Background: SAMHD1 is a host antiviral component that regulates cellular dNTP levels.
Results: We report a tight kinetic interplay between SAMHD1 level and the ability of HIV-1 to replicate in MDMs.
Conclusion: Very fast kinetics of SAMHD1 degradation by Vpx is mechanistically tied with HIV-1 DNA synthesis in MDMs.
Significance: The observed temporal relationship is central to better understanding HIV-1 infection in MDMs.
Keywords: DNA Replication, Enzyme Degradation, HIV, Macrophages, Nucleotide, SAMHD1, dNTPs
Abstract
Recently, SAMHD1 has come under intense focus as a host anti-HIV factor. SAMHD1 is a dNTP triphosphohydrolase, which leads to the regulation of DNA metabolism in host cells. HIV-2/SIV (simian immunodeficiency virus) viral protein x (Vpx) has been shown to promote the degradation of SAMHD1. In this study, we examine the kinetics of SAMHD1 degradation, the increase in the dNTP pool level, and the efficiency of proviral DNA synthesis in Vpx+ virus-like particle (VLP)-treated monocyte-derived macrophages (MDMs). Our results indicate a very close temporal link with a reduction in SAMHD1 detected within the first few hours of Vpx+ VLP treatment. This loss of SAMHD1 is followed by a significant increase in cellular dNTP levels by 8 h after Vpx+ VLP addition, ultimately leading to the enhancement of the HIV proviral DNA synthesis rate and HIV infection in MDMs. Finally, the pretreatment of MDMs with the Vpx+ VLPs, which is a widely used protocol, displayed identical proviral DNA synthesis as compared with MDMs co-treated with Vpx+ VLP and HIV vector. These findings further indicate that Vpx degradation of SAMHD1 is sufficiently rapid to enable appropriate progression of reverse transcription in MDMs, even when present at the time of infection. Overall, this study demonstrates a tight interplay between SAMHD1 level, dNTP levels, and HIV proviral DNA synthesis kinetics in MDMs.
Introduction
Sterile α motif domain and HD domain-containing protein 1 (SAMHD1)3 has been linked to Aicardi-Goutières syndrome (1), which is a hereditary neurodegenerative disorder with similarities to systemic lupus erythematosus. The SAM domain has been reported to contribute to developmental regulation (2), whereas the HD domain is widely found in many proteins (3). SAMHD1, along with TREX1, has been suggested to regulate host cellular DNA metabolism and could also be an innate antiviral response (1). Two recent studies by Goldstone et al. (4) and Powell et al. (5) have demonstrated that SAMHD1 is a dNTP triphosphohydrolase. Several recent studies by Hrecka et al. (6) and Laguette et al. (7), as well as another study by Berger et al. (8), have revealed that the HIV-2/SIV accessory protein, Vpx, counteracts SAMHD1, a newly identified antiviral HIV restriction factor, specifically in macrophages and dendritic cells. It was hypothesized that SAMHD1 may delay the HIV/SIV reverse transcription step by lowering cellular dNTP pools.
In 2004, for the first time, we reported that human primary monocyte-derived macrophages (MDMs) harbor ∼100 times lower cellular dNTP pools (20–40 nm) than activated CD4+ T cells (1–5 μm) (9). These dNTP pool levels were further validated recently using LC-MS/MS analysis (10). Collectively, these data suggest that the reduced dNTP pool in MDMs is a biochemical host factor that restricts HIV-1 reverse transcription. Indeed, the series of recent studies indicated above, including our recent study (11), confirmed that SAMHD1 is responsible for the low dNTP concentrations found in MDMs. However, after our study, we immediately investigated how tightly regulated the three key reported molecular events are: 1) degradation of SAMHD1, 2) increase in dNTP levels, and 3) HIV-1 proviral DNA synthesis kinetics influenced by Vpx treatment in MDMs. These events essentially encompass the complete mechanistic parameters of the dNTP-HIV replication landscape in nondividing cells. Our laboratory has the capability to measure dNTP pools in nondividing cells such as MDMs using the HIV-1 RT-based dNTP assay (9), which is sensitive enough to detect the low dNTP levels and not ribonucleoside triphosphates in MDMs. Therefore, we simultaneously monitored these three molecular events after Vpx treatment in MDMs.
EXPERIMENTAL PROCEDURES
Ethics Statement
These experiments used primary human primary monocytes obtained from human buffy coats (New York Blood Services, Long Island, NY). These are preexisting materials that are publicly available, and there is no subject-identifying information associated with the cells. As such, the use of these samples does not represent human subjects research because: 1) materials were not collected specifically for this study, and 2) we are not able to identify the subjects.
Cells
Primary human monocytes were isolated from the peripheral blood buffy coats by positive selection using MACS® CD14+ beads as described previously (12). Monocytes were maturated into MDMs in the presence of 5 ng/ml human GM-CSF (Miltenyi Biotec) treated at days 0 and 2 of maturation. MDMs were used at day 7 of maturation for experiments. RPMI medium (10% FBS with antibiotics) was replaced before transducing with virus-like particles (VLPs) and D3-HIV GFP vectors. For quantitative real-time PCR (qPCR) 2LTR circle copy number analysis, 1 million cells/well (6-well plates) were treated with VLPs (145 ng of p27/well), and total DNA was harvested using a Promega genomic purification kit. Two different VLP conditions were used: 2 h of pretreatment and co-treatment when treating with D3-HIV GFP vector (2.2 × 105 ng of p24/well, roughly a multiplicity of infection 0.1). For Western blot analysis and dNTP analysis, 2 million cells (3-cm dishes) were treated with 290 ng of p27/dish.
VLP Generation
Due to the requirement for large amounts of VLPs to concomitantly treat samples from the same donor for all the different assays, we subsequently modified the protocol from Berger et al. (8). Six T225 flasks containing 293T cells were transfected with 40 μg of pVpx− VLP or pVpx+ VLP (kindly provided by Drs. Florence Margottin-Goguet and Nathaniel Landau) and 20 μg of pVSVg at a ratio of 1 μg of DNA to 5 μl of polyethylenimine (1 mg/ml; PEI). The following day, medium were discarded and replaced with fresh DMEM medium (5% FBS and antibiotics). On days 2–4 after transfection, the medium was collected and replaced with fresh medium. On the day of collection, medium was centrifuged at 1200 rpm for 5 min to remove cells. Supernatant was subsequently filtered through a 0.45-μm membrane (Corning Inc.). Thirty milliliters of supernatant were overlaid on top of 5 ml of a 25% sucrose cushion (25% (w/v) sucrose, 10 mm Tris-HCl, pH 7.5), 0.1 m NaCl and 1 mm EDTA). VLPs were concentrated at 28,000 rpm for 90 min by ultracentrifugation. Supernatant was aspirated, and pellets were suspended in 700 μl of serum-free DMEM. Aliquots (50 μl) were stored at −80 °C. Each harvest was processed that day. The p27 antigen level was determined using an ELISA kit (Advanced BioScience Laboratories, Inc., Rockville MD).
D3-HIV GFP Vector Generation
D3-HIV GFP plasmid encodes the HIV-1 NL4-3 genome with the enhanced GFP gene in place of the HIV-1 nef gene and has a deleted envelope (9). To generate virus, 293T cells in T225 flasks were transfected with 40 μg of pD3-HIV and 20 μg of pVSV-g plasmids using PEI in 37 ml of DMEM medium/flask. At day 1 of HIV-1 production, media were discarded and replaced with fresh DMEM medium. At day 2, medium was harvested and replaced with fresh DMEM medium. The medium was centrifuged at 2500 rpm for 7 min to remove cellular debris and then stored at 4 °C in a T75 flask. Day 3 medium was harvested and processed as described for day 2. D3-HIV GFP virus was concentrated using ultracentrifugation (22,000 rpm for 2 h in a SW28 rotor). Viral pellets were DNase I-digested for 1 h at 37 °C. Afterward, debris was removed by centrifugation (14,000 rpm for 5 min). Sample aliquots were frozen at −80 °C until used. The level of p24 antigen was determined using an ELISA kit (Advanced BioScience Laboratories Inc.). Samples were analyzed for GFP expression at 24 or 48 h after transduction using an Accuri C6 flow cytometer (BD Biosciences). Data files were analyzed using the FlowJo software (TreeStar).
HIV-1 RT-based dNTP Assay
The nucleotide incorporation assay employs a 19-mer DNA template (3′-CAGGGAGAAGCCCGCGGTN-5′). The template is annealed to a 5′ end 32P-labeled 18-mer DNA primer (5′-GTCCCTGTTCGGGCGCCA-3′) for the HIV-1 RT-based assay used for this reaction, and the standard curves made with known amounts of dNTPs were used to covert the percentage of primer extension into the dNTP quantity used in each reaction as described previously (13). MDMs were collected and lysed with 60% cold methanol. Samples were incubated at 95 °C for 3 min. Cellular debris was cleared by centrifugation at 14,000 rpm. Supernatant was dried in a SpeedVac. Pellets were resuspended in 20 μl of reaction buffer (50 mm Tris-HCl, pH 8.0, and 10 mm MgCl2). Two microliters of the sample extract were used in the primer extension assay. Reactions were resolved on 14% urea-PAGE. Data were captured using a PMI instrument (Bio-Rad) and then analyzed using the Quantity One software (Bio-Rad). The amounts of dNTPs per cell and the macrophage cell volume were used to calculate cellular dNTP concentration as described.
Western Blot Analysis
Samples were processed in radioimmune precipitation buffer containing 1 μm DTT, 10 μm PMSF, 10 μl/ml phosphatase inhibitor (Sigma), and 10 μl/ml protease inhibitor (Sigma). The cells were sonicated with three 5-s pulses to ensure compete lysis. Cellular debris was removed by 15,000 rpm centrifugation for 10 min. Supernatants were stored at −80 °C before use. Cell lysates (20 μg) were resolved on 8% SDS-PAGE. Proteins were transferred to nitrocellulose membrane. The membrane was blocked with 2% nonfat milk in TBST for 1 h followed by the addition of primary SAMHD1 antibody (Abcam) and incubation overnight at 4 °C. The next day, the membrane was washed (3×, 20 min with TBST) and stained with donkey anti-rabbit HRP for 1 h at room temperature. Membrane was washed (3× with TBST) and developed using the SuperSignal West Femto Kit (Thermo Scientific). The membrane was stripped and reprobed for actin. Images were captured using the Bio-Rad ChemiDoc Imager. Data were normalized to actin as a loading control.
RESULTS
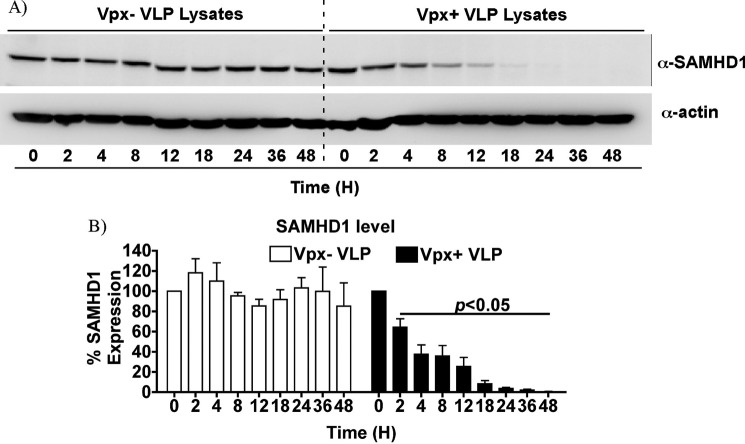
Primary human MDMs from three donors were treated with VLPs with and without Vpx. MDMs were harvested for analysis at the indicated time points: 0, 2, 4, 8, 12, 18, 24, 36, and 48 h. We chose this intense sampling scheme to investigate the interplay among three key reported molecular events: 1) degradation of SAMHD1, 2) increase in dNTP levels, and 3) HIV-1 proviral DNA synthesis kinetics for each donor tested. A representative Western blot for SAMHD1 and actin is displayed (Fig. 1A). As can clearly be observed, Vpx− VLPs did not promote SAMHD1 degradation. However, by 4 h after Vpx+ VLP treatment, a noticeable decrease in SAMHD1 protein level was detected. Data compilations for the three independent donors are shown in Fig. 1B and show significant reduction in SAMHD1 level at 2 h. Moreover, the SAMHD1 level continued to decrease to undetectable levels by 48 h after Vpx+ VLP addition.
FIGURE 1.
Western blot analysis of MDM lysates. A, a representative Western blot is shown for SAMHD1 and actin expression levels. Two million MDMs were treated with VLPs with and without Vpx (145 ng p27/million cells), and then harvested at the indicated time points. B, data are graphed as the mean and S.E. for three independent donors. Data were normalized to actin, used as a loading control. Student's t test analysis was performed to determine statistical significance (p < 0.05) for the different time points.
Next, we examined changes in cellular dNTP levels for MDMs using the highly sensitive HIV RT-based assay (9). A representative gel for the HIV RT-based primer extension assay is shown in Fig. 2A. The top panel shows the controls: no dNTPs and a sample with exogenous dNTPs added to have no extension (Primer) and extension (Primer +1), respectively. MDM extracts from Vpx− VLPs treatment (Fig. 2A, top) show only moderate amounts of extended products over the time course. Importantly, this is in contrast to MDM extracts from Vpx+ VLP treatment (Fig. 2A, bottom), which show much more extended products at 4 h after VLP treatment. The data were quantitated and displayed in Fig. 2B as a graph for the three independent donors. We detected a significant difference (p < 0.05) with the dNTP levels starting 8 h after Vpx+ VLPs treatment. Importantly, the dNTP levels rapidly increased 8–18 h after treatment before reaching a plateau. Moreover, dNTP levels increased concomitantly with SAMHD1 degradation. This supports previous studies indicating that SAMHD1 degradation by Vpx directly results in a subsequent increase in cellular dNTP levels in MDMs (6, 7, 11, 14).
FIGURE 2.
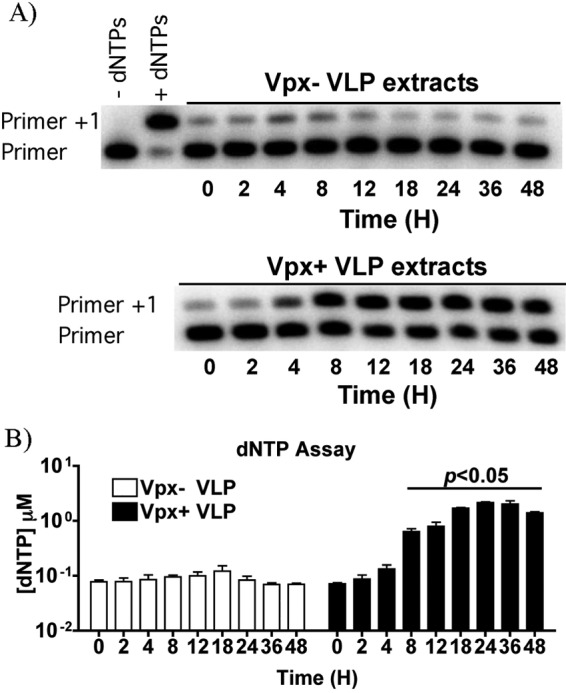
HIV RT-based primer extension assay. A, a representative HIV-1 RT-based primer extension gel is shown. Extension products are indicated by Primer +1 lane. Extracts from Vpx− VLPs are shown at the top, whereas Vpx+ VLPs extracts are below. B, data collected are graphed as mean and S.E. for three independent donors. Student's t test analysis was performed to determine statistical significance between the different time points.
Next, because the dNTP abundance is directly related with DNA polymerization kinetics, we examined the time-dependent proviral DNA synthesis kinetics by measuring 2LTR circle copy numbers using the qPCR technique. 2LTR circles are defective proviral DNA products that have failed to integrate into the host genome. Moreover, they are commonly used as a surrogate to monitor late proviral synthesis steps in the HIV life cycle (15). Previous studies have used pretreatment of target cell populations, macrophages, and dendritic cells before transducing them with HIV vectors (6–8, 11, 16, 17). Indeed, as shown in Fig. 3A, the pretreatment of MDMs with the Vpx+ VLPs accelerated proviral DNA synthesis tracking with the increase in dNTP levels.
FIGURE 3.
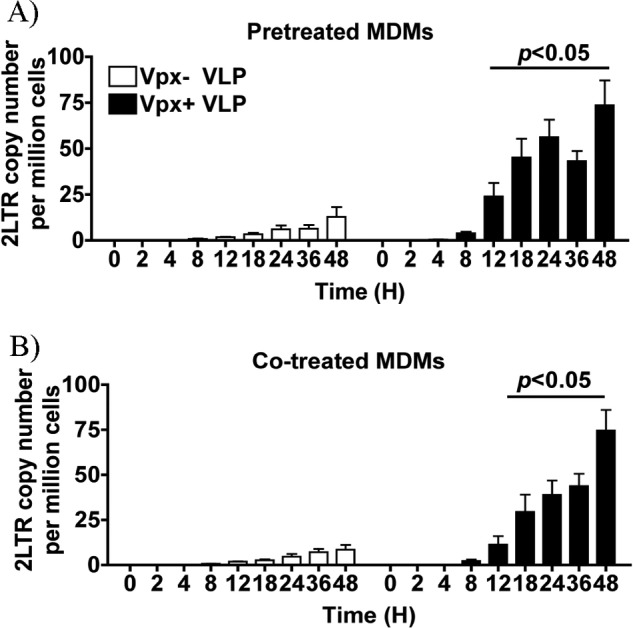
qPCR analysis for 2LTR circle copy numbers. A, MDMs were pretreated for 2 h with VLPs before the addition of D3-HIV GFP virus. At the indicated time points, cells were collected and processed for total DNA. qPCR for 2LTR circle copy numbers were performed in duplicate for three independent donors. B, MDMs were co-treated with VLPs and D3-HIV GFP virus. Data are plotted as mean and S.E., and paired Student's t test analysis was performed to determine statistical significance.
The 2-h pretreatment with the VLPs is a commonly used protocol to investigate the impact of Vpx on lentiviral vector transduction. Next, we compared the proviral DNA synthesis kinetics in MDMs with either the co-treatment with both VLPs and HIV vector or the 2-h pretreatment. As depicted in Fig. 3, A and B, both the pretreatment and the co-treatment with Vpx+ VLPs led to enhanced 2LTR circle copy numbers as compared with the MDMs treated with Vpx− VLPs. Both Vpx+ treatment groups were significantly higher than Vpx− treatment groups at 12 h after D3-HIV GFP addition and continuously increased even up to 48 h. Both Vpx+ VLPs treatment groups reached similar 2LTR copy numbers by 48 h. We also examined GFP expression by FACS and found comparable frequency of GFP+ cells in both the pretreated and the co-treated Vpx+ VLP groups (supplemental Fig. 1). Collectively, our data presented in this study strongly support that the accelerated reverse transcription kinetics is induced by the tight interplay between SAMHD1 degradation and dNTP pool increase, which greatly improves the overall success rate of proviral DNA synthesis completion in nondividing cells.
DISCUSSION
Vpx is believed to have arisen from gene duplication from Vpr (18). Although sequence similarities exist between Vpr and Vpx, they have different cellular functions (19–21). Vpx promotes the DCAF1 E3 ubiquitin ligase-mediated degradation of SAMHD1 (6, 7). Our recent study revealed that the Vpx-mediated degradation of SAMHD1 causes an increase in the cellular dNTP pool size (11), which, in turn, allows HIV-1 reverse transcriptase (RT) to efficiently carry out reverse transcription of the viral RNA genome into double-stranded proviral DNA. Moreover, a series of our extensive studies revealed that cellular dNTP concentration is critical for efficient reverse transcription by HIV-1 RT (9, 10, 22–26), especially in nondividing HIV target cell types.
Recently, Sunseri et al. (27) have shown that a modified Vpx can be incorporated into HIV-1 via p6. Although in this system, Vpx competes with Vpr for packaging into the virion, the authors found enhancement of transduction efficiency. Moreover, Vpx proteins from different lentiviruses, SIV versus HIV-2, have different activities in promoting transduction of MDM and dendritic cell populations, with SIV being more effective at enhancing transduction. Alternatively, the amount of Vpx being packaged within the virion could also be the reason for this observation because packaging of HIV-2 Vpx is reduced as compared with SIV of macaque Vpx (27). This may be of importance when trying to directly compare SIV versus HIV-2 or HIV-1 infection in animal models and humans. Moreover, Vpr is known to be released by infected cells and may negatively influence bystander cells (28). Vpx may also be released from infected cells and influence bystander cells. However, a putative model would be that Vpx, unlike Vpr, would promote infection of macrophage and dendritic cells, both nondividing cells, by down-regulating SAMHD1 expression at the site of infection. In addition to SAMHD1 degradation by Vpx, we believe that intracellular dNTP biosynthesis is required. Briefly, dNTP biosynthesis occurs through ribonucleotide reductase (RNR), the de novo synthesis pathway, and the salvage pathway. In eukaryotes, RNR is a heterodimeric enzyme composed of R1 and either R2 or p53R2 subunits (29). This heterodimeric complex catalyzes the conversion of ribose nucleoside diphosphates into deoxynucleoside diphosphate (30) and is highly regulated within the cell by the expression of the R2 subunits, cellular localization of subunits, and substrate feedback control mechanisms (31–34). Deoxynucleoside diphosphates require an additional phosphorylation step to become dNTPs, which then serve as a substrate for DNA polymerases and HIV-1 RT. R2 is mainly expressed during the S phase of the cell cycle (35), whereas p53R2 is induced upon DNA damage (36). We have not detected an increase in either R2 or p53R2 levels within the Vpx+ VLP-treated MDMs (data not shown). However, the functional activity of this complex still remains to be measured at this time. We speculate that Vpx may be influencing the RNR and kinase activities concomitantly with promoting SAMHD1 degradation to rapidly increase cellular dNTP levels in nondividing cells.
This work was supported, in whole or in part, by National Institutes of Health Grants AI049781 (to K. B.), F31 GM095190 to (W. D.) and F32 AI089079–01A1 (to J. A. H.).

This article contains supplemental Fig. 1.
- SAMHD1
- sterile α motif domain and HD domain-containing protein 1
- Vpx
- viral protein x
- Vpr
- viral protein r
- VLP
- virus-like particle
- MDM
- monocyte-derived macrophage
- SIV
- simian immunodeficiency virus
- qPCR
- quantitative real-time PCR
- TBST
- Tris-buffered saline with Tween
- RNR
- ribonucleotide reductase.
REFERENCES
- 1. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., Crow Y. J. (2009) Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schultz J., Ponting C. P., Hofmann K., Bork P. (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6, 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aravind L., Koonin E. V. (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23, 469–472 [DOI] [PubMed] [Google Scholar]
- 4. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 5. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutières syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic and myeloid cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger G., Durand S., Goujon C., Nguyen X. N., Cordeil S., Darlix J. L., Cimarelli A. (2011) A simple, versatile, and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 6, 806–816 [DOI] [PubMed] [Google Scholar]
- 9. Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., Kim B. (2004) Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chugh P., Bradel-Tretheway B., Monteiro-Filho C. M., Planelles V., Maggirwar S. B., Dewhurst S., Kim B. (2008) Akt inhibitors as an HIV-1-infected macrophage-specific antiviral therapy. Retrovirology 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weiss K. K., Chen R., Skasko M., Reynolds H. M., Lee K., Bambara R. A., Mansky L. M., Kim B. (2004) A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry 43, 4490–4500 [DOI] [PubMed] [Google Scholar]
- 14. Goujon C., Arfi V., Pertel T., Luban J., Lienard J., Rigal D., Darlix J. L., Cimarelli A. (2008) Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 82, 12335–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butler S. L., Johnson E. P., Bushman F. D. (2002) Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76, 3739–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pertel T., Reinhard C., Luban J. (2011) Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tristem M., Marshall C., Karpas A., Petrik J., Hill F. (1990) Origin of vpx in lentiviruses. Nature 347, 341–342 [DOI] [PubMed] [Google Scholar]
- 19. Casey L., Wen X., de Noronha C. M. (2010) The functions of the HIV1 protein Vpr and its action through the DCAF1·DDB1·Cullin4 ubiquitin ligase. Cytokine 51, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Marzio P., Choe S., Ebright M., Knoblauch R., Landau N. R. (1995) Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 69, 7909–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ueno F., Shiota H., Miyaura M., Yoshida A., Sakurai A., Tatsuki J., Koyama A. H., Akari H., Adachi A., Fujita M. (2003) Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect 5, 387–395 [DOI] [PubMed] [Google Scholar]
- 22. Jamburuthugoda V. K., Chugh P., Kim B. (2006) Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J. Biol. Chem. 281, 13388–13395 [DOI] [PubMed] [Google Scholar]
- 23. Kennedy E. M., Daddacha W., Slater R., Gavegnano C., Fromentin E., Schinazi R. F., Kim B. (2011) Abundant noncanonical dUTP found in primary human macrophages drives its frequent incorporation by HIV-1 reverse transcriptase. J. Biol. Chem. 286, 25047–25055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen L. A., Daddacha W., Rigby S., Bambara R. A., Kim B. (2012) Altered strand transfer activity of a multiple drug-resistant human immunodeficiency virus type 1 reverse transcriptase mutant with a dipeptide fingers domain insertion. J. Mol. Biol. 415, 248–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Cor-Hosmer S. K., Daddacha W., Kelly Z., Tsurumi A., Kennedy E. M., Kim B. (2012) The impact of molecular manipulation in residue 114 of human immunodeficiency virus type 1 reverse transcriptase on dNTP substrate binding and viral replication. Virology 422, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Cor-Hosmer S. K., Daddacha W., Kim B. (2010) Mechanistic interplay among the M184I HIV-1 reverse transcriptase mutant, the central polypurine tract, cellular dNTP concentrations, and drug sensitivity. Virology 406, 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sunseri N., O'Brien M., Bhardwaj N., Landau N. R. (2011) Human immunodeficiency virus type 1 modified to package simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 85, 6263–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muthumani K., Montaner L. J., Ayyavoo V., Weiner D. B. (2000) Vpr-GFP virion particle identifies HIV-infected targets and preserves HIV-1Vpr function in macrophages and T-cells. DNA Cell Biol. 19, 179–188 [DOI] [PubMed] [Google Scholar]
- 29. Stubbe J. (2000) Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol. 10, 731–736 [DOI] [PubMed] [Google Scholar]
- 30. Herrick J., Sclavi B. (2007) Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 63, 22–34 [DOI] [PubMed] [Google Scholar]
- 31. Elledge S. J., Zhou Z., Allen J. B., Navas T. A. (1993) DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15, 333–339 [DOI] [PubMed] [Google Scholar]
- 32. Larsson K. M., Jordan A., Eliasson R., Reichard P., Logan D. T., Nordlund P. (2004) Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat. Struct. Mol. Biol. 11, 1142–1149 [DOI] [PubMed] [Google Scholar]
- 33. Jordan A., Reichard P. (1998) Ribonucleotide reductases. Annu. Rev. Biochem. 67, 71–98 [DOI] [PubMed] [Google Scholar]
- 34. Hofer A., Crona M., Logan D. T., Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elledge S. J., Zhou Z., Allen J. B. (1992) Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem. Sci. 17, 119–123 [DOI] [PubMed] [Google Scholar]
- 36. Guittet O., Håkansson P., Voevodskaya N., Fridd S., Gräslund A., Arakawa H., Nakamura Y., Thelander L. (2001) Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J. Biol. Chem. 276, 40647–40651 [DOI] [PubMed] [Google Scholar]