Abstract
Observations that genome-wide DNA hypomethylation induces genomic instability and tumors in animals caution against the indiscriminate use of demethylating agents, such as 5-aza-2′-deoxycytidine (5-Aza-dC). Using primary mouse embryonic fibroblasts harboring a lacZ mutational reporter construct that allows the quantification and characterization of a wide range of mutational events, we found that in addition to demethylation, treatment with 5-Aza-dC induces γ-H2AX expression, a marker for DNA breaks, and both point mutations and genome rearrangements. To gain insight into the source of these mutations we first tested the hypothesis that the mutagenic effect of 5-Aza-dC may be directly mediated through the DNA methyltransferase 1 (DNMT1) covalently trapped in 5-Aza-dC-substituted DNA. Knock-down of DNMT1 resulted in increased resistance to the cytostatic effects of 5-Aza-dC, delayed onset of γ-H2AX expression and a significant reduction in the frequency of genome rearrangements. There was no effect on the 5-Aza-dC-induced point mutations. An alternative mechanism for 5-Aza-dC-induced demethylation and genome rearrangements via activation-induced cytidine deaminase (AID) followed by base excision repair (BER) was found not to be involved. That is, 5-Aza-dC treatment did not significantly induce AID expression and inhibition of BER did not reduce the frequency of genome rearrangements. Thus, our results indicate that the formation of DNMT1 adducts is the prevalent mechanism of 5-Aza-dC-induced genome rearrangements, although hypomethylation per se may still contribute. Since the therapeutic effects of 5-Aza-dC greatly depend on the presence of DNMT1, the expression level of DNA methyltransferases in tumors may serve as a prognostic factor for the efficacy of 5-Aza-dC treatment.
Keywords: DNA methylation, 5-aza-2′-deoxycytidine, mutations, genome rearrangements, DNMT1
Introduction
Hypermethylation of cytosines has been identified as a critical factor in transcriptional repression of tumor suppressor genes (1). Therefore, inhibition of DNA methylation could be an effective cancer treatment. 5-Aza-2′-deoxycytidine (5-Aza-dC) has significant demethylating activity and is currently approved by the FDA to treat myelodysplastic syndromes. 5-Aza-dC is incorporated into nucleic acids of proliferating cells as a cytidine analog and prevents methylation at CpG sites of newly synthesized DNA by irreversible covalent binding to DNA methyltransferase 1 (DNMT1), the major methylation maintenance enzyme, eventually depleting the cellular pool of this enzyme (2). This makes the maintenance of the genomic DNA methylation status less efficient and leads to global hypomethylation. Thus, the therapeutic effect of 5-Aza-dC could be due to re-expression of growth suppressor genes in the tumor cells or toxicity of the covalently bound DNMT1 to 5-Aza-dC-substituted DNA.
The most important long-term effects of 5-Aza-dC are mutations, which can arise during processing of 5-Aza-dC induced DNA lesions. Interestingly, the hypomethylation itself, caused by 5-Aza-dC, has been speculated to cause genetic instability in form of gains or losses of whole chromosomes (3). Moreover, studies with mouse embryonic stem cells deficient for DNMT1, leading to pronounced hypomethylation, demonstrated increased level of genome rearrangements, the latter measured by the frequency of inactivation of the endogenous reporter gene HPRT and an integrated thymidine kinase transgene (4).
While studies on the mutagenicity of 5-Aza-dC have been performed using different models and different mutation detection methods (5-10), the utilized approaches were not able to comprehensively detect both point mutations and genome rearrangements. Here we investigated the mutagenic effects of 5-Aza-dC using embryonic fibroblasts from a mouse model harboring a lacZ-plasmid reporter construct that allows the detection of a broad range of mutations and discriminate between point mutations and genome rearrangements (11). We confirm that 5-Aza-dC is highly mutagenic and capable of inducing point mutations as previously shown. However, we also show that the drug causes a substantial number of genome rearrangements in primary mouse fibroblasts. We subsequently show that although 5-Aza-dC is a potent demethylating agent, it is the DNMT1 DNA adducts but not the hypomethylation per se that cause genome rearrangements possibly via induction of DNA double-strand breaks.
Results
Mutagenesis, DNA methylation and growth in 5-Aza-dC-treated MEFs
Although there are numerous reports on the mutagenicity of 5-Aza-dC, the exact mechanism of the mutagenic effects and the spectrum of induced mutations still remain unclear. To examine 5-Aza-dC-induced mutation frequency and spectra in mammalian cells, we used mouse embryonic fibroblasts (MEFs) from a transgenic mouse harboring a chromosomally integrated lacZ reporter gene construct that can be recovered in E. coli. Mutation frequencies are determined on a per locus basis, i.e., as the ratio of inactivated lacZ genes over the total number of lacZ copies recovered from a given DNA sample. Downstream analysis of the mutant lacZ genes allowed us to determine the class of mutation, i.e., point mutation or genomic rearrangement, that led to the inactivation of the lacZ gene (11, 12).
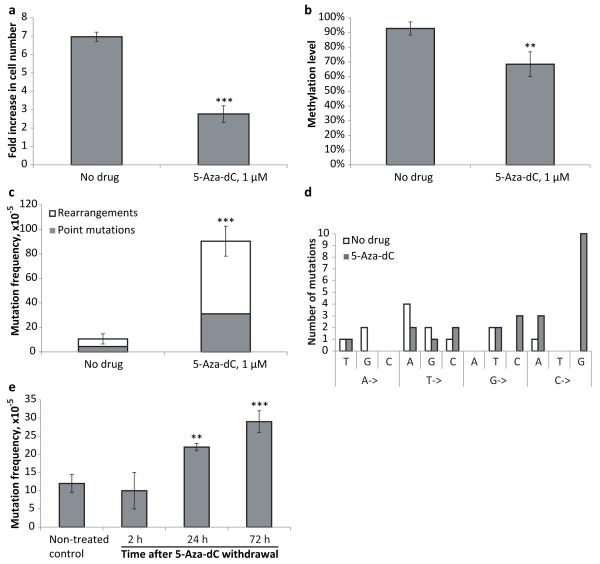
We first tested if 1 μM 5-Aza-dC, the concentration previously reported to have a virtually maximal cytotoxic effect on mouse ES cells (13), was still effective when used with our model system. We found that the proliferation of transgenic lacZ MEFs cultured in the presence of 1 μM 5-Aza-dC was significantly suppressed (Figure 1a). However, cells were still able to divide, thereby incorporating the cytidine analog. This was indicated by a significant decrease in DNA methylation level of the lacZ reporter gene, which is normally heavily methylated, after 3 days of growth in the presence of 5-Aza-dC (Figure 1b). Under these conditions 5-Aza-dC has a significant mutagenic effect, as indicated by an almost 10-fold increase of the lacZ mutation frequency (Figure 1c). The majority of the mutations were genome rearrangements (66%), slightly higher than the rearrangement fraction of the spontaneous mutations in the untreated control cells (59%). While these results confirm that 5-Aza-dC induces point mutations, they also indicate that genome rearrangements are the major component of the mutation spectrum. Analysis of 5-Aza-dC-induced point mutations revealed that almost 42% of them were C->G transversions, whereas in control samples no apparent bias towards any particular type of substitution was detected (Figure 1d). Rearrangements in this system are lacZ mutants with one breakpoint in the lacZ gene and the other breakpoint outside the reporter on the same chromosome (deletions or inversions) or another chromosome (translocations). Only illegitimate recombinations can be detected with this system and extensive homology at the breakpoints has never been reported (14).
Figure 1.
(a) Treatment with 5-Aza-dC inhibits MEF growth. The number of cells was determined after 72 hours of treatment with 5-Aza-dC; fresh medium containing the drug was added every 24 hours. (b) Treatment with 5-Aza-dC leads to DNA hypomethylation. (c) Treatment with 5-Aza-dC induces point-mutations and genome rearrangements in MEFs. (d) Analysis of spontaneous and 5-Aza-dC-induced point mutations. (e) Pulse-chase analysis of mutation frequency upon treatment with 5-Aza-dC. Cultured MEFs were subjected to a short, 1 hour pulse of 5-Aza-dC administration (5 μM), after which the medium was replaced and the cells further cultured for 2, 24, and 72 hours, when mutation frequencies were analyzed. All samples in all experiments (except where it is noted specifically) were assayed in triplicate with at least three independent experiments performed. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
To rule out the possibility that a large fraction of the observed mutations had originated in E. coli during the rescue process we tested the mutagenic effect of a short, one hour incubation with 5-Aca-dC. While this would give rise to partially 5-Aza-dC-substituted DNA, in mammalian cells the time period would be too short to fix the damage into mutations. As shown in Figure 1e, longer incubation times are required for elevated mutation frequencies to appear. This indicates that 5-Aza-substituted DNA does not by itself gives rise to mutations in E. coli but requires processing in mammalian cells.
Down-regulation of DNMT1 attenuates the ability of 5-Aza-dC to induce genomic rearrangements
DNMT1 can covalently bind to 5-Aza-dC at the hemi-methylated CpG sites. To directly test for a possible effect of DNMT1 on 5-Aza-dC-induced DNA mutations we analyzed lacZ mutagenicity of 5-Aza-dC in MEFs with normal DNMT1 expression level and in cells with down-regulated DNMT1 by specific siRNA (DNMT1-KD MEFs). A comparison of the DNMT1 mRNA levels in control MEFs and DNMT1-KD MEFs revealed a sharp (>90%) decrease in the amount of DNMT1 transcripts 24 hours after transfection with targeting siRNA. The mRNA levels of DNMT1 remained significantly (>75%) down-regulated for at least 72 hours after the transfection (data not shown).
The administration of 5-Aza-dC began 72 hours after lacZ MEFs were transfected with either DNMT1 or scrambled, i.e. non-targeting, siRNA. To ensure that cells were actively dividing at the moment of the 5-Aza-dC treatment and were able to incorporate the drug, cells were re-plated 48 hours after transfection, i.e. 24 hours before the first administration of 5-Aza-dC. The lacZ MEFs were cultured in medium supplemented with 5-Aza-dC for 72 hours, with fresh medium containing the drug added every 24 hours. Cells were then collected and mutation frequencies measured.
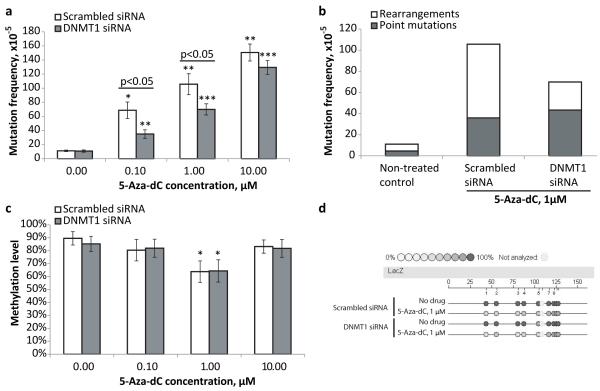
Similar to the results shown in Figure 1c, treatment with 5-Aza-dC (1 μM) led to an about 10-fold increase in the mutation frequency in MEFs transfected with scrambled siRNA (Figure 2a). Here we tested several concentrations of 5-Aza-dC, but even at the lowest concentration of 0.1 μM, a 6-fold increase in the mutation frequency as compared to the non-treated MEFs was observed. Overall, the mutation frequency correlated with the amount of 5-Aza-dC applied. However, the pattern of correlation was non-linear, with a tendency of the mutation frequency to plateau at the higher doses of the drug. That is, the administration of 1 μM 5-Aza-dC, or 10% of the maximal concentration tested, was able to produce ~70% of maximal mutation frequency.
Figure 2.
(a) Down-regulation of DNMT1 attenuates 5-Aza-dC mutagenicity. (b) Down-regulation of DNMT1 inhibits generation of 5-Aza-dC-associated genome rearrangements in MEFs. (c) Effects of different doses of 5-Aza-dC and DNMT1 down-regulation on DNA methylation level. (d) Representative epigram of the methylation analysis of the lacZ gene of transgenic lacZ MEFs. All samples in all experiments (except where it is noted specifically) were assayed in triplicate and at least three independent experiments were performed. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
The administration of 5-Aza-dC to DNMT1-KD MEFs also caused a dose-dependent increase in mutation frequency. However, the mutagenic effects of 5-Aza-dC were significantly attenuated in DNMT1-KD MEFs as compared to scrambled siRNA treated MEFs, at lower doses (0.1 and 1 μM) but not as much at higher dose (10 μM) (Figure 2a). This may suggest that there are two components in the mechanism of the 5- Aza-dC mutagenicity – DNMT1-dependent and DNMT1-independent, where the latter become prevalent upon administration of higher doses of the drug. Of note, the DNMT1 knock-down per se did not have any noticeable effect on the level of spontaneous mutations in the non-treated MEFs.
Depletion of DNMT1 also led to changes in the mutation spectra following the 5- Aza-dC treatment (Figure 2b). Specifically, there was a significant reduction in the frequency of genomic rearrangements in DNMT1-KD MEFs (27×10−5 or 38% of all mutations) as compared to MEFs treated with scrambled siRNA (70×10−5 or 66% of all mutations), while the frequency of point mutations remained the same between DNMT1-KD and control MEFS, 43×10−5 and 36×10−5 respectively. The spectra of spontaneous mutations was not affected by DNMT1 knock-down and remained similar to that observed in cells transfected with scrambled siRNA.
To find out if there was a correlation between 5-Aza-dC induced hypomethylation and mutagenicity we explored the effect of 5-Aza-dC administration on methylation status of control and DNMT1-KD MEFs. The observed levels of demethylation upon treatment with 1 μM 5-Aza-dC were similar in control and DNMT1-KD MEFs (Figures 2c and 2d). Of note, the transient down-regulation of the DNMT1 expression also did not cause any detectable decline in the methylation level of the lacZ gene in the absence of 5-Aza-dC (Figures 2c and 2d). The absence of a demethylating effect at the high (10 μM)dose of 5-Aza-dC is most likely due to the toxicity, which fully inhibits growth and therefore incorporation of the cytidine analog into DNA.
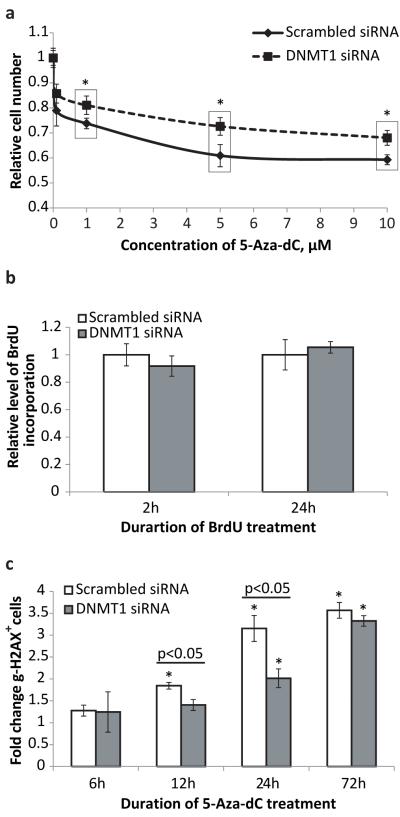
DNMT1-KD MEFs were significantly more resistant to the cytotoxic/cytostatic effects of 5-Aza-dC at all tested doses than MEFs with normal levels of DNMT1 expression (Figure 3a). This protective effect was not due to DNMT1 affecting cell proliferation rate since the rate of BrdU uptake was similar in control MEFs and in DNMT1-KD MEFs (Figure 3b).
Figure 3.
(a)DNMT1 down-regulation attenuates cytotoxic/cytostatic effects of 5-Aza-dC on MEFs. Dose-dependent effect is shown. The number of cells was determined after 72 hours of treatment with 5-Aza-dC at the concentrations indicated; fresh medium containing the drug was added every 24 hours. The data were normalized to non-treated control cells; n=12 for each data point; asterisk (*) designates statistically significant difference with scrambled siRNA controls (* - p<0.05) (b) DNMT1 down-regulation has a marginal effect on MEF proliferation rate detected only after short-term application of BrdU. Longer BrdU treatment revealed no difference in the uptake of labeling agent between control and DNMT1 KD cells. The data was normalized to control cells transfected with scrambled siRNA; n=35 for each data point. (c) DNMT1 down-regulation delays onset of γ-H2AX expression induced by 5-Aza-dC treatment. The data are presented as a fold increase relative to non-treated control. All samples in all experiments (except where it is noted specifically) were assayed in triplicate with at least three independent experiments were performed. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
5-Aza-dC associated DNA damage response is reduced in the cells with down-regulated DNMT1 expression
The significant effect of DNMT1 down-regulation on the induction of genome rearrangements but not point mutations by 5-Aza-dC led us to test whether 5-Aza-dC induces DNA double strand breaks (DSBs), an essential step in the formation of genome rearrangements. MEFs were treated with 1 μM of 5-Aza-dC for 6, 12, 24 and 72 hours and stained for the phosphorylated form of histone H2AX (γ-H2AX), a marker of DNA breaks, both double- and single-strand (15, 16). The flow-cytometric analysis revealed that 5-Aza-dC treatment increases the fraction of the γ-H2AX positive cells in a time-depedent manner (Figure 3c). The down-regulation of DNMT1 by siRNA delayed the onset of and attenuated the DNA damaging effect of 5-Aza-dC up to 24 hours. However, the protective effect of DNMT1 down-regulation completely vanished when 5-Aza-dC was applied for 72 hours, most likely because γ-H2AX expression reflects the current levels of DNA damage and the transiently down-regulated expression of DNMT1 is recovered by 72 hours, which is equivalent to 6 days after siRNA transfection.
5-Aza-dC-induced mutagenesis is not mediated by AID-BER pathway
A possible alternative mechanism of both 5-Aza-dC mutagenicity and 5-Aza-dC associated hypomethylation is through activation-induced cytosine deaminase (AID), previously considered to be a B cell-specific factor (17). AID deaminates 5-methylcytosines or cytosines, generating T-G mismatches or U-G pairs in DNA, respectively (18). These are substrates for DNA glycosylases which initiate base excision repair (BER) pathway, analogous to their functions in Class Switch Repair (19). Recently, genotoxic stress has been shown to induce AID expression, which is actively involved in generation of DSBs at the tumor translocation sites, and knock-down of AID resulted in inhibition of DSBs generation and subsequent tumor translocation (20). We first tested whether the genotoxic stress induced by 5-Aza-dC treatment leads to upregulated expression of AID.
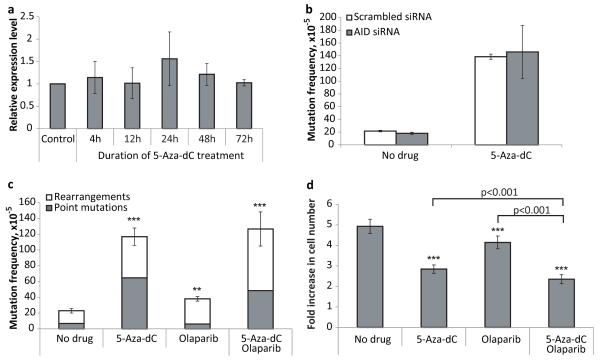
Analysis of the AID expression level in the MEFs treated with 5-Aza-dC (1 μM, 72 hours) did not reveal any significant changes in comparison with non-treated control (Figure 4a). Conversely, when tested if down-regulation of the AID expression by siRNA (more than 70% of suppression by 72 hours after transfection) would affect the 5-Aza-dC-induced mutagenesis, no difference was detected (Figure 4b). We then directly tested for the possible involvement of BER in 5-Aza-dC-induced genome rearrangements by comparing the mutation frequency and spectrum in the absence and presence of the PARP inhibitor, olaparib. PARP inhibition has been shown to inhibit BER (21). Cells were treated with either olaparib (500 nM), 5-Aza-dC (1 μM), or both for 3 consecutive days. We found that olaparib alone has a modest mutagenic effect on the MEFs (Figure 4c). However, there was no significant effect of combined 5-Aza-dC and olaparib treatment on mutation frequency or spectra. We also found that olaparib alone led to a statistically significant suppression of the cell growth (Figure 4d). The effect of combined application of 5-Aza-dC and olaparib on cell growth/survival was more profound than effect of either drug administered alone. However, the combined treatment did not reveal any synergy between olaparib and 5-Aza-dC – the joint effect was rather an arithmetical sum of their cytostatic/cytotoxic traits. Taken together, these results indicate that AID-initiated BER does not affect 5-Aza-dC-induced mutations and does not reduce the level of genome rearrangements.
Figure 4.
(a) The relative level of AID mRNA in MEFs treated with 5-Aza-dC (1 μM). (b) The effect of AID down-regulation on mutagenic properties of 5-Aza-dC (c) The effect of PARP-1 inhibitor olaparib on mutagenic properties of 5-Aza-dC. (d) The effect of PARP-1 inhibitor olaparib on cytotoxic/cytostatic properties of 5-Aza-dC. Data shown as average ±SD; asterisk (*) designates statistically significant difference with corresponding control (* - p<0.05; ** - p<0.01; *** - p<0.001).
Discussion
While the induction of point mutations by 5-Aza-dC has been described previously, our present data show for the first time that the drug also induces a large number of genome rearrangements, a much more toxic type of mutation. Interestingly, while in the past it has been speculated that genome rearrangements could be a consequence of DNA hypomethylation, which is also a consequence of 5-Aza-dC treatment, our present results highlight the role of DNA double-strand breaks induced by DNMT1 covalently bound to this nucleoside analog. Indeed, DNMT1 knock-down significantly reduces the number of genome rearrangements, the level of γ-H2AX expression and the toxicity after treatment of MEFs with 5-Aza-dC. Point mutations, however, were not affected.
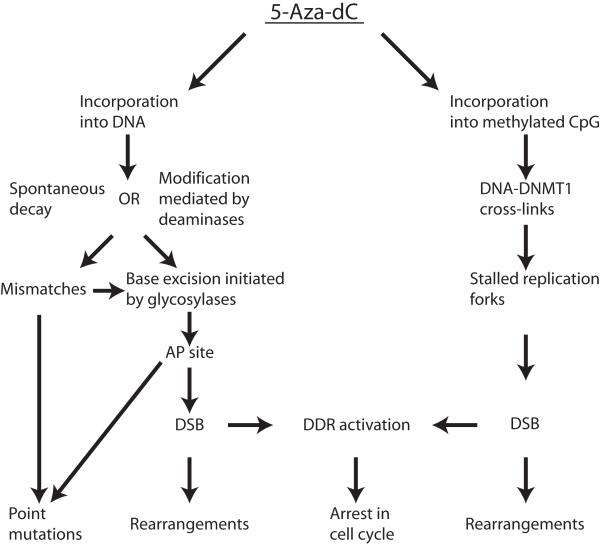
The mechanism of 5-Aza-dC mutagenicity is complex and includes several steps (Figure 5). First, due to the inherent instability of this compound in neutral aqueous solutions (22), 5-Aza-dC incorporated into DNA is also prone to spontaneous decay or can be metabolized by cytidine deaminases (23). Next, the resulting open ring species or 5-azauracil bases are subject to elimination by glycosylase through an activated base excision repair mechanism (BER). BER transiently generates abasic sites which can initiate formation of DSBs (24) and may lead to an observed accumulation of DSBs. The emerging DSBs can be converted into genome rearrangements. The point mutations could be created by the erroneous insertion of dAMP across from 5-Aza-dC-mediated abasic sites (25) or due to the ability of open ring remnants of 5-Aza-dC to pair with cytosine (7). Our data confirm the observation by Jackson-Grusby et al (7) that C->Gtransversion is a signature point-mutation induced by 5-Aza-dC, suggesting that the latter mechanism of generation of point mutation is the prevalent one.
Figure 5.
Schematic depiction of the possible mechanism of 5-Aza-dC mediated growth inhibition and mutagenicity.
Concurrently, when 5-Aza-dC is incorporated into a methylated CpG site, the nucleoside analogue may bind irreversibly to DNMT1, the enzyme responsible for maintenance of the methylation status, forming bulky adducts which prevent the progression of replication forks (26) and induce the ATM/ATR signaling pathway (27). The repair of such DNA-protein cross-links involves generation of double-strand breaks (27, 28) and consequently increases the risk of genome rearrangements. The latter scenario entirely depends on the presence of DNMT1 and we demonstrated a significant decline in the frequency of genome rearrangements upon treatment with 5-Aza-dC, as well as a corresponding delay in the onset of 5-Aza-dC induced DSBs in cells with down-regulated DNMT1 expression. Of note, we did not find a reduction in the number of 5-Aza-induced point mutations in cells after DNMT1 knock-down. This refutes the suggestion by Jackson-Grusby et al (7) that 5-Aza-dC-induced point mutations are mediated by DNMT1.
Alternatively, 5-Aza-dC mutagenicity could be associated with hypomethylation induced by the drug. Genome-wide hypomethylation in ES cells deficient for DNMT1 has been shown to lead to elevated mutation frequency (4). Although we found that 5-Aza-dC treatment at certain doses significantly decreases the level of DNA methylation, this decrease was not correlated with mutagenic effects of the drug. While at increased doses of up to 10 μM 5-Aza-dC did induce more mutations (Figure 2a), there was no effect on methylation level, which was found not different from the untreated control (Figure 2c). Moreover we demonstrated that down-regulation of DNMT1 suppressed the mutagenic effect of 5-Aza-dC without affecting methylation level of the cellular DNA. While we cannot rule out the possibility that 5-Aza-dC-induced hypomethylation leads to increased genome instability on the longer term, our findings do not suggest that hypomethylation and mutagenic effects of 5-Aza-dC are coupled.
One possible alternative hypothetical mechanism of 5-Aza-dC associated demethylation and mutagenesis, which we also investigated, involves the possible activation of the AID enzyme, induced by the genotoxic stress (20). The activated AID deaminates cytosines and (at much lower efficiency (29)), methyl-cytosines, and converts them into uracil and thymine, respectively. The erroneous repair of the resulting mismatches could explain accumulation of both point mutations and rearrangements as well as hypomethylation. However, we did not find any evidence for the up-regulation of AID expression in the MEFs treated with 5-Aza-dC at the same conditions that showed demethylation and accumulation of mutations. We also did not observe any effect of AID knock-down or inhibition of BER on the mutagenicity of 5-Aza-dC, which make the possible involvement of AID in the generation of genome rearrangements or 5-Aza-dC- induced hypomethylation unlikely.
Similar to its mutagenicity, the cytostatic effects of 5-Aza-dC treatment stem, to a large extent, from the ability of the drug to induce DNA lesions, such as DNA-protein bulky adducts, mismatches and abasic sites. This initiates a DNA damage response leading to cell-cycle arrest and growth inhibition. Formation of the DNMT1-CpG bulky adducts leading to stalled replication forks at the methylated CpG sites is a parallel mechanism potentiating 5-Aza-dC mediated growth inhibition. This mechanism should greatly depend on the level of DNMT1 expression and we found that the inhibitory effect of 5-Aza-dC on cell growth is significantly attenuated after down-regulation of DNMT1 expression.
Although the biological effects of 5-Aza-dC are dose-dependent we found that there is a plateau where further increases of applied dose lead to a minimal response in mutation frequency, growth suppression or DSB generation. The majority of nucleoside analogues exercise their biological effects by interfering with DNA replication (30) and 5-Aza-dC is not an exception. It is conceivable, therefore, that a correlation between the intensity of 5-Aza-dC stimulation and cellular response can be observed only while cells are capable of replicating their DNA.
BER is considered as a primary repair mechanism coping with 5-Aza-dCs. It is conceivable, therefore, that interventions targeting BER may affect therapeutic properties of 5-Aza-dC as an antitumor drug. The poly-ADP ribose polymerase 1 (PARP-1) is believed to play an important role in the repair of DNA strand breaks (31). Consequently, chemical inhibitors of PARP-1 were shown to have a substantial antitumor activity in a certain genetic background and also to potentiate the activity of some other antitumor drugs (32). However, we found that application of the PARP-1 inhibitor olaparib has no effect on mutagenic or cytostatic properties of 5-Aza-dC. Although the presence of active PARP-1 accelerates repair of the DNA SSB (33) the depletion of PARP-1 does not completely block the BER pathway, at least for certain types of damage (34, 35). Our findings suggest that ADP-ribosylation is not essential for repair of 5-Aza-dC associated DNA lesions. To clarify the role of BER in the repair of 5-Aza-dC induced damages further investigation needs to be performed employing more direct approaches affecting this repair mechanism.
Based on our results we propose the following mechanism of 5-Aza-dC-mediated growth inhibition and mutagenicity (Figure 5). Depending on the DNA milieu, there are two possible ways of dealing with incorporated 5-Aza-dC. The first mechanism (Figure 5, left branch) is employed in all instances and leads to growth suppression and accumulation of both point mutations and genome rearrangements. The second mechanism is employed only when the drug is incorporated into a methylated CpG site (Figure 5, right branch). This concurrent mechanism depends entirely on the presence of DNMT1 and also leads to inhibition of cell growth through induction of the DNA damage response, but is able to produce only one type of mutations: genomic rearrangements.
The antineoplastic activity of 5-Aza-dC is believed to have two different components (36, 37). Treatment with 5-Aza-dC has the ability to reactivate expression of epigenetically silenced tumor-suppressing genes by inhibiting the methylation of newly synthesized DNA thereby converting the genome of the actively dividing cells into a hypomethylated state (38, 39). Alternatively, 5-Aza-dC may act as a classic genotoxic drug, initiating a DNA damage response (40). While 5-Aza-dC is a potent methylation inhibitor, it is clear that many of cellular end-points associated with 5-Aza-dC treatment are methylation independent and instead depend on the induction of DNA lesions. The ability of 5-Aza-dC to induce DNA DSBs is greatly influenced by the level of DNMT1 in affected cells and our studies, as well as those of others (7, 13), confirming that cells depleted for DNMT1 are less susceptible to 5-Aza-dC’s toxic and mutagenic effects. These results suggest that the particular effectiveness of 5-Aza-dC in the treatment of myelodysplastic syndromes may be due to a heightened expression of DNMT1 and DNMT3a in the bone marrow of patients diagnosed with MDS (41). Therefore, it is conceivable that the expression profile of methyltransferases could constitute an effective prognostic factor for response to 5-Aza-dC treatment.
Materials and Methods
DNA methylation assay
For DNA methylation assay sodium bisulfite treatment was performed on 800 ng of DNA using the EZ DNA Methylation-Direct™ Kit (Zymo Research, CA, USA) following the manufacturer’s standard protocol. DNA methylation analysis was conducted following bisulfite-PCR amplification using the Sequenom EpiTYPER system (Sequenom Inc, CA, USA) as described elsewhere (42). This technique employs base-specific cleavage followed by MALDI-TOF mass spectrometry in which the size ratio of the cleaved products provides quantitative methylation estimates for CpG sites within a target region (43). Bisulfite primers, designed for the lacZ region using the Sequenom EpiDesigner, were the following: 5′-GGTAGTATTAGGGGAAAATTTTATTTATT-3′ (forward) and 5′-GGATTGGTTTGAATTGTTAGTTGG-3′ (reverse). The forward primer was tagged with a 10-mer (5′-AGGAAGAGAG-3′) to balance the PCR and the reverse primer contained a T7-promoter tag (5′ -CAGTAATACGACT CACTATAGGGAGAAGGCT-3′) for in vitro transcription. Bisulfite-PCR amplification was conducted using the HotStarTaq Master Mix Kit (Qiagen, CA, USA) under the following conditions: 95°C for 15 minutes, then 40 cycles of 94°C for 30 seconds, 64°C for 30 seconds and 72°C for 30 seconds, followed by 72°C for 10 minutes for the final extension. Analysis of the obtained data was performed using the EpiTYPER software.
Cell culturing and drugs application
MEFs were obtained from D13.5 embryos of lacZ transgenic mice and maintained in 10% CO2 and 3% O2 atmosphere at 37°C in DMEM (GIBCO, CA, USA) supplemented with 10% FBS (GIBCO). Medium with 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma, MO, USA) was prepared at the time of application and applied every 24 hours for the number of days indicated.
Mutation analysis and mutant classification
Mutation analysis was performed using the lacZ reporter assay as described previously (11). Briefly, reporter plasmids were released from genomic DNA of lacZ MEFs by digestion with HindIII restriction endonuclease, circularized, electroporated into E.coli galB-, ΔlacZ amp, and plated in selective and non-selective conditions. A mutation frequency was determined as the number of beta galactosidase defective versus wild-type bacterial colonies. Analysis of mutation spectra was performed as described previously (11). Briefly, 48 E.coli colonies containing mutated plasmids were individually picked and grown overnight for each sample. The plasmid fragment containing a mutated lacZ gene was PCR-amplified and the type of mutation was determined based on the restriction pattern after digestion with AvaI restriction endonuclease.
DNA double-strand break detection and quantification
DNA DSBs were detected by assessment of γ-H2AX expression (i.e. phosphorylated histone H2AX). Cells were trypsinized, fixed/permeabilized using the BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit (BD Biosciences, CA, USA) and stained with mouse anti-H2AX (pS139) antibodies conjugated with Alexa Fluor 488 (BD Biosciences) for 30 min at 37°C (1:50 in Perm/Wash buffer). The percentage of γ-H2AX positive cells was measured using a Guava EasyCyte Plus flow-cytometer (Millipore, MA, USA).
BrdU assay
Cells were cultured in 96-well plates in the presence of BrdU (10 ng/ml; Sigma) for 2 or 24 hours, 72 hours after transfection with siRNA. The amount of incorporated BrdU was determined using Cell Proliferation ELISA Kit (Roche Diagnostics, IN, USA) according to the manufacturer’s recommendations.
DNMT1 and AID knock-down experiments
For knock-down experiments MEFs were transfected with scrambled siRNA, DNMT1 siRNA and AID siRNA (ON-TARGETplus SMARTpool, Thermo Fisher Scientific, CO, U S A ) . T h e s e q u e n c e s o f s i R N A s t a r g e t i n g D N M T 1 w e r e : GGUAGAGAGUUACGACGAA, AAGCAAUUCAUGACGAGAA, GGUCGUGAGUGUUCGGGAA, GCUGGGAGAUGGCGUCAUA. The sequences of siRNAs targeting AID were: CCUGUAUACACGACCGUUA, CCUUGUACGAAGUCGAUGA, GGUGAUGAACCUCCGGAUU, ACGCUUUGCCCAACGAAAU. The ON-TARGETplus Non-targeting Pool was used as a control. The siRNA electroporation was performed using the Nucleofector device (Lonza Walkersville Inc., MD, USA) according to the manufacturers recommendations. Total RNA was extracted with ZR-Duet DNA/RNA MiniPrep kit (Zymo Research, CA, USA). The relative RNA expression level was determined using StepOne Plus Real-Time PCR system (Life Technologies Corp., CA, USA). Primers for real-time PCR were: AAATTCTGTCCGGCTAACCA (AID forward), CACGTGTGACATTCCAGGAG (AID reverse), AAGAATGGTGTTGTCTACCGAC (DNMT1 forward), CATCCAGGTTGCTCCCCTTG (DNMT1 reverse), AACTTTGGCATTGTGGAAGG (GAPDH forward), GGATGCAGGGATGATGTTCT (GAPDH reverse).
Cell proliferation assay
Relative cell number was measured with CyQUANT NF Cell Proliferation Assay Kit (Invitrogen, CA, USA) according to the manufacturer’s recommendations. For RNAi experiments cells were plated into 96-well plates 48 hours after transfection with either scrambled or DNMT1 siRNA at a density of 3000 cells/well. After 24 hours cells were administered medium with 5-Aza-dC and cultured for 72 hours with fresh medium containing the drug added every 24 hours. For PARP-1 inhibition experiments cells were plated into 96-well plates at a density of 3000 cell/well. After 24 hours cells were administered medium with different compounds for 72 hours with fresh medium containing the drug(s) added every 24 hours.
Statistical analysis
The p-values were calculated using a two-sample t-test (Microsoft Excel software package). Each experiment was repeated a minimum of 3 times; representative results are shown.
Acknowledgments
This work is supported by National Institute of Aging Grant P01 AG017242 to JV and YS; R01 AG024391 to YS. CT is a recipient of Ellison/AFAR postdoctoral fellowship.
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007 Feb 23;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6993–7. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci U S A. 1997 Mar 18;94(6):2545–50. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998 Sep 3;395(6697):89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 5.Amacher DE, Turner GN. The mutagenicity of 5-azacytidine and other inhibitors of replicative DNA synthesis in the L5178Y mouse lymphoma cell. Mutat Res. 1987 Jan;176(1):123–31. doi: 10.1016/0027-5107(87)90259-4. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez R, Frady A, Zhang XY, Varela M, Ehrlich M. Preferential induction of chromosome 1 multibranched figures and whole-arm deletions in a human pro-B cell line treated with 5-azacytidine or 5-azadeoxycytidine. Cytogenet Cell Genet. 1997;76(3-4):196–201. doi: 10.1159/000134548. [DOI] [PubMed] [Google Scholar]
- 7.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci U S A. 1997 Apr 29;94(9):4681–5. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelecsenyi Z, Spencer DL, Caspary WJ. Molecular analysis of 5-azacytidine-induced variants in mammalian cells. Mutagenesis. 2000 Jan;15(1):25–31. doi: 10.1093/mutage/15.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Landolph JR, Jones PA. Mutagenicity of 5-azacytidine and related nucleosides in C3H/10T 1/2 clone 8 and V79 cells. Cancer Res. 1982 Mar;42(3):817–23. [PubMed] [Google Scholar]
- 10.Zimmermann FK, Scheel I. Genetic effects of 5-azacytidine in Saccharomyces cerevisiae. Mutat Res. 1984 Jan;139(1):21–4. doi: 10.1016/0165-7992(84)90116-7. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AM, Busuttil RA, Rodriguez A, Cabrera C, Lundell M, Dolle ME, et al. Detection and analysis of somatic mutations at a lacZ reporter locus in higher organisms: application to Mus musculus and Drosophila melanogaster. Methods Mol Biol. 2007;371:267–87. doi: 10.1007/978-1-59745-361-5_20. [DOI] [PubMed] [Google Scholar]
- 12.Boerrigter ME, Dolle ME, Martus HJ, Gossen JA, Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995 Oct 19;377(6550):657–9. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 13.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolle ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002 Nov;12(11):1732–8. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998 Mar 6;273(10):5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 16.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011 Jun 15;25(12):1320–7. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010 Oct 1;24(19):2107–14. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011 Sep 16;146(6):866–72. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009 Dec 11;139(6):1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masaoka A, Horton JK, Beard WA, Wilson SH. DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst) 2009 Nov 2;8(11):1290–9. doi: 10.1016/j.dnarep.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogstad DK, Herring JL, Theruvathu JA, Burdzy A, Perry CC, Neidigh JW, et al. Chemical decomposition of 5-aza-2′-deoxycytidine (Decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chem Res Toxicol. 2009 Jun;22(6):1194–204. doi: 10.1021/tx900131u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chabot GG, Bouchard J, Momparler RL. Kinetics of deamination of 5-aza-2′-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol. 1983 Apr 1;32(7):1327–8. doi: 10.1016/0006-2952(83)90293-9. [DOI] [PubMed] [Google Scholar]
- 24.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22475–80. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, et al. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010 May 19;29(10):1738–47. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007 Sep 1;67(17):8248–54. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- 27.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009 Oct 2;284(40):27065–76. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008 Jan;28(2):752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, et al. Methylation protects cytidines from AID-mediated deamination. Mol Immunol. 2005 Mar;42(5):599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Sampath D, Rao VA, Plunkett W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene. 2003 Dec 8;22(56):9063–74. doi: 10.1038/sj.onc.1207229. [DOI] [PubMed] [Google Scholar]
- 31.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010 Apr;10(4):293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur J Cancer. 2010 Jan;46(1):9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007 Aug;27(15):5597–605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000 Oct 15;28(20):3887–96. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allinson SL, Dianova II, Dianov GL. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim Pol. 2003;50(1):169–79. [PubMed] [Google Scholar]
- 36.Ferguson AT, Vertino PM, Spitzner JR, Baylin SB, Muller MT, Davidson NE. Role of estrogen receptor gene demethylation and DNA methyltransferase.DNA adduct formation in 5-aza-2′deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997 Dec 19;272(51):32260–6. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- 37.Hoglund A, Nilsson LM, Forshell LP, Maclean KH, Nilsson JA. Myc sensitizes p53-deficient cancer cells to the DNA-damaging effects of the DNA methyltransferase inhibitor decitabine. Blood. 2009 Apr 30;113(18):4281–8. doi: 10.1182/blood-2008-10-183475. [DOI] [PubMed] [Google Scholar]
- 38.Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002 Oct 15;100(8):2957–64. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 39.Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16(17):2075–85. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- 40.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001 Apr;59(4):751–7. [PubMed] [Google Scholar]
- 41.Langer F, Dingemann J, Kreipe H, Lehmann U. Up-regulation of DNA methyltransferases DNMT1, 3A, and 3B in myelodysplastic syndrome. Leuk Res. 2005 Mar;29(3):325–9. doi: 10.1016/j.leukres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35(18):e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):15785–90. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]