K., Chan L., “Adenovirus-mediated overexpression of microsomal triglyceride transfer protein (MTP): mechanistic studies on the role of MTP in apolipoprotein B-100 biogenesis,” Biochemistry 38, 7532–7544 (1999). 10.1021/bi9904196 [DOI] [PubMed] [Google Scholar]
- 24.Liao W., Chan L., “Tunicamycin induces ubiquitination and degradation of apolipoprotein B in HepG2 cells,” Biochem. J. 353, 493–501 (2001). 10.1042/0264-6021:3530493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J., et al. , “A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells,” Science 265, 808–811 (1994). 10.1126/science.7914033 [DOI] [PubMed] [Google Scholar]
- 26.Takasawa S., et al. , “Requirement of calmodulin-dependent protein kinase II in cyclic ADP-ribose-mediated intracellular mobilization,” J. Biol. Chem. 270, 30257–30259 (1995). 10.1074/jbc.270.51.30257 [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y., et al. , “Characterization of the structure and function of a new mitogen-activated protein kinase (p38beta),” J. Biol. Chem. 271, 17920–17926 (1996). 10.1074/jbc.271.30.17920 [DOI] [PubMed] [Google Scholar]
- 28.Wang X. S., et al. , “Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase,” J. Biol. Chem. 272, 23668–23674b (1997). 10.1074/jbc.272.38.23668 [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., et al. , “Novel homologues of CSBP/p38 MAP kinase: activation, substrate specificity and sensitivity to inhibition by pyridinyl imidazoles,” Biochem. Biophys. Res. Commun. 235, 533–538 (1997). 10.1006/bbrc.1997.6849 [DOI] [PubMed] [Google Scholar]
- 30.Goedert M., et al. , “Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases,” EMBO J 16, 3563–3571 (1997). 10.1093/emboj/16.12.3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanash S. M., Baik C. S., Kallioniemi O., “Emerging molecular biomarkers--blood-based strategies to detect and monitor cancer,” Nat. Rev. Clin. Oncol. 8, 142–150 (2011). 10.1038/nrclinonc.2010.220 [DOI] [PubMed] [Google Scholar]
- 32.Donzella V., Crea F., “Optical biosensors to analyze novel biomarkers in oncology,” J. Biophotonics 4, 442–452 (2011). 10.1002/jbio.201000123 [DOI] [PubMed] [Google Scholar]
- 33.Fogh J., Wright W. C., Loveless J. D., “Absence of HeLa cell contamination in 169 cell lines derived from human tumors,” J. Natl. Cancer Inst. 58, 209–214 (1977). [DOI] [PubMed] [Google Scholar]
- 34.Hamilton T. C., et al. , “Characterization of a human ovarian carcinoma cell line (NIH:OVCAR-3) with androgen and estrogen receptors,” Cancer Res. 43, 5379–5389 (1983). [PubMed] [Google Scholar]
- 35.Provencher D. M., et al. , “Characterization of four novel epithelial ovarian cancer cell lines,” In Vitro Cell. Dev. Biol. Anim. 36, 357–361 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Sinna G. A., et al. , “Characterization of two human ovarian carcinoma cell lines,” Gynecol. Oncol. 7, 267–280 (1979). 10.1016/0090-8258(79)90104-5 [DOI] [PubMed] [Google Scholar]
- 37.Karlan B. Y., et al. , “Glucocorticoids stabilize HER-2/neu messenger RNA in human epithelial ovarian carcinoma cells,” Gynecol. Oncol. 53, 70–77 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Magnusson R., et al. , “Resonant photonic biosensors with polarization-based multiparametric discrimination in each channel,” Sensors 11, 1476–1488 (2011). 10.3390/s110201476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnusson R., Wang S. S., “New principle for optical filters,” Appl. Phys. Lett. 61, 1022–1024 (1992). 10.1063/1.107703 [DOI] [Google Scholar]
- 40.Wawro D. D., et al. , “Optical fiber endface biosensor based on resonances in dielectric waveguide gratings,” Proc. SPIE 3911, 86–94 (2000). 10.1117/12.384889 [DOI] [Google Scholar]
- 41.Kikuta H., et al. , “Refractive index sensor with a guided-mode resonant grating filter,” Proc. SPIE 4416, 219–222 (2001). 10.1117/12.427053 [DOI] [Google Scholar]
- 42.Cunningham B., et al. , “Colorimetric resonant reflection as a direct biochemical assay technique,” Sens. Actuators B Chem. 81, 316–328 (2002). 10.1016/S0925-4005(01)00976-5 [DOI] [Google Scholar]
- 43.Thermo Fisher Scientific, “Spike-and-recovery and linearity-of-dilution assessment,” Thermo Scientific Tech Tip #58, TR0058.1, http://www.piercenet.com/files/TR0058-Spike-and-Recovery.pdf.
- 44.Jin H., et al. , “Snail is critical for tumor growth and metastasis of ovarian carcinoma,” Int. J. Cancer 126, 2102–2111 (2010). [DOI] [PubMed] [Google Scholar]
- 45.L’Esperance S., et al. , “Gene expression profiling of paired ovarian tumors obtained prior to and following adjuvant chemotherapy: molecular signatures of chemoresistant tumors,” Int. J. Oncol. 29, 5–24 (2006). [PubMed] [Google Scholar]
- 46.Sengupta S., et al. , “Lysophosphatidic acid downregulates tissue inhibitor of metalloproteinases, which are negatively involved in lysophosphatidic acid-induced cell invasion,” Oncogene 26, 2894–2901 (2007). 10.1038/sj.onc.1210093 [DOI] [PubMed] [Google Scholar]
- 47.Imura M., et al. , “Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 ovarian cancer cell lines,” Cancer Lett. 241, 213–220 (2006). 10.1016/j.canlet.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 48.Dhillon V. S., Aslam M., Husain S. A., “The contribution of genetic and epigenetic changes in granulosa cell tumors of ovarian origin,” Clin. Cancer Res. 10, 5537–5545 (2004). 10.1158/1078-0432.CCR-04-0228 [DOI] [PubMed] [Google Scholar]
- 49.Ksiazek K., et al. , “Senescent peritoneal mesothelial cells promote ovarian cancer cell adhesion: the role of oxidative stress-induced fibronectin,” Am. J. Pathol. 174, 1230–1240 (2009). 10.2353/ajpath.2009.080613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franke F. E., et al. , “Association between fibronectin expression and prognosis in ovarian carcinoma,” Anticancer Res. 23, 4261–4267 (2003). [PubMed] [Google Scholar]
- 51.Grammatikakis I. E., et al. , “Fibronectin plasma levels in gynecological cancers,” J. BUON 15, 122–126 (2010). [PubMed] [Google Scholar]
- 52.Zhang Z., et al. , “Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer,” Cancer Res. 64, 5882–5890 (2004). 10.1158/0008-5472.CAN-04-0746 [DOI] [PubMed] [Google Scholar]
- 53.Hogdall C., et al. , “A novel proteomic biomarker panel as a diagnostic tool for patients with ovarian cancer,” Gynecol. Oncol. 123, 308–313 (2011). [DOI] [PubMed] [Google Scholar]