Background: How multiple receptors for phagocytosis contribute to host immunity against bacterial infection has been elusive.
Results: Drosophila phagocytes use two receptors, integrin and Draper, to effectively recognize and engulf Staphylococcus aureus.
Conclusion: Dual recognition system exists in Drosophila for phagocytic removal of pathogenic bacteria.
Significance: Study of cellular immune response is important for understanding host defense against infectious diseases.
Keywords: Bacteria, Drosophila, Infectious diseases, Innate Immunity, Phagocytosis
Abstract
Integrin βν, one of two β subunits of Drosophila integrin, acts as a receptor in the phagocytosis of apoptotic cells. We here examined the involvement of this receptor in defense against infection by Staphylococcus aureus. Flies lacking integrin βν died earlier than control flies upon a septic but not oral infection with this bacterium. A loss of integrin βν reduced the phagocytosis of S. aureus and increased bacterial growth in flies. In contrast, the level of mRNA of an antimicrobial peptide produced upon infection was unchanged in integrin βν-lacking flies. The simultaneous loss of integrin βν and Draper, another receptor involved in the phagocytosis of S. aureus, brought about a further decrease in the level of phagocytosis and accelerated death of flies compared with the loss of either receptor alone. A strain of S. aureus lacking lipoteichoic acid, a cell wall component serving as a ligand for Draper, was susceptible to integrin βν-mediated phagocytosis. In contrast, a S. aureus mutant strain that produces small amounts of peptidoglycan was less efficiently phagocytosed by larval hemocytes, and a loss of integrin βν in hemocytes reduced a difference in the susceptibility to phagocytosis between parental and mutant strains. Furthermore, a series of experiments revealed the binding of integrin βν to peptidoglycan of S. aureus. Taken together, these results suggested that Draper and integrin βν cooperate in the phagocytic elimination of S. aureus by recognizing distinct cell wall components, and that this dual recognition system is necessary for the host organism to survive infection.
Introduction
Phagocytosis is a cellular immune response by which cells foreign to host organisms are incorporated and digested by immune cells called phagocytes (1, 2). In innate immunity, phagocytosis plays a front-line role in the host defense against invasive microorganisms. The importance of phagocytosis relative to humoral immune responses in innate immunity, such as the production of antimicrobial substances, has been controversial and in Drosophila melanogaster appears to depend on infectious state: the two immune responses sometimes cooperate with each other to prevent infectious diseases (3, 4) or act differentially depending on the stage of infection (5) and the kind of microorganism (6). Another important issue is whether the occurrence of phagocytosis is necessary for the subsequent induction of humoral responses, with reports both for (7–10) and against (11, 12) this notion. To resolve these issues, it is necessary to deepen our understanding of the mechanism of phagocytosis, and in particular, how invading microorganisms are recognized and engulfed by phagocytes.
It is widely appreciated that the fundamental mechanism of immune response is common among species from Drosophila melanogaster and Caenorhabditis elegans, animals frequently used as models in the study of innate immunity (13), to mice and humans (14–16). The use of Drosophila provides the advantage that genetically tractable experiments are feasible using whole animals infected by either injury or feeding with microorganisms (14). There are three types of Drosophila blood cells, i.e. hemocytes, which play distinct roles in innate immunity, namely, plasmatocytes, crystal cells, and lamellocytes, with plasmatocytes responsible for the phagocytic elimination of invading microorganisms (15–17). In mammals, the phagocytosis of bacteria is accomplished mostly with the aid of a serum component called opsonin that connects target cells and phagocytes. Both opsonin-dependent and -independent mechanisms of phagocytosis are likely to exist in Drosophila, but the former is less clearly understood (18). In contrast, a number of candidate receptors for the phagocytosis of bacteria have been reported, which are presumed to directly recognize bacterial components, including peptidoglycan recognition protein LC (19); members of the scavenger receptor family of proteins such as Peste (20), dSR-CI (21), and Croquemort (22); and the Nimrod family of proteins including Eater (6, 23, 24), Nimrod C1 (25) and Draper (26). However, the identity of putative cell wall components serving as ligands for these receptor proteins is largely obscure. Nevertheless, it is anticipated that a multiple recognition system for the phagocytic removal of bacteria exists in Drosophila to protect host organisms from infection with pathogenic microorganisms. Croquemort (27, 28) and Draper (29, 30) were originally found to act as receptors in the phagocytosis of apoptotic cells in Drosophila, suggesting that common receptors are used in the phagocytic elimination of apoptotic cells and invading bacteria in Drosophila. We recently reported that integrin βν, a β subunit of Drosophila integrins, is required for the phagocytic elimination of apoptotic cells in Drosophila embryos (31). In the present study, we examined whether this subunit is also involved in host defense against microorganisms.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Fly Stocks, and Antibody
The strain RN4220 was used as wild-type S. aureus and the parental strain for M0674 and TS2901. The strain M0674 does not express ltaS that codes for polyglycerolphosphate synthase, an enzyme responsible for the synthesis of polyglycerolphosphate of lipoteichoic acid, and thus lacks lipoteichoic acid (32). The strain TS2901 possesses a temperature-sensitive murB that codes for UDP-N-acetylenolpyruvylglucosamine reductase, an enzyme responsible for the synthesis of peptidoglycan, and produces a reduced level of peptidoglycan at a non-permissive temperature (33). Complementation of TS2901 with wild-type murB was done according to standard procedures using the plasmid pND50 as a vector (33). The strains W3110 and 168 were used as wild-type Escherichia coli and Bacillus subtilis, respectively. The bacteria were cultured at 30 °C (S. aureus strains M0674 and TS2901) or 37 °C (all other S. aureus, E. coli, and B. subtilis strains) with Luria-Bertani medium supplemented with glucose and antibiotics wherever required. When the cultures reached full growth, the bacteria were harvested, washed with PBS, and used in the experiments. The following lines of Drosophila were used in this study: w1118 (Bloomington Drosophila Stock Center, Indiana University, Bloomington, IN), betaInt-nu1 (34), betaInt-nu2 (34), drprΔ5 (29), elmoKO (35), and Rac1J11 Rac2Δ (4) (Bloomington Drosophila Stock Center). Some of the fly lines were used after changing balancers, and other lines were generated through the mating of existing flies. The anti-integrin βν antiserum was raised by immunizing rats with an extracellular region (amino acid positions 6–697 with the amino terminus numbered 1) of integrin βν that had been expressed in E. coli as a protein fused to GST at the amino terminus and purified to homogeneity. Antigen specificity of this antibody was confirmed in Western blotting (supplemental Fig. S1). Anti-GST mAb and HRP-conjugated anti-mouse IgG antibody were purchased from Millipore and GE Healthcare, respectively.
Bacterial Infection, and Assays for Bacterial Growth and Antimicrobial Peptide Production
The injection of bacteria into the abdomen of male adult flies, reminiscent of a septic infection, was done according to established procedures (37) with modifications (26). Briefly, flies 3–7 days after eclosion (15–20 flies per vial, 1–3 vials in each experiment) were injected with 50 nl of PBS containing given numbers of bacteria (1 × 103 per fly except for the analysis of phagocytosis, see below). For the oral infection, adult flies (13–15 flies per vial, 3 vials in each experiment) that had been dehydrated for 2 h without food were maintained in a vial containing filter paper soaked with 0.3 ml of 1% (w/v) sucrose including about 1010 bacteria or 50 mm H2O2 according to a published procedure (38). The flies infected with bacteria were maintained at 29 °C until use. The growth of bacteria in flies was analyzed by determining the colony-forming activity of injected bacteria as described previously (26). Briefly, homogenates of infected flies were plated at serial dilutions on an agar medium, and the number of colonies that appeared after incubation was expressed as cfu per fly. The level of mRNA of drosomycin was determined by quantitative RT-PCR as described previously (26). In brief, total RNA was prepared from about 100 infected flies and used as a template in RT to synthesize cDNA. The cDNA was then used as a template in real-time PCR for amplifying the sequences corresponding to mRNA of drosomycin and ribosomal protein 49, and the levels of the signals derived from drosomycin mRNA were normalized against that from ribosomal protein 49 mRNA, which served as an internal control. The DNA oligomers used as primers in real-time PCR were: 5′-CGTGAGAACCTTTTCCAATATGATG-3′ (forward) and 5′-TCCCAGGACCACCAGCAT-3′ (reverse) for drosomycin; and 5′-GACGCTTCAAGGGACAGTATCTG-3′ (forward) and 5′-AAACGCGGTTCTGCATGAG-3′ (reverse) for ribosomal protein 49.
Assay for Phagocytosis
An assay for the phagocytosis of S. aureus in vivo and in vitro was carried out as described previously (26). Briefly, for the assay in vivo male adults were injected with S. aureus (1 × 106 per fly), which had been labeled with FITC or 5-carboxyfluorescein, succinimidyl ester (Molecular Probes), maintained at 29 °C for 2 h, administered with PBS containing 0.4% (w/v) trypan blue to quench fluorescence derived from bacteria left unengulfed, and examined under a fluorescence microscope after 30 min. The level of phagocytosis was determined based on the size of clusters of fluorescent materials, which corresponded to aggregates of hemocytes that had engulfed bacteria. For the assay in vitro, larval hemocytes were prepared essentially according to established procedures (37) with modifications (26). In brief, hemolymph was collected from wandering third-instar larvae, placed on Teflon-coated glass slides, and incubated on ice for 10 min so that isolated hemocytes adhered to the slides. The hemocyte cultures were incubated with FITC-labeled bacteria (hemocytes:targets = 1 : 500) or fluorescence-labeled latex beads (2.0 μm in diameter; Molecular Probes) (hemocytes:targets = 1 : 200) at 25 °C for 10–30 min, washed with PBS, enclosed with 0.4 mm sodium acetate buffer (pH3.5) containing 0.4% trypan blue, and examined by fluorescence microscopy. The ratio of hemocytes containing target particles and the number of target particles contained in 100 phagocytes were determined and exhibited as “phagocytosing hemocytes” and “engulfed targets,” respectively.
Assays for Binding of Peptidoglycan to Hemocytes, and Integrin to Bacteria and Peptidoglycan
To examine the binding of peptidoglycan to hemocytes, larval hemocytes (1 × 104) were incubated for 10 min at 25 °C with an insoluble preparation of peptidoglycan (20 μg), which had been prepared from wild-type S. aureus (39) and labeled with FITC. The cells were then washed, fixed, and examined by fluorescence microscopy. For the analysis of the binding of integrin βν to S. aureus, a mixture of GST-fused integrin βν (10 pmol) used as an antigen for generating the antibody and GST alone (10 pmoles) was incubated with strains of S. aureus (1 × 108) for 15 min at room temperature, and the bacteria were collected by centrifugation, washed with PBS, lysed, and analyzed by Western blotting with anti-GST mAb, essentially as described previously (31). Binding of integrin βν to peptidoglycan was tested using insoluble and soluble fractions of peptidoglycan. A mixture of GST-fused integrin βν (10 pmol) and GST (10 pmol) was incubated with insoluble fractions of peptidoglycan prepared from E. coli (5 μg) (40) or S. aureus (wild-type strain) (40 μg) for 5 min at room temperature, and peptidoglycan was precipitated by centrifugation, washed with PBS, and analyzed by Western blotting using anti-GST mAb. To examine the binding of integrin βν to soluble peptidoglycan preparations, dishes of a 96-well culture container (MS-8496F; Sumitomo Bakelite, Tokyo, Japan) were coated with peptidoglycan (4 μg per well), which had been prepared from wild-type S. aureus (39), and increasing amounts of GST-fused integrin βν or GST were added in triplicate. The dishes were incubated for 3 h at room temperature, washed, successively supplied with anti-GST mAb and HRP-conjugated anti-mouse IgG antibody, and subjected to a colorimetric reaction using o-phenylenediamine as a substrate, and the amount of the reaction products was determined by measuring A490.
Data Processing and Statistical Analysis
Results from quantitative analyses are expressed as the mean ± S.D. of the data from at least three independent experiments, unless otherwise stated in the text. Other data are representative of at least three independent experiments that yielded similar results. Statistical analyses were performed using Student's t test, and p values of less than 0.05 were considered significant and are indicated in the figures.
RESULTS
Role for Integrin βν in Phagocytosis of S. aureus by Drosophila hemocytes
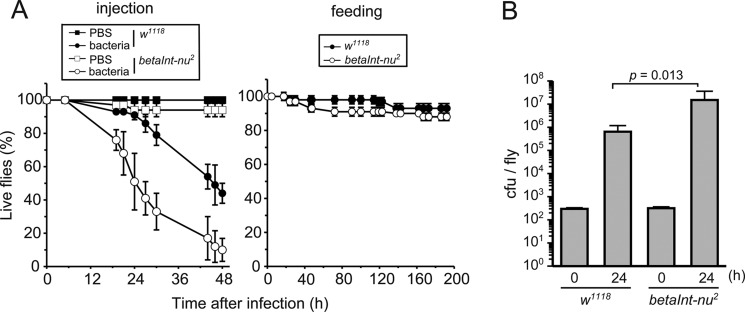
We first examined the role of integrin βν in the defense of Drosophila against S. aureus. For this purpose, adult flies of an integrin βν-lacking mutant (betaInt-nu2) were subjected to an abdominal or oral infection with S. aureus, and the ratio of live flies was determined (Fig. 1A). As we showed previously (26), the abdominal injection of the bacterium led to the death of control flies (w1118) while feeding flies with bacteria-containing meals did not have a lethal effect. Under these conditions, a loss of integrin βν accelerated the death of flies that had received the abdominal infection but did not influence the consequence of the oral infection with or without superoxide (data not shown), which enhances the killing of flies orally infected with S. aureus (38). There was no difference in survival between control flies and the integrin βν-lacking mutant that had received an injection with PBS, the solvent for bacteria. These results indicated the involvement of integrin βν in the mechanism to combat invading S. aureus. We then examined if a loss of integrin βν helps the growth of S. aureus in adult flies. The number of colony-forming bacteria recovered from flies 24 h after the infection was determined and compared between the mutant and control flies. More colony-forming bacteria were obtained from integrin βν-lacking flies (Fig. 1B), suggesting the inhibitory effect of integrin βν on bacterial growth in adult flies.
FIGURE 1.
Requirement of integrin βν for host defense against septic infection with S. aureus. Adult flies of control (w1118) (closed symbols) and integrin βν-deficient (betaInt-nu2) (open symbols) lines were infected with wild-type S. aureus (strain RN4220) by injury (injection at the abdomen) (left in A and B) or feeding (right in A), and subjected to an analysis of fly survival (A) and bacterial growth (B) at the indicated periods. The square symbols in the left panel of A denote the results for flies injected with solvent (PBS) alone. In A, data from one of three independent experiments that yielded similar results are presented. In B, cfu values at 24 h relative to those at 0 h were compared between w1118 and betaInt-nu2. Genotype of the fly line analyzed is w; betaInt-nu2 (betaInt-nu2).
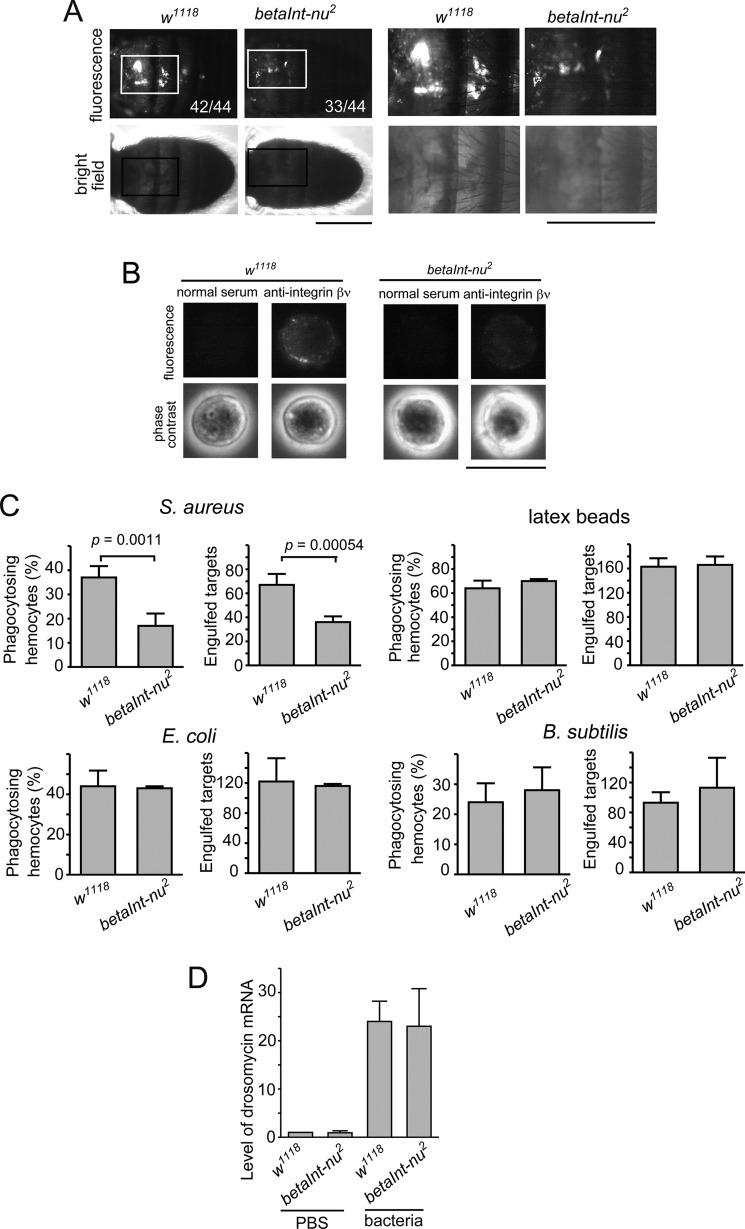
We next examined which type of immune response, cellular or humoral, integrin βν participates in. An assay for phagocytosis, a cellular response, was first conducted in vivo using fluorescence-labeled S. aureus injected into the abdomen of adult flies. We found that the size of fluorescent clusters, a hallmark of the engulfment of bacteria by hemocytes, was smaller in integrin βν-lacking flies than in control flies (Fig. 2A). To further confirm this, we determined the phagocytic activity of hemocytes isolated from third-instar larvae in vitro. When larval hemocytes were first immunocytochemically analyzed for the presence of integrin βν, most (>95%) cells were positive for punctate signals around surfaces, which were almost lost in the null mutant (Fig. 2B). The level of phagocytosis of S. aureus with integrin βν-lacking hemocytes was about half of that with control hemocytes while latex beads were phagocytosed equally by either hemocyte preparation (Fig. 2C). A decreased level of phagocytosis upon the loss of integrin βν was replicated in experiments both in vivo and in vitro with another null allele of betaInt-nu (betaInt-nu1) (data not shown). In contrast, E. coli and B. subtilis were effectively phagocytosed by larval hemocytes regardless of the absence of integrin βν (Fig. 2C), suggesting that hemocytes do not use this integrin subunit for phagocytosing these bacteria. The level of mRNA of drosomycin, an antimicrobial peptide produced as an innate immune response to bacterial infection via the Toll pathway (15), was not altered by the loss of integrin βν (Fig. 2D). These results indicated that integrin βν is required for the phagocytic elimination of invading S. aureus, most likely as a receptor for phagocytosis, but not for the production of antimicrobial peptides.
FIGURE 2.
Requirement of integrin βν for phagocytosis but not humoral response after septic infection with S. aureus. A, adult flies of control (w1118) and integrin βν-deficient (betaInt-nu2) lines were injected with FITC-labeled wild-type S. aureus and analyzed for the level of phagocytosis in vivo. Phase contrast and fluorescence views of the same microscopic fields are shown as vertically aligned panels. Parts of fly abdomen containing clusters of fluorescent materials, indicative of phagocytosis by hemocytes, are shown with squares, and their magnified views are exhibited at the right. The number of flies that gave the fluorescent clusters similar in size to that shown here is indicated in the panels (the denominator is the total number of flies analyzed). Scale bars, 0.5 mm. B, larval hemocytes isolated from the indicated fly lines were subjected to immunocytochemistry with anti-integrin βν or control rat serum. Phase contrast and fluorescence views of the same microscopic fields are shown as vertically aligned panels. Scale bar, 10 μm. C, an assay for phagocytosis in vitro was conducted with fluorescence-labeled wild-type S. aureus, latex beads, E. coli, or B. subtilis as targets and larval hemocytes prepared from the indicated fly lines as phagocytes. D, adult flies of the indicated lines were injected with wild-type S. aureus (bacteria) or solvent alone (PBS). After 18 h, flies were subjected to quantitative RT-PCR for the amount of drosomycin mRNA, which is shown relative to that in the control experiment (w1118 injected with PBS) as 1.
Independent Actions of Integrin βν and Draper in Defense against S. aureus
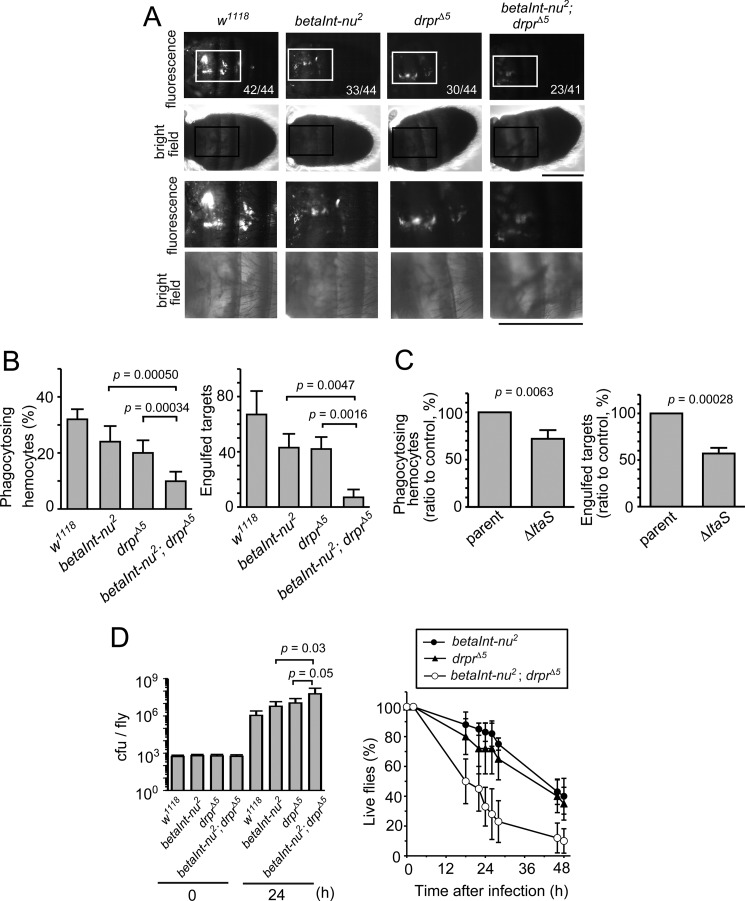
We previously reported that Draper acts as a receptor in the phagocytosis of S. aureus in Drosophila (26). The functional relationship of integrin βν with Draper was determined because integrin βν also likely acts as a receptor for phagocytosis. We determined the level of phagocytosis with flies lacking both integrin βν and Draper, and compared it to that with flies deficient in either protein alone. In an assay in vivo, the level of phagocytosis in double-mutant flies seemed to be somewhat lower than that in single-mutants (Fig. 3A). This was more significant when larval hemocytes prepared from mutant flies were examined for their phagocytic activity in vitro. The level of phagocytosis by hemocytes lacking either integrin βν or Draper was 60–70% of that by control hemocytes while it was reduced to about 30% of the control level upon a simultaneous loss of the two receptors (Fig. 3B). These results indicated that both integrin βν and Draper are required for a maximal level of the phagocytosis of S. aureus by Drosophila hemocytes. The S. aureus gene ltaS codes for an enzyme responsible for the synthesis of glycerolphosphate repeat of lipoteichoic acid, and bacteria that do not express ltaS lack lipoteichoic acid (32). Draper requires lipoteichoic acid for the efficient phagocytosis of S. aureus: hemocytes of control flies phagocytose ltaS-deficient S. aureus less effectively than parental bacteria while Draper-lacking hemocytes almost equally phagocytose these two bacterial strains at a lower level (26). We found that hemocytes lacking integrin βν required the presence of lipoteichoic acid in S. aureus for a maximal level of phagocytosis (Fig. 3C), suggesting that the integrin βν-mediated phagocytosis of S. aureus does not depend on lipoteichoic acid. The fact that Draper recognizes lipoteichoic acid as a ligand for phagocytosis (26) suggested that integrin βν recognizes a cell wall component(s) of S. aureus other than lipoteichoic acid. We next examined the cooperation between the two proteins in defense mechanisms besides phagocytosis. The level of bacterial growth was higher in the double-mutant than single-mutants (left panel in Fig. 3D), and flies lacking both integrin βν and Draper died earlier than those deficient in the expression of either protein (right panel in Fig. 3D). The above-described data collectively showed that integrin βν and Draper cooperate with each other for flies to survive infection with S. aureus.
FIGURE 3.
Independent actions of integrin βν and Draper in defense against S. aureus. A, adult flies of control (w1118), integrin βν-deficient (betaInt-nu2), and Draper-deficient (drprΔ5) lines, and a double mutant for betaInt-nu and drpr (betaInt-nu2; drprΔ5) were injected with FITC-labeled wild-type S. aureus and analyzed for the level of phagocytosis in vivo. The data with betaInt-nu2 and drprΔ5 fly lines are a duplicate of those shown in Fig. 2A. The micrographs at the bottom two rows are magnified views of the area with the squares in micrographs at the top two rows. The data are presented as in Fig. 2A. Scale bars, 0.5 mm. B, an assay for phagocytosis in vitro was conducted with FITC-labeled wild-type S. aureus as targets and larval hemocytes prepared from the indicated fly lines as phagocytes. C, an assay for phagocytosis in vitro was conducted with FITC-labeled wild-type (parent) or LtaS-deficient (ΔltaS) S. aureus as targets and larval hemocytes prepared from integrin βν-deficient fly lines as phagocytes. The level of phagocytosis is shown relative to that with wild-type bacteria taken as 100. Data from one of three independent experiments with similar results are presented. D, adult flies of betaInt-nu2, drprΔ5, and betaInt-nu2; drprΔ5 lines were injected with wild-type S. aureus and analyzed for bacterial growth (left) and fly survival (right) at the indicated time points. In the left panel, cfu values at 24 h relative to those at 0 h were compared between the fly lines. Genotypes of the fly lines analyzed are w; +; drprΔ5 (drprΔ5) and w; betaInt-nu2; drprΔ5 (betaInt-nu2; drprΔ5).
Role for Peptidoglycan in Integrin βν-mediated Phagocytosis of S. aureus
It is likely that a cell wall component other than lipoteichoic acid acts as a ligand for integrin βν. In a previous study, we determined the level of phagocytosis of various S. aureus strains with defects in the synthesis of cell walls, and found teichoic acid and glycolipids but not lipoproteins to be required for efficient phagocytosis (26). Lipoteichoic acid was later found to serve as a ligand for Draper (26), and the involvement of peptidoglycan was yet to be examined. We therefore wondered if peptidoglycan is involved in the integrin βν-mediated phagocytosis of S. aureus. A strain of S. aureus possessing a temperature-sensitive murB (strain TS2901) produces a reduced level of peptidoglycan at a non-permissive temperature (33). This mutant strain behaved as a less effective target for phagocytosis than the parental strain in an assay in vivo (Fig. 4A) as well as in vitro (Fig. 4B). Complementation of the mutant strain with the wild-type gene almost completely restored this defect (Fig. 4, A and B). These results indicated that the expression of murB is necessary for S. aureus to be effectively phagocytosed by Drosophila hemocytes. We then examined the phagocytosis of the murB-mutant and parental strains by hemocytes prepared from integrin βν-lacking and control (w1118) flies (Fig. 4C). The murB-mutant strain was still less effectively phagocytosed than the parental strain by hemocytes lacking integrin βν. However, a difference in the susceptibility to phagocytosis between the two S. aureus strains was significantly smaller in experiments using integrin βν-lacking hemocytes compared with that with control hemocytes. These results suggested the involvement of peptidoglycan in the integrin βν-mediated phagocytosis of S. aureus.
FIGURE 4.
Identification of S. aureus murB required for integrin βν-mediated phagocytosis. A, adult flies of the control (w1118) line were injected with 5-carboxyfluorescein-labeled wild-type (parent) or murB mutant (murBts) S. aureus and analyzed for the level of phagocytosis in vivo. The mutant strain was also analyzed for susceptibility to phagocytosis with (murB) or without (vector) complementation of the wild-type gene. The data are aligned and presented as in Fig. 3A. Scale bars, 0.5 mm. B, an assay for phagocytosis in vitro was conducted with FITC-labeled S. aureus as targets and larval hemocytes prepared from control (w1118) lines as phagocytes. The bacteria used are wild-type (parent) and murB mutant (murBts) strains with (murB) and without (vector) complementation of the wild-type gene. C, an assay for phagocytosis in vitro was conducted with FITC-labeled wild-type (parent) and murB mutant (murBts) S. aureus as targets and larval hemocytes prepared from control (w1118) and integrin βν-deficient (betaInt-nu2) lines as phagocytes. The extent of phagocytosis is shown relative to that with wild-type bacteria taken as 100. D, larval hemocytes prepared from the indicated fly lines were incubated with FITC-labeled peptidoglycan prepared from wild-type S. aureus, washed, and examined by fluorescence microscopy. Left, fluorescence, bright field, and overlaid views of the same microscopic fields are shown as vertically aligned panels. Right, number of hemocytes bound by peptidoglycan was determined and is shown relative to the total number of hemocytes analyzed. Scale bar, 50 μm. E, binding of the extracellular region of integrin βν to S. aureus strains (left), insoluble peptidoglycan (PGN) (middle), and soluble peptidoglycan (right) was examined. A mixture of GST-fused integrin βν and GST at an equal molar ratio was incubated with bacteria (left) or insoluble peptidoglycan (middle) and centrifuged, and the resulting precipitates were subjected to Western blotting with anti-GST antibody. The open and closed arrowheads indicate the positions of GST-fused integrin βν and GST, respectively. In the right panel, GST-fused integrin βν (GST-βν) (open circles) and GST (closed circles) were incubated in culture dishes coated with soluble peptidoglycan prepared from parental S. aureus, and the amount of GST proteins attached to dishes was immunochemically determined. Data from one of three independent experiments that yielded similar results are presented.
We next asked if integrin βν binds peptidoglycan. To test the binding of peptidoglycan to hemocytes, a fraction of the cell wall rich in peptidoglycan was labeled with FITC and incubated with larval hemocytes, and the hemocytes were recovered by centrifugation, washed, and examined by fluorescence microscopy. While intense signals were detectable with control hemocytes, the level of the signal with integrin βν-lacking hemocytes was only marginal (Fig. 4D), suggesting the integrin βν-mediated binding of peptidoglycan to hemocytes. We next examined the binding of integrin βν to S. aureus strains. Bacteria, parental, or murB mutant strain, were incubated with a mixture of GST-fused recombinant protein corresponding to the extracellular region of integrin βν and GST alone at an equal molar ratio, precipitated by centrifugation, lysed, and analyzed by Western blotting for the presence of the GST proteins. The signal corresponding to GST-fused integrin βν was more intense than that of GST alone using the parental S. aureus strain, but this was not the case in the experiment using the murB mutant (left panel in Fig. 4E). To directly analyze the binding of integrin βν to peptidoglycan, insoluble preparations of peptidoglycan obtained from S. aureus (parental strain) and E. coli were incubated with a mixture of GST-fused integrin βν and GST, precipitated, and subjected to Western blotting with anti-GST antibody. We found that more GST-fused integrin βν was recovered than GST alone using S. aureus peptidoglycan while recovery of the two GST proteins did not significantly differ using peptidoglycan prepared from E. coli (middle panel in Fig. 4E). Finally, we analyzed the binding of integrin βν to a soluble preparation of peptidoglycan in a solid-phase assay. GST-fused integrin βν showed dose-dependent adhesion to culture dishes coated with S. aureus peptidoglycan, but GST alone did not give appreciable binding (right panel in Fig. 4E). We were unable to examine the binding of integrin βν to a soluble preparation of E. coli peptidoglycan that caused extremely high background in this assay. These results collectively indicated a physical association between integrin βν and peptidoglycan of S. aureus.
Involvement of Rac but Not Elmo in Integrin βν-mediated Phagocytosis of S. aureus
We next addressed an event located downstream of integrin βν for the induction of the phagocytosis of S. aureus. There are two partly overlapping pathways for the induction of the phagocytosis of apoptotic cells in C. elegans, namely, CED-1-CED-6/CED-7-CED-10 and CED-2-CED-5-CED-12-CED-10, which are presumably conserved among species (41, 42). Draper appears to be located at the furthest point upstream of the former pathway in the phagocytosis of apoptotic cells (43) and degenerated neural axons (44). This suggests the involvement of the latter pathway in the integrin βν-mediated phagocytosis of S. aureus although this is not clear in the integrin βν-mediated phagocytosis of apoptotic cells (31). Drosophila counterparts of C. elegans CED-12 and CED-10 are Elmo and Rac, respectively (41, 42). There are three RacGTPases; namely, Rac1, Rac2, and Mig2-like, besides Rho and Cdc42 that together constitute the Drosophila RhoGTPase family of protein. We examined the involvement of Rac1 and Rac2 in the phagocytosis of S. aureus because previous reports showed that these two GTPases are necessary for the cellular immune response by Drosophila hemocytes: Rac2 is required for the phagocytosis of bacteria (45), and both Rac1 and Rac2 are required for the encapsulation of wasp eggs (46, 47).
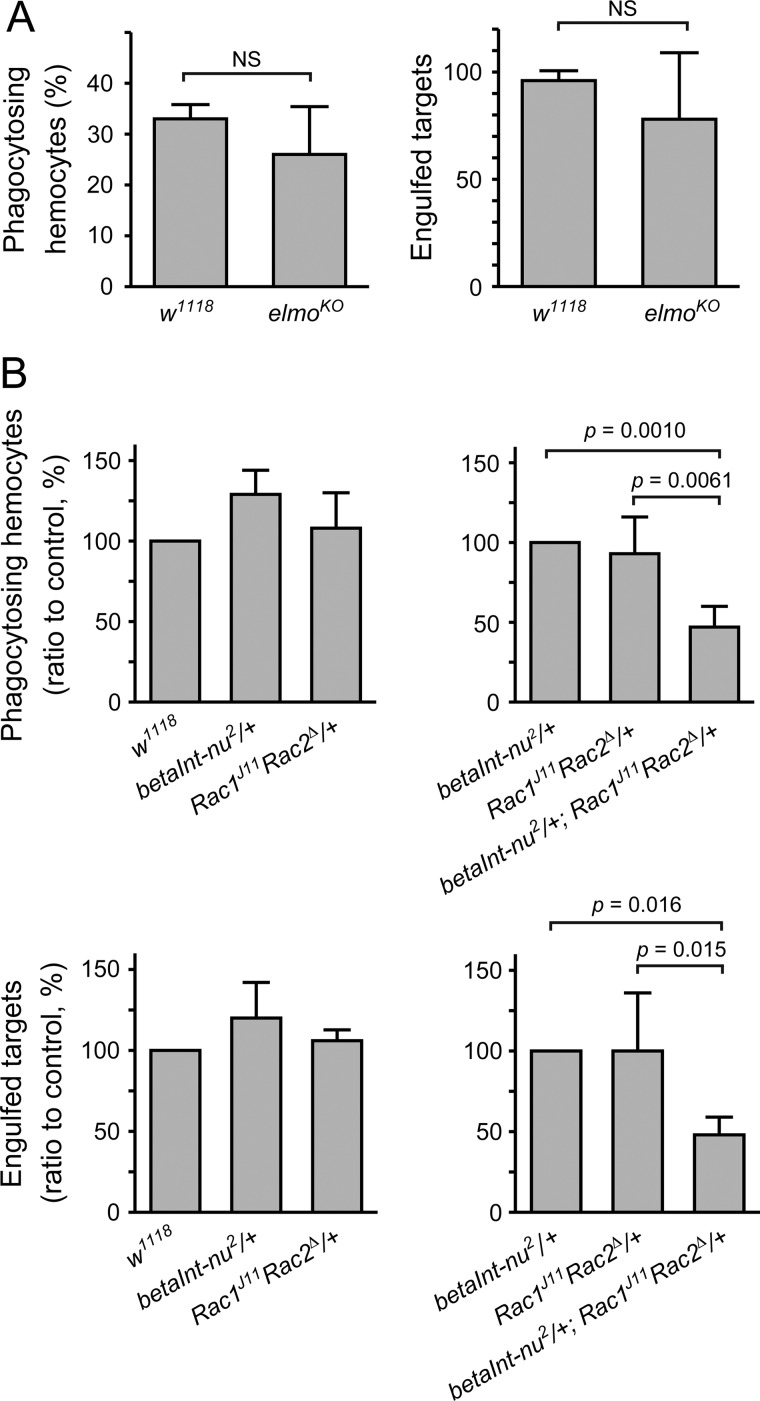
We first examined the involvement of Elmo, a Drosophila homologue of CED-12 and a component of a guanine nucleotide exchange factor specific for the small G protein Rac (48). However, larval hemocytes lacking the expression of Elmo showed phagocytic activity comparable to those prepared from control flies (Fig. 5A), suggesting that Elmo is dispensable. In contrast, hemocytes isolated from flies heterozygous for mutant alleles of both betaInt-nu and Rac showed a decreased level of phagocytosis compared with those from a heterozygote of either mutation (Fig. 5B), indicating a genetic interaction between the two genes. These results suggested that Rac1/Rac2 is contained in a signaling pathway located downstream of integrin βν for the induction of the phagocytosis of S. aureus.
FIGURE 5.
Genetic interaction between betaInt-nu and rac1/rac2 in phagocytosis of S. aureus. An assay for phagocytosis in vitro was conducted with FITC-labeled wild-type S. aureus as targets and hemocytes prepared from larvae of control flies (w1118) (A), elmo null flies (elmoKO) (A), flies heterozygous for betaInt-nu2 (betaInt-nu2/+) (B), flies heterozygous for rac1 and rac2 (Rac1J11 Rac2Δ/+) (B), or flies trans-heterozygous for betaInt-nu2, rac1, and rac2 (betaInt-nu2/Rac1J11 Rac2Δ) (B) as phagocytes. Genotypes of the fly lines used are elmoKOFRT40, 42/elmoKOFRT40, 42 (elmoKO) and y1 w*; Rac1J11 Rac2Δ FRT2A/TM6B, Tb+ (Rac1J11 Rac2Δ).
DISCUSSION
In the present study, we found that betaInt-nu coding for βν, a β subunit of Drosophila integrins, participates in the phagocytosis of Gram-positive S. aureus by hemocytes. We previously reported that Draper, another membrane-transverse protein of Drosophila hemocytes, is involved in the phagocytosis of the same bacterium (26). Here we showed that Draper and integrin βν cooperate in the defense against S. aureus to confer flies prolonged survival after a septic infection with the bacterium. The two receptors appeared to recognize distinct cell wall components as ligands for phagocytosis: Draper binds to lipoteichoic acid (26) while integrin βν does peptidoglycan. This suggests the presence of a dual recognition system by hemocytes for the efficient removal of S. aureus by phagocytosis. Signaling pathways located downstream of the receptors remain to be clarified, but they both seemed to require Rac, a small G protein involved in the phagocytosis of apoptotic cells (41, 42), suggesting that the two pathways overlap at least partly. Integrin βν seemed not required for the phagocytosis of E. coli and B. subtilis by larval hemocytes. The structure of peptidoglycan of S. aureus differs from that of these two bacteria species particularly in the composition of peptide, suggesting that a peptide portion of peptidoglycan is involved, either directly or indirectly, in the recognition by integrin βν. In fact, our data suggested that integrin βν does not effectively recognize peptidoglycan of E. coli.
Integrins exist as heterodimers of two trans-membrane proteins named α and β (49–51). There are five α (αPS1, 2, 3, 4, and 5) and two β (βPS and βν) subunits for Drosophila integrins (52, 53). It is unknown at present which of the five α subunits forms a heterodimer with βν and acts as a receptor for the phagocytosis of S. aureus. The other β subunit of Drosophila integrin, βPS, has been shown to be required for the encapsulation by lamellocytes of larvae of the wasp Leptopilina boulardi (54), and the small G protein Rac1 is located downstream of βPS for the induction of encapsulation (55). Rac is thus likely responsible for the induction of a change of cell shape and cell motility so that hemocytes accomplish cellular immune reactions. Integrins of other insect species have been reportedly involved in the phagocytosis of bacteria: larval hemocytes of Ceratitis capitata, a Mediterranean fruit fly, seem to engulf E. coli in vitro in a manner mediated by βPS (56, 57); and the β subunit BINT2 of the mosquito Anopheles gambiae is required for the phagocytosis of E. coli but not S. aureus both in vivo (58) and in vitro (59). These findings indicate that insect integrins in general play roles in the defense against invasive microorganisms.
The three receptors for the phagocytosis of bacteria in Drosophila, namely, Croquemort (27, 28), Draper (29, 30), and integrin βν (31), were originally found as receptors responsible for the phagocytic elimination of apoptotic cells. Another example of the participation of a receptor in the phagocytosis of both apoptotic cells and bacteria comes from mammals. There is a mammalian receptor for phagocytosis, named brain angiogenesis inhibitor 1, which is required for the phagocytic elimination of dying cells in a manner mediated by the recognition of phosphatidylserine exposed at the surface of target cells (60). This receptor was recently reported to be responsible for the phagocytosis of Salmonella typhimurium, a Gram-negative bacterium, by a murine macrophage cell line, recognizing the cell wall component LPS (36). It is therefore possible that some receptors for phagocytosis target both altered self-tissues and invading microorganisms as part of innate immune responses. In any case, it remains to be solved how such receptors recognize apparently distinct structures present at the surface of apoptotic cells and bacteria. Our results indicate that integrin βν recognizes peptidoglycan at the surface of S. aureus. Other phagocytosis receptors of Drosophila such as peptidoglycan recognition protein LC (19) and Eater (24) have been suggested to use peptidoglycan as a ligand for the phagocytosis of bacteria. Taken together, there might exist a multiple recognition system for the effective removal of invading bacteria by phagocytosis in Drosophila; that is, multiple receptors with distinct ligand-specificity in the phagocytosis of a single bacterium as well as multiple receptors with the same ligand-specificity in the phagocytosis of different bacteria species containing the same structure.
Acknowledgments
We thank Kenji Kurokawa, Yasuhiko Matsumoto, and Motoyuki Sugai for the bacterial strains, and Nicholas Brown, Marc Freeman, Pernille Rørth, Bloomington Drosophila Stock Centre, and Kyorin-Fly for the fly lines. Yukichika Tabuchi, Yumi Hashimoto, Yoichi Osada, and Kazuki Takeuchi are thanked for their contribution to this study at the initial stage.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science (Grant Numbers 20570127 and 23570160, to A. S.; 22370049, to Y. N.), Hayashi Memorial Grant for Female Scientists (to A. S.), a grant from Astellas Foundation for Research on Metabolic Disorders (to A. S.), a grant from Danone Foundation (to A. S.), and an institutional research grant from Kanazawa University (to A. S.).

This article contains supplemental Fig. S1.
REFERENCES
- 1. Aderem A., Underhill D. M. (1999) Mechanism of phagocytosis in macrophages. Annu. Rev. Immunol. 17, 593–623 [DOI] [PubMed] [Google Scholar]
- 2. Stuart L. M., Ezekowitz R. A. (2005) Phagocytosis: elegant complexity. Immunity 22, 539–550 [DOI] [PubMed] [Google Scholar]
- 3. Braun A., Hoffmann J. A., Meister M. (1998) Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc. Natl. Acad. Sci. U.S.A. 95, 14337–14342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elrod-Erickson M., Mishra S., Schneider D. (2000) Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 10, 781–784 [DOI] [PubMed] [Google Scholar]
- 5. Haine E. R., Moret Y., Siva-Jothy M. T., Rolff J. (2008) Antimicrobial defense and persistent infection in insects. Science 322, 1257–1259 [DOI] [PubMed] [Google Scholar]
- 6. Nehme N. T., Quintin J., Cho J. H., Lee J., Lafarge M.-C., Kocks C., Ferrandon D. (2011) Relative roles of the cellular and humoral responses in the Drosophila host defense against three Gram-positive bacterial infections. PLoS. ONE. 6, e14743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basset A., Khush R. S., Braun A., Gardan L., Boccard F., Hoffmann J. A., Lemaitre B. (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. U.S.A. 97, 3376–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foley E., O'Farrell P. H. (2003) Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan C. A., Delaney J. R., Schneider D. S., Anderson K. V. (2007) Psidin is required in Drosophila blood cells for both phagocytic degradation and immune activation of the fat body. Curr. Biol. 17, 67–72 [DOI] [PubMed] [Google Scholar]
- 10. Nehme N. T., Liégeois S., Kele B., Giammarinaro P., Pradel E., Hoffmann J. A., Ewbank J. J., Ferrandon D. (2007) A model of bacterial intestinal infections in Drosophila melanogaster. PLoS. Pathog. 3, e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charroux B., Royet J. (2009) Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 9797–9802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Defaye A., Evans I., Crozatier M., Wood W., Lemaitre B., Leulier F. (2009) Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J. Innate Immun. 1, 322–334 [DOI] [PubMed] [Google Scholar]
- 13. Mylonakis E., Aballay A. (2005) Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect. Immun. 73, 3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khush R. S., Lemaitre B. (2000) Genes that fight infection: what the Drosophila genome says about animal immunity. Trends Genet. 16, 442–449 [DOI] [PubMed] [Google Scholar]
- 15. Lemaitre B., Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 16. Strand M. R. (2008) The insect cellular immune response. Insect Sci. 15, 1–14 [Google Scholar]
- 17. Fauvarque M. O., Williams M. J. (2011) Drosophila cellular immunity: a story of migration and adhesion. J. Cell Sci. 124, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 18. Stuart L. M., Ezekowitz R. A. (2008) Phagocytosis and comparative innate immunity: learning on the fly. Nat. Rev. Immunol. 8, 131–141 [DOI] [PubMed] [Google Scholar]
- 19. Rämet M., Manfruelli P., Pearson A., Mathey-Prevot B., Ezekowitz R. A. (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 [DOI] [PubMed] [Google Scholar]
- 20. Philips J. A., Rubin E. J., Perrimon N. (2005) Drosophila RNAi screen reveals CD36 family member required for Mycobacterial infection. Science 309, 1251–1253 [DOI] [PubMed] [Google Scholar]
- 21. Rämet M., Pearson A., Manfruelli P., Li X., Koziel H., Göbel V., Chung E., Krieger M., Ezekowitz R. A. (2001) Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity 15, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 22. Stuart L. M., Deng J., Silver J. M., Takahashi K., Tseng A. A., Hennessy E. J., Ezekowitz R. A., Moore K. J. (2005) Response to Staphylococcus aureus required CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 170, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kocks C., Cho J. H., Nehme N., Ulvila J., Pearson A. M., Meister M., Strom C., Conto S. L., Hetru C., Stuart L. M., Stehle T., Hoffmann J. A., Reichhart J.-M., Ferrandon D., Rämet M., Ezekowitz R. A. B. (2005) Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123, 335–346 [DOI] [PubMed] [Google Scholar]
- 24. Chung Y. S., Kocks C. (2011) Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor Eater. J. Biol. Chem. 286, 26524–26532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurucz E., Márkus R., Zsámboki J., Folkl-Medzihradszky K., Darula Z., Vilmos P., Udvardy A., Krausz I., Lukacsovich T., Gateff E., Zettervall C. J., Hultmark D., Andó I. (2007) Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17, 649–654 [DOI] [PubMed] [Google Scholar]
- 26. Hashimoto Y., Tabuchi Y., Sakurai K., Kutsuna M., Kurokawa K., Awasaki T., Sekimizu K., Nakanishi Y., Shiratsuchi A. (2009) Identification of lipoteichoic acid as a ligand for Draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J. Immunol. 183, 7451–7460 [DOI] [PubMed] [Google Scholar]
- 27. Franc N. C., Dimarcq J. L., Lagueux M., Hoffmann J., Ezekowitz R. A. (1996) Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4, 431–443 [DOI] [PubMed] [Google Scholar]
- 28. Franc N. C., Heitzler P., Ezekowitz R. A. B., White K. (1999) Requirement for Croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284, 1991–1994 [DOI] [PubMed] [Google Scholar]
- 29. Freeman M. R., Delrow J., Kim J., Johnson E., Doe C. Q. (2003) Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38, 567–580 [DOI] [PubMed] [Google Scholar]
- 30. Manaka J., Kuraishi T., Shiratsuchi A., Nakai Y., Higashida H., Henson P., Nakanishi Y. (2004) Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J. Biol. Chem. 279, 48466–48476 [DOI] [PubMed] [Google Scholar]
- 31. Nagaosa K., Okada R., Nonaka S., Takeuchi K., Fujita Y., Miyasaka T., Manaka J., Ando I., Nakanishi Y. (2011) Integrin βν-mediated phagocytosis of apoptotic cells in Drosophila embryos. J. Biol. Chem. 286, 25770–25777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oku Y., Kurokawa K., Matsuo M., Yamada S., Lee B. L., Sekimizu K. (2009) Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J. Bacteriol. 191, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishida S., Kurokawa K., Matsuo M., Sakamoto K., Ueno K., Kita K., Sekimizu K. (2006) Identification and characterization of amino acid residues essential for the active site of UDP-N-acetylenolpyruvylglucosamine reductase (MurB) from Staphylococcus aureus. J. Biol. Chem. 281, 1714–1724 [DOI] [PubMed] [Google Scholar]
- 34. Devenport D., Brown N. H. (2004) Morphogenesis in the absence of integrins: mutation of both Drosophila β subunits prevents midgut migration. Development 131, 5405–5415 [DOI] [PubMed] [Google Scholar]
- 35. Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., Fulga T. A., Rørth P. (2007) Two distinct modes of guidance signaling during collective migration of border cells. Nature 448, 362–365 [DOI] [PubMed] [Google Scholar]
- 36. Das S., Owen K. A., Ly K. T., Park D., Black S. G., Wilson J. M., Sifri C. D., Ravichandran K. S., Ernst P. B., Casanova J. E. (2011) Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avet-Rochex A., Bergeret E., Attree I., Meister M., Fauvarque M.-O. (2005) Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa. Cell. Microbiol. 7, 799–810 [DOI] [PubMed] [Google Scholar]
- 38. Ha H. M., Oh C. T., Bae Y. S., Lee W. J. (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 [DOI] [PubMed] [Google Scholar]
- 39. Park J. W., Kim C. H., Kim J. H., Je B. R., Roh K. B., Kim S. J., Lee H. H., Ryu J. H., Lim J. H., Oh B. H., Lee W. J., Ha N. C., Lee B. L. (2007) Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. U.S.A. 104, 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu Y., Park J. W., Kwon H. M., Hwang H. O., Jang I. H., Masuda A., Kurokawa K., Nakayama H., Lee W. J., Dohmae N., Zhang J., Lee B. L. (2010) Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan. J. Biol. Chem. 285, 32937–32945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lettre G., Hengartner M. O. (2006) Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 7, 97–108 [DOI] [PubMed] [Google Scholar]
- 42. Kinchen J. M., Ravichandran K. S. (2007) Journey to the grave: signaling events regulating removal of apoptotic cells. J. Cell Sci. 120, 2143–2149 [DOI] [PubMed] [Google Scholar]
- 43. Kuraishi T., Nakagawa Y., Nagaosa K., Hashimoto Y., Ishimoto T., Moki T., Fujita Y., Nakayama H., Dohmae N., Shiratsuchi A., Yamamoto N., Ueda K., Yamaguchi M., Awasaki T., Nakanishi Y. (2009) Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J. 28, 3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Awasaki T., Tatsumi R., Takahashi K., Arai K., Nakanishi Y., Ueda R., Ito K. (2006) Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 [DOI] [PubMed] [Google Scholar]
- 45. Avet-Rochex A., Perrin J., Bergeret E., Fauvarque M. O. (2007) Rac2 is a major actor of Drosophila resistance to Pseudomonas aeruginosa acting in phagocytic cells. Genes Cells 12, 1193–1204 [DOI] [PubMed] [Google Scholar]
- 46. Williams M. J., Ando I., Hultmark D. (2005) Drosophila melanogaster Rac2 is necessary for a proper cellular immune response. Genes Cells 10, 813–823 [DOI] [PubMed] [Google Scholar]
- 47. Williams M. J., Wiklund M. L., Wikman S., Hultmark D. (2006) Rac1 signalling in the Drosophila larval cellular immune response. J. Cell Sci. 119, 2015–2024 [DOI] [PubMed] [Google Scholar]
- 48. Côté J. F., Vuori K. (2007) GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giancotti F. G., Ruoslahti E. (1999) Integrin signaling. Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 50. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 51. Caswell P. T., Vadrevu S., Norman J. C. (2009) Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10, 843–853 [DOI] [PubMed] [Google Scholar]
- 52. Brown N. H., Gregory S. L., Martin-Bermudo M. D. (2000) Integrins as mediators of morphogenesis in Drosophila. Dev. Biol. 223, 1–16 [DOI] [PubMed] [Google Scholar]
- 53. Brower D. L. (2003) Platelets with wings: the maturation of Drosophila integrin biology. Curr. Opin. Cell Biol. 15, 607–613 [DOI] [PubMed] [Google Scholar]
- 54. Irving P., Ubeda J. M., Doucet D., Troxler L., Lagueux M., Zachary D., Hoffmann J. A., Hetru C., Meister M. (2005) New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 7, 335–350 [DOI] [PubMed] [Google Scholar]
- 55. Xavier M. J., Williams M. J. (2011) The Rho-family GTPase Rac1 regulates integrin localization in Drosophila immunosurveillance cells. PLoS. ONE. 6, e19504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Foukas L. C., Katsoulas H. L., Paraskevopoulou N., Metheniti A., Lambropoulou M., Marmaras V. J. (1998) Phagocytosis of Escherichia coli by insect hemocytes requires both activation of the Ras/mitogen-activated protein kinase signal transduction pathway for attachment and β3 integrin for internalization. J. Biol. Chem. 273, 14813–14818 [DOI] [PubMed] [Google Scholar]
- 57. Mamali I., Lamprou I., Karagiannis F., Karakantza M., Lampropoulou M., Marmaras V. J. (2009) A β integrin subunit regulates bacterial phagocytosis in medfly haemocytes. Dev. Comp. Immunol. 33, 858–866 [DOI] [PubMed] [Google Scholar]
- 58. Moita L. F., Wang-Sattler R., Michel K., Zimmermann T., Blandin S., Levashina E. A., Kafatos F. C. (2005) In vitro identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity 23, 65–73 [DOI] [PubMed] [Google Scholar]
- 59. Moita L. F., Vriend G., Mahairaki V., Louis C., Kafatos F. C. (2006) Integrins of Anopheles gambiae and a putative role of a new β integrin, BINT2, in phagocytosis of E. coli. Insect Biochem. Mol. Biol. 36, 282–290 [DOI] [PubMed] [Google Scholar]
- 60. Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., Ravichandran K. S. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]