Background: MicroRNAs (miRNAs) are post-transcriptional regulators involved in the regulation of gene expression.
Results: miR-449 and miR-34b/c function redundantly in male germ cells.
Conclusion: CREMτ-SOX5-mediated miR-449 expression regulates male germ cell development by targeting the E2F-pRb pathway.
Significance: Upstream regulators of miR-449 expression and a redundant role between miR-449 and miR-34b/c in the control of male germ cell development were revealed.
Keywords: Cell Cycle, Gene Knock-out, Gene Regulation, MicroRNA, Spermatogenesis, miR-449
Abstract
MicroRNAs (miRNAs) mainly function as post-transcriptional regulators and are involved in a wide range of physiological and pathophysiological processes such as cell proliferation, differentiation, apoptosis, and tumorigenesis. Mouse testes express a large number of miRNAs. However, the physiological roles of these testicular miRNAs remain largely unknown. Using microarray and quantitative real time PCR assays, we identified that miRNAs of the microRNA-449 (miR-449) cluster were preferentially expressed in the mouse testis, and their levels were drastically up-regulated upon meiotic initiation during testicular development and in adult spermatogenesis. The expression pattern of the miR-449 cluster resembled that of microRNA-34b/c (miR-34b/c) during spermatogenesis. Further analyses identified that cAMP-responsive element modulator τ and SOX5, two transcription factors essential for regulating male germ cell gene expression, acted as the upstream transactivators to stimulate the expression of the miR-449 cluster in mouse testes. Despite its abundant expression in testicular germ cells, miR-449-null male mice developed normally and exhibited normal spermatogenesis and fertility. Our data further demonstrated that miR-449 shared a cohort of target genes that belong to the E2F transcription factor-retinoblastoma protein pathway with the miR-34 family, and levels of miR-34b/c were significantly up-regulated in miR-449-null testes. Taken together, our data suggest that the miR-449 cluster and miR-34b/c function redundantly in the regulation of male germ cell development in murine testes.
Introduction
Spermatogenesis is a complex process through which male germ line stem cells differentiate into male gametes called spermatozoa. Spermatogenesis can be divided into three phases based upon the major cellular events occurring during this process: mitotic (spermatogonial proliferation and differentiation), meiotic (chromosomal reduction in spermatocytes through meiosis), and haploid (spermatid differentiation into spermatozoa) (1). These cellular events require precise spatiotemporal expression of specific protein-coding genes, which are tightly controlled at both the transcriptional and post-transcriptional levels (2, 3). One example is the initiation of the meiotic phase of spermatogenesis, which requires male germ cells to exit from the mitotic cell cycle and the timely expression of meiotic genes (4). During the subsequent postmeiotic development, translation of a large number of mRNAs is uncoupled with their transcription due to an earlier transcriptional cessation occurring at step 9 spermatids in mouse testes (2). Translational repression of mRNAs that are transcribed in spermatocytes and/or early spermatids (steps 1–8) must be achieved through a post-transcriptional regulatory mechanism, which still remains elusive. MicroRNAs (miRNAs),4 a class of short (20–25-nucleotide) non-coding RNA, are well documented as critical post-transcriptional regulators of gene expression by inducing target mRNA degradation (perfect complementarity) and translational repression (incomplete match) (3). Primary miRNA transcripts are transcribed from the host genome by RNA polymerase II and are further processed by two RNase III enzymes, DROSHA and DICER, into mature miRNAs, which are eventually incorporated into an RNA-induced silencing complex to exert inhibitory effects on their target mRNAs (5). The miR-449 cluster was first identified in embryonic mouse brain (6) and whole mouse embryos (7) by comparative genomic prediction and small RNA cloning through sequencing (8, 9). The miR-449 cluster consists of three members, miR-449a, miR-449b, and miR-449c, in mice and humans. They are mapped to the second intron of Cdc20b gene, thus forming an miRNA cluster, and they are highly conserved among different species. Given that the three miR-499 members are transcribed simultaneously and their expression and seed sequences are all the same, for simplicity, we collectively called them miR-449 from here on. In human and Xenopus lung tissues, miR-449 members are able to induce epithelial differentiation by directly repressing the Delta/Notch pathway, whereas their depletion leads to defects in pulmonary epidermis differentiation, and are thus suggested to direct cell cycle exit and epidermis differentiation (10). In addition, up-regulation of miR-449 has also been reported during myoblast differentiation (11) and in the epithelial cell layer of choroid plexus in the brain (12). Recently, in an assay screening for E2F1-responsive miRNAs, miR-449 was shown to be robustly up-regulated and, in turn, attenuated E2F1 activity by inhibiting positive cell cycle regulators such as CDK6 and CDC25A, leading to cell cycle arrest and apoptosis (13). Interestingly, another conserved miRNA family, miR-34, which consists of miR-34a, miR-34b, miR-34c, has been demonstrated to structurally resemble those of the miR-449 members. Coincidentally, miR-34c has been shown previously to be highly enriched in germ cells and enhance germinal phenotypes (14). Despite similarities in their sequences and involvement in germ cell development, miR-449 and miR-34 families appear to have distinct functions and may well be under differential regulation because previous studies have shown that E2F1 can provoke the activity of miR-449 members in a p53-independent manner, whereas p53 can up-regulate the expression of miR-34 family members (15, 16).
In the present study, we report that miR-449 is preferentially expressed in the murine testis with the highest levels in meiotic (spermatocytes) and postmeiotic (spermatids) male germ cells and that testes-specific transcription factors cAMP-responsive element modulator τ (CREMτ) and SOX5 mediate meiotic and postmeiotic expression of miR-449 by binding to two highly conserved cis-elements of the Cdc20b/miR-449 cluster. Although no discernible phenotype was observed in miR-449 cluster knock-out (KO) mice, our data suggest that miR-34b/c could compensate for the absence of miR-449, and both miRNA families function redundantly by targeting the E2F-pRb pathway.
EXPERIMENTAL PROCEDURES
Animals
miR-449 cluster KO mice were generated, bred, and housed in the Shanghai Research Center for Model Organisms. Wild type C57/BL6 mice were purchased from Shanghai SLAC Laboratory Animal Corp. (Shanghai, China) and housed in a temperature- and humidity-controlled room with a 12-h light-dark cycle at Shanghai Jiao Tong University School of Medicine. All animal procedures were approved by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University School of Medicine.
Microarray Analyses
Total RNA was extracted from snap frozen testis samples using TRIzol reagent (Invitrogen) as described previously (17). For small RNA isolation, the mirVana miRNA Isolation kit (Ambion) was used to enrich RNA fractions shorter than 200 nucleotides. miRNA microarray analyses on postnatal day 7 (P7) and P60 testis samples were performed at CapitalBio Corp. (Beijing, China) according to the manufacturer's procedures (17). Each group of small RNA samples was labeled with Cy3 or Cy5 fluorescent dyes, respectively, followed by hybridization on CapitalBio Corp. miRNA arrays in which an individual oligonucleotide probe was coupled on chemically modified glass slides in triplicate. Raw data of digital signals from the scanned fluorescence readings were normalized and analyzed using the significance analysis of microarrays (SAM) software (Stanford University, Stanford, CA) to determine the significantly differential expression of miRNAs between P7 and P60 testes (-fold change >2). To identify genes differentially expressed between wild type and miR-449−/− mouse testes, Illumina Mouse WG-6 v2.0 Expression BeadChip microarray analyses were performed following the manufacturer's protocols. Briefly, the RNA samples extracted from both wild type and miR-449−/− testes were first amplified using the Illumina TotalPrep RNA Amplification kit. Microarray hybridization, scanning, and analyses were performed based on Illumina's instructions. Data were then background-corrected and normalized to internal control prior to statistical analyses performed by the “limma” package software.
Cell Culture, Transfection, and Cell Proliferation
GC-1 (ATCC catalog number CRL-2053) and GC-2 (ATCC catalog number CRL-2196) cell lines were purchased from ATCC. All GC-1, GC-2, HEK293, and HeLa cell lines were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) under 5% CO2 at 37 °C. For luciferase reporter assay, HEK293 cells were transiently transfected with miR-449 precursor (pre-miR-449) mimics with putative wild type or mutated 3′-UTR constructs using Lipofectamine 2000 as described under “Luciferase Reporter Assay.” For cell proliferation assays, GC-2 cells were transfected with pre-miR-449 (50 nm) (Ambion) in 96-well plates using Lipofectamine 2000, whereas a scrambled miRNA precursor (Ambion/Applied Biosystems) was used as a negative control. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were performed at 0, 24, 48, and 72 h post-transfection, respectively, to determine the relative number of surviving cells following the manufacture's protocol. siCREMτ and siSOX5 were synthesized in GeneChem (Shanghai, China) and transfected into GC-2 cells according to the manufacturer's protocols. All experiments were performed in biological triplicates.
Histology and Immunohistochemistry
Testes from wild type, miR-449+/−, and miR-449−/− mice were collected and fixed in Bouin's fixative for >12 h followed by embedding in paraffin. Paraffin sections (5-μm thickness) were further processed for eosin solution staining followed by hematoxylin counterstaining. Immunohistochemistry was carried out as described previously (18).
miRNA in Situ Hybridization
In situ hybridization was performed as described previously (19) with minor modifications. Briefly, cryosections (10 μm) were hybridized with digoxigenin-labeled locked nucleic acid-modified oligonucleotide probes specific for mmu-miR-449a and mmu-miR-34a (Exiqon; 15 nm) or scrambled control probes (Exiqon; 15 nm) at 53 °C overnight. Following sequential washing, biotinylated anti-digoxigenin antibody (Boster, China) was applied, and subsequently alkaline phosphatase-conjugated streptavidin complex was added for incubation at 37 °C. Hybridization signals were finally visualized with Fast Red Substrate (Dako) through an alkaline phosphatase-mediated color reaction, whereas the cell nuclei were counterstained by DAPI (Vector Laboratories).
RT-PCR and miRNA Real Time PCR
All RNA extraction procedures were carried out using TRIzol reagent (Invitrogen) as described (19). A two-step method was exploited for general reverse transcription-PCR. For miRNA-specific real time PCR, TaqMan® MicroRNA Assay kits (Applied Biosystems) were used as follows: mmu-miR-449a, Assay ID 001030; mmu-miR-449b, Assay ID 001667; mmu-miR-34a, Assay ID 000426; mmu-miR-34b, Assay ID 001065; mmu-miR-34c, Assay ID 000428; mmu-miR-184, Assay ID 000485; mmu-miR-204, Assay ID 000508; mmu-miR-127, Assay ID 000452; mmu-miR-154, Assay ID 000477; mmu-miR-376a, Assay ID 001069; mmu-miR-376b, Assay ID 001070; mmu-miR-379, Assay ID 001138; mmu-miR-411, Assay ID 001075; and RNU6B control, Assay ID 001093. All quantitative real time PCR experiments were carried out according to the manufacturer's instructions. Briefly, for each miRNA assay, total RNA was first reverse transcribed into cDNA using TaqMan MicroRNA Reverse Transcription kit (part number 4366596) in a 15-μl reaction volume at 42 °C followed by miRNA-specific PCR amplification in a total reaction volume of 20 μl in an Applied Biosystems 7500 PCR System. Small nucleolar RNU6B was amplified in parallel for normalization. All PCRs were performed in triplicate. Data were normalized against RNU6B to determine the relative -fold changes by the Fast 7500 Real-Time System software.
Luciferase Reporter Assay
For miRNA target validation, the 3′-UTRs of Ccnd1, Bcl2, E2f2, E2f3, and Myc were PCR-amplified using mouse genomic DNA and cloned into the XbaI-FseI site downstream of pGL3.0 control reporter vector (Promega), respectively. Constructs containing mutated miRNA binding sites (seed sequence) in 3′-UTRs were generated using the QuikChange site-directed mutagenesis kit (Stratagene). HEK293 cells were cultured in 24-well plates in DMEM supplemented with 10% FBS at 37 °C. For each miRNA target assay, HEK293 cells were co-transfected with wild type 3′-UTR or mutated 3′-UTR constructs (200 ng/well) and pre-miR-449a mimics (20 nm) using Lipofectamine 2000 reagent in a 24-well plate (Invitrogen). pRL-TK (Promega; 50 ng/well) Renilla luciferase was transfected for normalization of transfection efficiency. Cells transfected with scrambled miRNA (a synthesized RNA duplex showing no homology to any murine mRNA sequence) served as negative controls. At 48 h post-transfection, cell lysates were analyzed for luciferase activity using the Dual-Glo luciferase assay kit (Promega) according to the manufacturer's protocol. Data were normalized against values of co-transfected Renilla luciferase. All experiments were performed in triplicate.
For Cdc20b promoter mapping analyses, PCR fragments of various lengths around the transcription start site (−4 to +0.2 kb) obtained by 5′ rapid amplification of cDNA ends as well as mutational fragments generated by the QuikChange site-directed mutagenesis kit (Stratagene) deficient in the core sequence of CREMτ and SOX5 binding sites were cloned into pGL3.0-Basic vector (Promega). The full-length open reading frames (ORFs) of mouse CREMτ and SOX5 mRNAs were inserted into pFLAG-CMV-4.0 vector in-frame to allow CREMτ and SOX5 protein expression, respectively. The fidelity of nucleotides for all constructs was validated by sequencing. GC-2 cells derived from mouse spermatocytes were cultured in DMEM supplemented with 10% FBS and transfected with constructs as well as an empty pGL3.0 control vector as described above. pRL-TK Renilla luciferase was used for normalization against transfection efficiency. The PCR primers used are listed in supplemental Table S1.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation was carried out using a ChIP Assay kit (Beyotime, catalog number P2078) according to manufacturer's instructions. Briefly, after preparation of single cell suspensions of adult murine testes using 100 μg/ml collagenase in DMEM/F-12 medium, single cells were fixed using 1% formaldehyde for 10 min at 37 °C followed by neutralization using 125 mm glycine. The cells were then lysed on ice in SDS lysis buffer (50 mm Tris-HCl, pH8.0, 10 mm EDTA, 150 mm NaCl, 1% SDS) supplemented with 1× proteinase inhibitor mixture (Roche Applied Science). Then the lysate was sonicated on ice (10-s pulse, 50-s break, six times, 20% amplitude; Sonics) to break the genome into 200–1000 bp in size. 2 μg of primary antibody (α-CREMτ and α-SOX5; Millipore) was added to 1 ml of Protein G-precleared lysate to allow precipitation of CREMτ- or SOX5-associated genomic DNA fragments by incubation at 4 °C overnight followed by washing with different washing buffers as follows: Low Salt Immune Complex Wash Buffer, one time; High Salt Immune Complex Wash Buffer, one time; LiCl Immune Complex Wash Buffer, one time; and Tris-EDTA Buffer, two times. The DNA components in the precipitation were finally extracted using a standard phenol-chloroform method.
Generation of miR-449 Cluster Knock-out Mice
An 11.7-kb genomic DNA fragment encompassing Cdc20b gene and miR-449 cluster was cloned into pBR322 vector. The PGK-Neo cassette flanked by a short loxp sequence was inserted to replace a ∼1.7-kb genomic fragment of the second intron of Cdc20b gene where the miR-449 cluster is localized. After validation by sequencing and linearization by NotI, the targeting vector was then electroporated into AB2.1 embryonic stem (ES) cells, which are derived from 129S7/SvEv strain mice. ES cells were cultured in a 96-well plate in M15 medium supplemented with G418 antibiotics. Among ∼200 clones screened for homologous recombination, five positive ES clones were identified by a long range PCR assay. Chimeras were generated by standard microinjection of C57BL/6J blastocysts with two positive miR-449 cluster-targeted ES clones, and the targeted ES cell incorporation was monitored by the ratio of agouti/black coat color marker. Male chimeras were then bred with wild type C57BL/6J females to generate heterozygous miR-449+/PGK-Neo mice. To delete the PGK-Neo cassette, miR-449+/PGK-Neo males were further crossed with a transgenic mouse line with ubiquitous expression of Flippase (The Jackson Laboratory, stock number 009086) to delete the PGK-Neo cassette flanked by two loxp sequences. The resultant heterozygous mutant mice (miR-449+/−) were intercrossed to produce homozygous null (miR-449−/−) mice. Mouse tail DNA was used for PCR genotyping. PCR primers are listed in supplemental Table S1.
Fertility Test and Computer-assisted Sperm Analysis
Adult miR-449+/− and miR-449−/− male mice were individually caged with C57BL/6 wild type females for 3 months. All males were on a C57BL/6:129S7/SvEv hybrid background. The number of litters and total pups born were recorded. Epididymal sperm from miR-449−/− and age-matched wild type (control) males were prepared for computer-assisted semen analysis as described previously (20). Briefly, the epididymis was dissected, and sperm inside were squeezed out with forceps. After incubation in human tubal fluid medium (500 μl per epididymis) at 37 °C for 30 min, sperm were subject to motility analyses using a Sperm Quality Analyzer (Hamilton Thorne graphic series, G70f). For each measurement, a suspension of spermatozoa was loaded into a microchamber slide with 100-μm depth (HTR1099, VitroCom Inc.). ∼300 spermatozoa were analyzed using the standard setting (30 frames at 60-Hz frequency and 37 °C) for the percentage of motility as well as progressive motility.
Bioinformatics
miRNA sequences were retrieved from miRBase (Release 17), and targets prediction was performed using TargetScan (Release 5.2) and miRanda (Release 2010). Conservation of the sequence was verified using UCSC Genome Browser. Sequence alignment was examined using Invitrogen Vector NTI Advance software and NCBI BLAST. Putative transcription factor binding sites were identified using TFsearch online tool and the Genomatix suite program (MatInspector).
Statistical Analysis
To globally evaluate the mRNA transcriptomic changes induced by the transfection of miRNAs (miR-449a/b and miR-34b/c), we tested the null hypothesis that the expression changes of mRNAs (possessing the predicted miRNA seed match) were equal to the distribution of mRNAs without predicted miRNA binding motifs using the non-parametric Wilcoxon rank sum test as described previously (21). Experiments were performed in triplicate, and data are represented as mean ± S.E. (n = 3). The results were analyzed by analysis of variance or Student's t test as appropriate. A difference with a p value less than 0.05 was considered as significant. *, p < 0.05; **, p < 0.01.
RESULTS
miR-449 Is Significantly Up-regulated with Progression of Postnatal Testicular Development
During postnatal testicular development in mice, spermatogonia represent the major male germ cell type at P7, whereas all three types of male germ cells, spermatogonia, spermatocytes, and spermatids, are present at P60. To unveil miRNAs that are significantly up-regulated in spermatocytes and spermatids, we conducted miRNA microarray analyses using P7 and P60 total testes. A total of 72 and 47 miRNAs were significantly enriched in P60 and P7 testes, respectively (supplemental Fig. S1A and Tables S3 and S4). To validate the microarray data, we performed quantitative real time PCR (qPCR) assays to determine expression levels of the top 12 up- or down-regulated miRNAs in P7 and P60 mouse testes. Consistent with our microarray data, similar changes were observed in qPCR analyses (supplemental Fig. S1B). These data are in agreement with several recent reports (17, 22–24) showing much higher levels of miR-449 in adult than in prepubertal mouse testes. Among miRNAs highly enriched in P60 compared with P7, miR-449 displayed the most drastic up-regulation with 67- and 253-fold increases by microarray and qPCR assays, respectively (supplemental Fig. S1B). Interestingly, we found that miR-34b/c, unlike miR-34a, were also significantly up-regulated with similar -fold changes in P60 testes (supplemental Fig. S1B), and this expression pattern is consistent with previous reports (14, 23). miR-184, which we have previously demonstrated to target nuclear receptor corepressor 2 in mouse testicular germ cells (19), also showed an apparent up-regulation in P60 testes.
miR-449 Is Preferentially Expressed in Mouse Testes and Localized to Spermatocytes and Spermatids
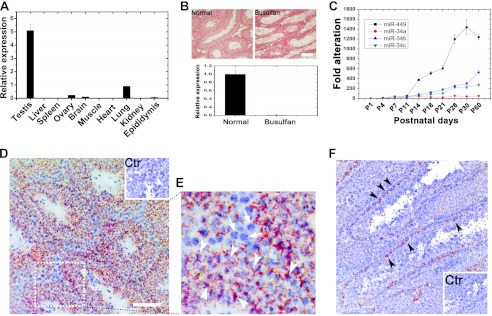
To further decipher the expression profile of miR-449 in mice, we next examined miR-449 expression in multiple mouse tissues and developing testes. The highest levels of miR-449 were detected in the testis, and yet notable levels were observed in lung, brain, ovary, and epididymis (Fig. 1A). The tissue distribution data are consistent with several recent studies (10, 25–28) suggesting that miR449 is preferentially expressed in the testis in mice.
FIGURE 1.
Expression profiles of miR-449 cluster and miR-34 family in mice. A, qPCR analyses of expression levels of miR-449 in 11 tissues collected from adult male mice. B, germ cell depletion through busulfan treatment followed by qPCR detection of miR-449. miR-449 was abundant in control (untreated wild type) testes, whereas miR-449 was undetectable in busulfan-treated (germ cell-depleted) testes, suggesting that miR-449 is exclusively expressed in germ cells. Scale bar, 50 μm. C, qPCR analyses of expression levels of miR-449 and members of the miR-34 family during postnatal testicular development. D, localization of miR-449 in the mouse testis using in situ hybridization. Hybridization signals are in red (Texas Red), and the nuclei were counterstained with hematoxylin (blue). The inset is a control. Scale bar, 70 μm. E, an enlarged image of the field framed be the dashed box in D. Specific miR-449a hybridization signals were mainly detectable in the cytoplasm of spermatocytes and spermatids (arrowheads), whereas no signals were fund in somatic cell types and spermatogonia (arrows). Scale bar, 50 μm. Ctr, control. F, localization of miR-34a in the murine testis by in situ hybridization. Hybridization signals (red) were only detected in spermatogonia. Cell nuclei (blue) were counterstained with hematoxylin (blue). The inset is a control. Scale bar, 50 μm. Data are represented as mean ± S.E. (n = 3).
Because the testis comprises both somatic cell types (e.g. Sertoli cells, Leydig cells, and peritubular myoid cells) and male germ cells at various developmental stages, we asked whether miR-449 exhibited cell type-specific expression in mouse testes. We selectively depleted testicular germ cells by treating mice with busulfan for 1 month as reported previously (29). We then performed qPCR to examine expression levels of miR-449 in normal and busulfan-treated (i.e. germ cell-depleted) testes (Fig. 1B). miR-449 was barely detectable in busulfan-treated testes, indicating that miR-449 is exclusively expressed in male germ cells in the testis (Fig. 1B). qPCR analyses using developing testes demonstrated that levels of miR-449 along with those of miR-34b and miR-34c were increasingly up-regulated with the progression of testicular development, whereas miR-34a levels were relatively constant throughout the testicular development (Fig. 1C). Levels of miR-449 sharply increased at P11, corresponding to the period of meiotic initiation (Fig. 1C). To determine the germ cell types that express miR-449, we performed in situ hybridization using testicular cryosections. Hybridization signals specific for miR-449 were detected mainly in pachytene spermatocytes and spermatids (Fig. 1, D and E, arrowheads), whereas no signals were detectable in somatic cell types (i.e. Sertoli and Leydig cells) (Fig. 1, D and E, arrows). We did not perform in situ hybridization assays for miR-34b/c because these two miRNAs have been previously shown to localize mainly to spermatocytes and spermatids (14). Given that miR-34a belongs to the miR-34 family, we examined miR-34a localization (Fig. 1F) in the testis. miR-34a appeared to be restricted to spermatogonia residing on the basal membrane of seminiferous tubules (Fig. 1F, arrowheads). This localization pattern of miR-34a is consistent with our qPCR results (Fig. 1C) because its levels showed minimal variation due to the constantly low number of spermatogonia in developing testes. The differential expression profiles and localization patterns between miR-34a and miR-34b/c are not surprising because although they possess similar sequences and thus constitute a miRNA family the miR-34a locus is far away from the miR-34b/c locus, and thus they may be under different regulation (30). Taken together, our data demonstrate that the up-regulated expression of miR-449 during testicular development coincides with meiotic initiation, and miR-449 is predominantly expressed in spermatocytes and spermatids during adult spermatogenesis. miR-34a is expressed in spermatogonia (Fig. 1), whereas miR-34b and miR-34c exhibit expression profiles similar to that of miR-449 during postnatal testicular development and spermatogenesis (14).
CREMτ and SOX5 Modulate Activity of miR-449 Cluster in Mouse Testes
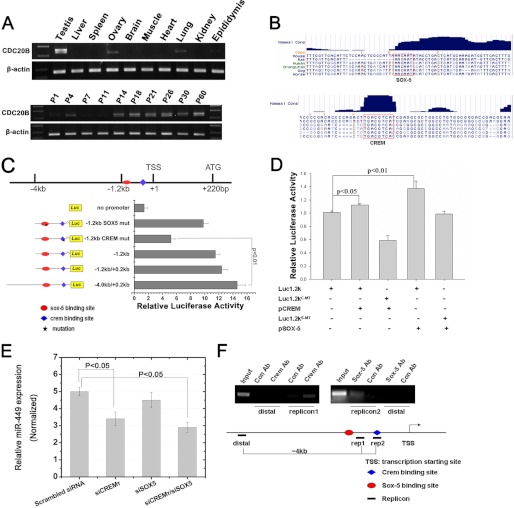
Recent studies have demonstrated that E2F1, a critical cell cycle regulator, directly activates the expression of miR-449a/b, which can subsequently impede phosphorylation of pRb through inhibiting CDC25A and CDK6, providing a negative feedback circuit involved in the E2F-pRb pathway (13, 27, 31). However, this pathway may not operate in the mouse testis because earlier studies have shown that although E2F1 mRNA is present throughout all spermatogenic cells in mouse testes E2F1 protein is solely present in spermatogonia due to the translational repression by the miR-17–92 cluster (32, 33). Given that miR-449 is mainly expressed in spermatocytes and spermatids (Fig. 2), it is highly unlikely that E2F1 is responsible for the regulation of miR-449 expression in mouse testes. To unveil upstream regulators responsible for the expression of the miR-449 cluster, we next conducted deletion mapping and ChIP assays to search for the conserved cis-elements located upstream of the miR-449 cluster and the potential transcriptional factors. The miR-449 cluster resides in the second intron of Cdc20b gene. Our qPCR analyses indicated that Cdc20b exhibited an expression profile similar to that of miR-449 in mouse multiple tissues and developing testes (Fig. 2A), suggesting a common mechanism of transcriptional regulation adopted by both Cdc20b and the miR-449 cluster in mouse testes that has been suggested in several previous reports (10, 25, 31). In silico analyses by scanning 4 kb upstream and 220 bp downstream of the transcription start site identified two highly conserved binding sites for CREMτ and SOX5, which were located at −50 and −1200 bp, respectively (Fig. 2, B and C). The transcription start site of Cdc20b gene as determined by 5′ rapid amplification of cDNA 5′ ends was located 220 bp upstream of its translational start site (Fig. 2C). To further define the precise promoter region, various luciferase reporter plasmids containing deletion mutants were constructed (Fig. 2C), and the luciferase reporter activity was analyzed in GC-2 cells. The minimal −1.2 kb fragment construct could activate the expression of the luciferase reporter as efficiently as the −4.0/+0.2 kb full-length luciferase construct (Fig. 2C). However, mutations in the putative CREMτ binding site led to dramatically reduced activity of the luciferase reporter in comparison with that of mutations in the putative SOX5 binding site (Fig. 2C). These data suggest that CREMτ rather than SOX5 modulates Cdc20b/miR-449 cluster activity in GC-2 cells. However, considering that the origin of the GC-2 cell line was spermatocytes in which SOX5 is not expressed (34), we further examined the activity of the −1.2 kb fragment constructs in the presence of ectopic expression of CREMτ or SOX5 plasmids. As shown in Fig. 2D, both CREMτ and SOX5 were able to efficiently stimulate luciferase activity individually in comparison with CREMτ/SOX5 binding site-mutated luciferase constructs, supporting the notion that both CREMτ and SOX5 cooperatively mediate the activity of the polycistronic Cdc20b/miR-449 gene. In addition, we knocked down mRNAs for CREMτ and SOX5 using two synthesized specific siRNAs in GC-2 cells. As shown in Fig. 2E, either individual or simultaneous knockdown of CREMτ and SOX5 could efficiently reduce miR-449 activity (Fig. 2E), whereas there was no significant effect in the case of individual knockdown of SOX5 (Fig. 2E), which was consistent with results described above (Fig. 2C). The lack of effects when only SOX5 was suppressed may be due to the CREMτ activity.
FIGURE 2.
CREMτ and SOX5 transactivate miR-449 expression by binding two conserved cis-elements upstream of Cdc20b. A, RT-PCR analyses of Cdc20b expression in multiple adult mouse tissues (upper panel) and developing mouse testes (lower panel). B, schematic representation of putative conserved CREMτ and SOX5 binding sites (highlighted with red boxes) in the promoter region of Cdc20b. C, mapping of the upstream transcriptional regulatory region of Cdc20b/miR-449 cluster by luciferase reporter assays. Luciferase plasmids containing a partial deletion or mutation in CREMτ and SOX5 binding sites were constructed as indicated. Normalized luciferase activity is represented as mean ± S.E. (n = 3). All groups were performed in triplicate. D, co-transfection luciferase reporter assays using the GC-2 cell line identified that both CREMτ (p < 0.05) and SOX5 (p < 0.01) proteins can efficiently activate the −1.2 kb-containing reporter construct. Various combinations of plasmids transfected into GC-2 cells are indicated below each group. Luc1.2kC-MT, luciferase construct containing a mutation in the candidate CREMτ binding site; Luc1.2kS-MT, luciferase reporter containing a mutation in the candidate SOX5 binding site. E, knockdown of CREMτ individually or knockdown of CREMτ and SOX5 simultaneously using siRNAs led to decreased levels of miR-449 in GC-2 cells (n = 3; p < 0.05). siCREMτ, siRNA designed against CREMτ mRNA; siSOX5, siRNA designed against SOX5. Small nucleolar RNU6B was concurrently amplified for data normalization. F, ChIP assays confirmed the binding of both CREMτ and SOX5 to the −1.2 kb promoter region upstream of Cdc20b gene. rep1, rep2, and distal represent three different PCR fragments designed against the genomic region as indicated. Luc, luciferase; mut, mutant; TSS, transcription start site; Ab, antibody; Con, control. Data are represented as mean ± S.E. (n = 3).
To further validate these findings, we performed ChIP-PCR assays using adult mouse testes. Three specific pairs of PCR primers were designed corresponding to three regions designated as rep1, rep2, and distal (Fig. 2F). Both antibodies for CREMτ and SOX5 were demonstrated to be specific by reabsorption assays (data not shown). As shown in Fig. 2F, both antibodies appeared to have pulled down corresponding genomic sequence within −1.2 kb, whereas PCR signals were undetectable using primers against distal genomic sequence (Fig. 2F), suggesting that a direct CREMτ/SOX5 binding site is located within the −1.2 kb genomic region in mouse testes in vivo. Furthermore, a full-length reporter plasmid (p1.2k-DsRed) containing the −1.2 kb cis-element was electroporated into seminiferous tubules of the mouse testes along with an empty pDsRed reporter control. At 36 h postelectroporation, obvious red fluorescence was observed in the testicular cryosections transfected with p1.2k-DsRed under fluorescent microscopy, whereas no fluorescence was detected in the pDsRed control (supplemental Fig. S2), suggesting a strong promoter activity of the −1.2 kb fragment in vivo. Taken together, these data show that in mouse testes CREMτ and SOX5 cooperatively mediate Cdc20b/miR-449 cluster activity during postnatal testicular development.
Generation and Characterization of miR-449 Cluster Knock-out Mice
To investigate the physiological role of miR-449 in vivo, we generated a global knock-out mouse line with a complete deletion of the miR-449 cluster (supplemental Fig. S3). To minimize the risk of interfering with the transcription and processing of its host Cdc20b gene, the targeting construct was designed to replace a small portion of the miR-449 cluster locus (∼1.7 kb) with a PGK-Neo cassette flanked by FRT sites (supplemental Fig. S3). This strategy allowed the subsequent deletion of the PGK-Neo cassette by breeding the miR-449+/PGK-Neo offspring with Flippase-expressing transgenic mice (35, 36), thus further generating the miR-449-null allele (supplemental Fig. S3). PCR analyses demonstrated that both alleles of miR-449 were deleted in the miR-449−/− mice (supplemental Fig. S3), and the presence of Cdc20b mRNA transcript in both miR-449−/− and wild type (WT) testes indicates that transcription of Cdc20b gene was not affected by the deletion of the miR-449 cluster (supplemental Fig. S3). Therefore, we successfully generated a universal miR-449 cluster KO mouse line.
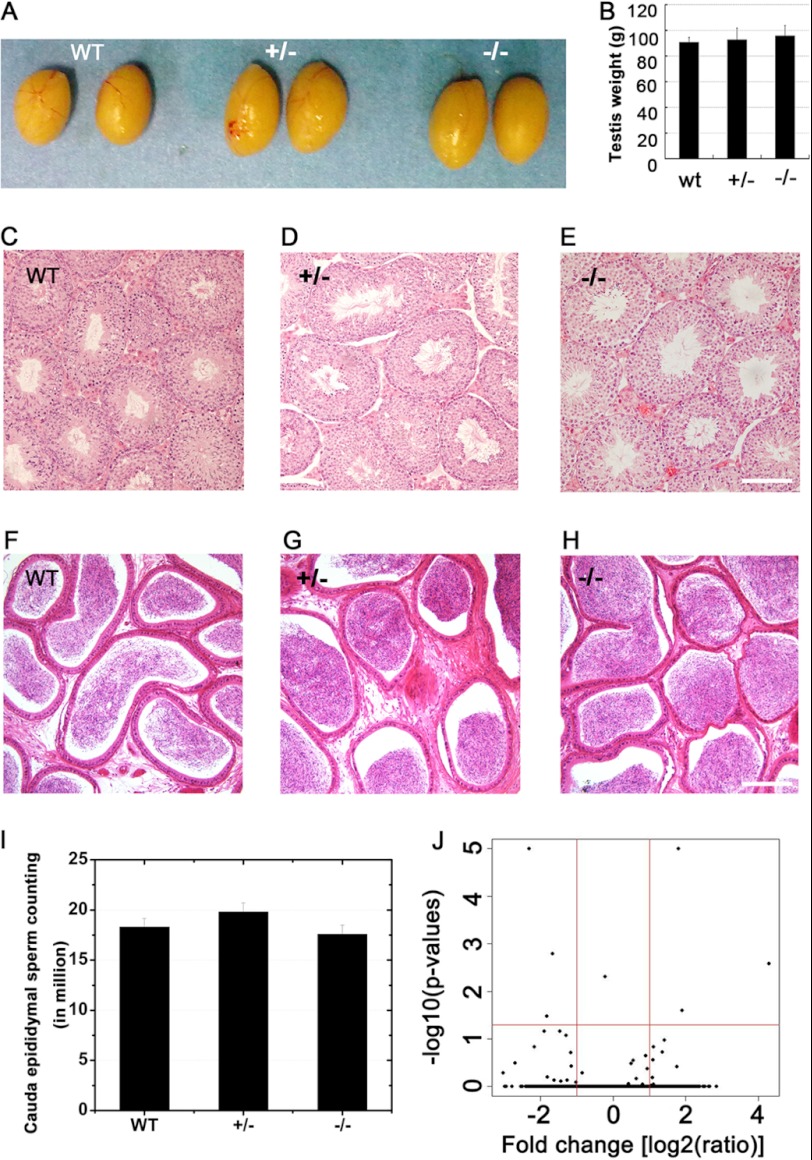
Male miR-449−/− pups developed normally without gross defects in any organs. Their testes also displayed the same size and weight as the heterozygous and wild type littermates (Fig. 3, A and B). Histological examination of the testes indicated normal spermatogenesis in miR-449−/− male mice (Fig. 3, C, D, and E), and abundant sperm were also present in miR-449−/− cauda epididymides (Fig. 3, F, G, and H). Furthermore, computer-assisted sperm analysis-based motility assays failed to find differences in sperm between KO mice and wild type controls (data not shown), and sperm counts were also comparable among the three genotypes (Fig. 3I). Terminal deoxynucleotidyltransferase (TdT)-mediated dUTP nick end labeling (TUNEL) assays revealed no difference in the number of apoptotic germ cells between miR-449−/− and wild type testes (data not shown).
FIGURE 3.
Lack of testicular phenotype in miR-449−/− mice. A, gross morphology of testes from adult male WT, miR-449+/−, and miR-449−/− mice. B, average testicular weight of adult WT, miR-449+/−, and miR-449−/− mice. C–E, hematoxylin-eosin-stained testicular sections of WT (C), miR-449+/− (D), and miR-449−/− (E) mice. Scale bar, 70 μm. F–H, hematoxylin-eosin-stained cauda epididymal sections of WT (C), miR-449+/− (D), and miR-449−/− (E) mice. Scale bar, 100 μm. I, cauda epididymal sperm counts in WT, miR-449+/−, and miR-449−/− male mice. Data are represented as mean ± S.E. (n = 3). J, volcano plot showing differentially expressed mRNAs between miR-449 KO and wild type testes determined by Illumina microarray analyses. The x axis represents -fold changes (KO versus WT) in log2 scale (red vertical line marks the 2-fold change). The y axis shows p value significance for each gene-specific analysis of variance in the −log10 scale (red horizontal line means p = 0.05). Only six genes (Cyp2c55, Gbf1, Itgad, Pbx4, Plk1, and Vav2) can be considered as significantly deregulated between KO and WT (fold change >2; p < 0.05). Data are represented as mean ± S.E. (n = 3).
To unveil potential effects of miR-449 deficiency on the mRNA transcriptome, we performed gene chip analyses using Illumina microarrays (Mouse WG-6 v2.0 Expression BeadChips). Microarray analyses revealed that the mRNA transcriptome in miR-449−/− testes differed from that of WT controls by only six mRNAs (of >25,000 mRNAs probed). The six differentially expressed genes with statistical significance (-fold change >2; p < 0.05) between WT and miR-449 knock-out testes included Cyp2c55, Gbf1, Itgad, Pbx4, Plk1, and Vav2, none of which is involved in the E2F-pRb pathway. This result suggests that the lack of miR-449 has no overall significant effects on mRNA expression.
Fertility tests by mating male miR-449−/− mice or their WT littermates (controls) with WT C57BL/6 females for 3 months revealed no obvious differences in the number of litters, total pups, and pups per litter (supplemental Table S2). These experiments were conducted using mice on a mixed C57BL/6:129S7/SvEv background. To see whether the genetic background had any effects on the phenotype, we then adopted two different mating strategies aimed at bringing the KO line onto a more pure C57 or 129 genetic background. One group of miR-449−/− mice was backcrossed with wild type C57BL/6 females, whereas another group was backcrossed with 129S7/SvEv to get F5 miR-449+/− mice. Among the F6 male miR-449−/− mice derived from intercrossing F5 miR-449+/−, all were fertile (four of four on C57BL/6 and five of five on 129S7/SvEv backgrounds), suggesting that the miR-449 cluster is dispensable for testicular development and male fertility in mice.
Functional Compensation by miR-34b/c upon Ablation of miR-449 Cluster
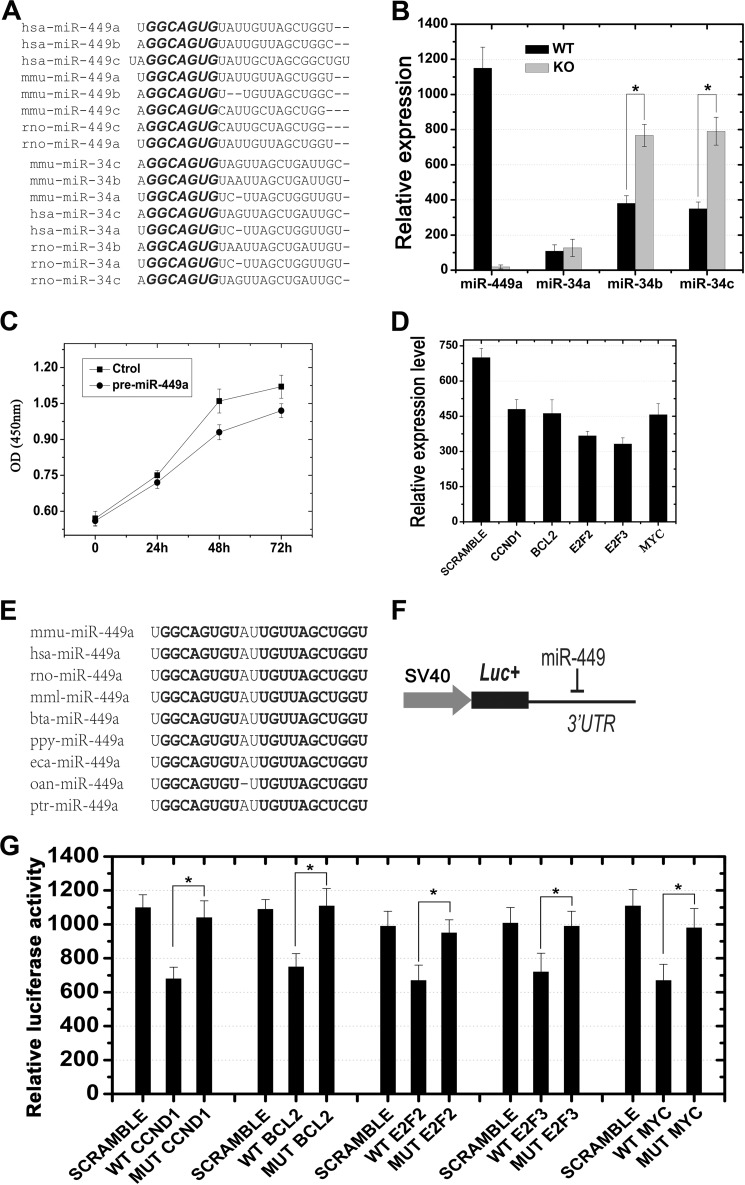
Previous studies have unraveled that multiple miRNAs may target the same mRNAs, and one mRNA can be targeted by multiple miRNAs (37). A complete lack of phenotypic defects in the miR-449−/− mice may result from a compensatory effect of miRNAs that can substitute for the roles of the miR-449 cluster. In silico analyses identified that members of the miR-449 cluster and the miR-34 family possess similar mature sequences and identical seed regions (Fig. 4A). Among the three members of the miR-34 family, miR-34a is transcribed from a separate locus different from that harboring miR-34b and miR-34c, which are transcribed as a single primary miR-34b/c and thus form an miRNA cluster (30). Our microarray- and qPCR-based expression profiling analyses revealed that miR-34b/c displayed similar expression and localization patterns during both testicular development and adult spermatogenesis (Fig. 1). These data point to a possibility that miR-34b/c may be the miRNAs that are compensating for the lack of miR-449 in those KO testes. Therefore, we examined the levels of miR-34 family members in WT and miR-449−/− mouse testes (Fig. 4B). As a control, no miR-449 transcripts were detected in miR-449−/− testes, whereas high levels of miR-449 were present in wild type testes (Fig. 4B). Interestingly, levels of miR-34b/c, rather than miR-34a, exhibited a significant up-regulation in miR-449−/− testes (p < 0.05) (Fig. 4B). This finding further supports the notion that up-regulation of miR-34b/c may have compensated for the absence of the miR-449 cluster in the KO mice. However, it remains unclear whether CREMτ and SOX5 are responsible for the up-regulation of miR-34b/c in miR-449-null testes.
FIGURE 4.
Shared mRNA targets between miR-449 and miR-34b/c and potential compensatory effects of miR-34b/c in miR-449 knock-out mice. A, high similarity of mature sequences between members of the miR-449 cluster and the miR-34 family from humans, mice, and rats. The conserved motif “GGCAGUG” located in the “seed region” of each miRNA is highlighted in bold. B, qPCR analyses showing that levels of miR-34b and miR-34c, rather than miR-34a, were notably up-regulated in miR-449 KO mice as compared with those in wild type. *, p < 0.05. C, cell proliferation assays showing a time-dependent decrease in total cell number of GC-1 cells transfected with pre-miR-449a compared with the scrambled transfection control (Ctrol). D, qPCR analysis of levels of mRNAs for CCND1, BCL2, E2F2, E2F3, and MYC in HeLa cells transfected with pre-miR-449a. The values were normalized against GAPDH endogenous control. E, highly conserved miR-449a sequences among nine species. Sequences highlighted in bold indicate identical nucleotides. F, schematic illustration of the luciferase reporter assay. The 3′-UTR sequence for each target gene was inserted downstream of the luciferase coding sequence in pGL3.0 control vector (Promega) as described previously (19). G, luciferase reporter assays demonstrated that miR-449 was capable of repressing expression of genes encoding factors belonging to the E2F-pRb pathway, including CCND1, BCL2, E2F2, E2F3, and MYC (n = 3; *, p < 0.05). SCRAMBLE, a non-sense double strand siRNA showing no homology to any mRNA in the murine genome. Data are represented as mean ± S.E. (n = 3).
Members of miR-449 Cluster and miR-34b/c Target Multiple Common mRNAs, Especially Those Involved in E2F-pRb Pathway
Recent data suggest that cell cycle regulators (e.g. CDC25A, CDK6, and CCNE2) and Notch pathway players (e.g. DLL1 and NOTCH1) that are involved in cellular differentiation are direct targets of miR-449 (10, 13, 28, 38). Similarly, several studies have revealed that a large number of cell cycle regulator genes are direct targets of miR-34 family members that are ubiquitously expressed in normal tissues/organs but commonly down-regulated in tumor cells possibly via epigenetic mechanisms (15, 30, 40, 41). To test whether miR-449 and miR-34b/c indeed share a common cohort of mRNA targets, we next investigated whether other experimentally validated miR-34 targets related to cell cycle regulation are also direct targets of the miR-449 cluster. We first introduced miR-449a mimics into the GC-1 cell line derived from spermatogonia of pubertal mouse testes along with scrambled miRNA controls. The cell proliferation assays revealed that miR-449a mimics could inhibit GC-1 cell growth at 48 and 72 h post-transfection compared with scrambled miRNA control (Fig. 4C). We then examined levels of mRNAs for several cell cycle regulators that have been previously validated to be direct targets of the miR-34 family (15, 30, 41, 42), including Ccnd1, Bcl2, E2f2, E2f3, and Myc (Fig. 4D). Levels of all of the five mRNAs were significantly down-regulated in GC-1 cells overexpressing miR-449a as compared with scrambled control, indicating that indeed these genes can be modulated by miR-449a in vitro (Fig. 4D and supplemental Fig. S4). To confirm whether these are direct targets for miR-449a, we performed Dual-Luciferase reporter assays in which 3′-UTRs with WT or mutant binding sites for the seed sequence of miR-449a (Fig. 4, E and F) were fused to the pGL3.0 control vector as described previously (19). Relative luciferase activities of all five WT 3′-UTR constructs were significantly lower than those in the scrambled miRNA controls, whereas mutant 3′-UTR constructs showed luciferase activities comparable with those in scrambled miRNA controls (Fig. 4G). These data suggest that these five mRNAs, which had been shown to be direct targets of miR-34b/c, can also be directly regulated by miR-449a in vitro.
Exhaustive Correlation Analysis of Genome-wide mRNA Transcriptomic Changes Induced by miR-449a/b or miR-34b/c
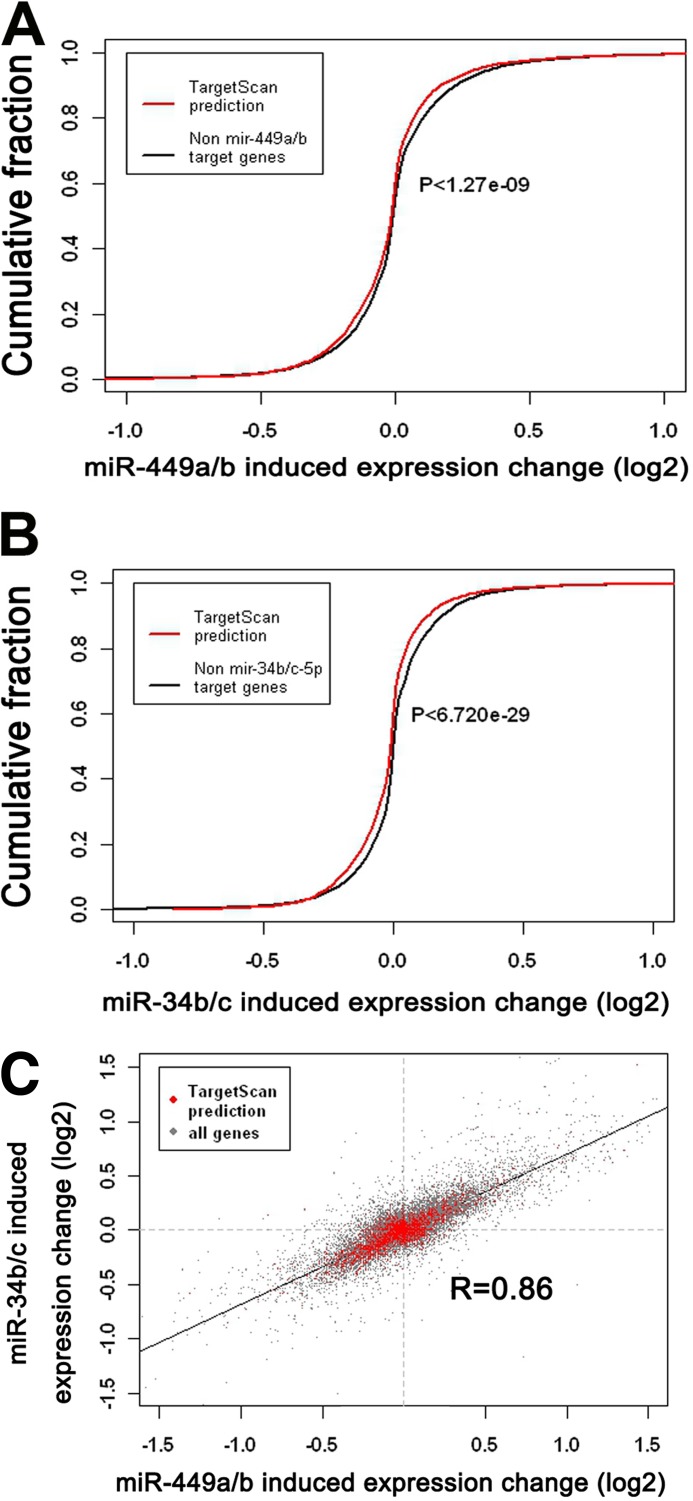
To further demonstrate that the miR-449 cluster and miR-34b/c play redundant roles, we conducted exhaustive correlation analysis of mRNA transcriptomes using deposited GEO data available in the Gene Expression Omnibus (accession number GSE22147) (10). Proliferating human airway mucociliary epithelial cells were individually transfected with Hsa-miR-449a/b or Hsa-miR-34b/c followed by microarray analyses at 48 h post-transfection (10). In silico analyses revealed that mRNAs possessing miR-449a/b target sites predicted by TargetScan were significantly down-regulated as compared with mRNAs without predicted target sites (p < 1.27e−09; two-tailed Wilcoxon rank sum test) (Fig. 5A). This case also applied to miR-34b/c, which also induced a significant down-regulation of their predicted mRNAs targets (p < 6.72e−29) upon introduction into human airway mucociliary epithelial cells (Fig. 5B). Importantly, the genome-wide mRNA transcriptomic alterations induced by transfection of miR-449a/b and miR-34b/c were also well correlated (Pearson's correlation coefficient r = 0.86) when only the predicted target mRNAs were compared (Fig. 5C), suggesting that miR-449a/b and miR-34b/c exert similar effects on mRNA transcriptomes in proliferating human airway mucociliary epithelial cells. These data are also further supported by a recent study showing that the shared seed sequences between miR-449b and miR-34a can elicit highly correlated genome-wide mRNA transcriptomic changes in gastric SNU638 cells (27).
FIGURE 5.
Exhaustive correlation analyses between miR-449a/b- and miR-34b/c-induced mRNA transcriptomic changes in human airway mucociliary epithelial cells. A, significant down-regulation (p < 1.27e−09; two-tailed Wilcoxon rank sum test) of mRNAs (red line) possessing predicted binding motifs for the miR-449a/b seed region as compared with mRNAs lacking miR-449a/b binding motifs (black line). B, significant down-regulation (p < 6.720e−29; two-tailed Wilcoxon rank sum test) of mRNAs (red line) possessing predicted binding motifs for the miR-34b/c seed sequence compared with mRNAs lacking miR-34b/c binding motifs (black line). C, mRNA transcriptomic changes induced by miR-449a/b are highly correlated with those by miR-34b/c. -Fold changes are represented as log2 scale. Pearson's correlation coefficient r = 0.86.
DISCUSSION
Production of fertilization-competent sperm relies on the precise spatiotemporal regulation of gene expression at both the transcriptional and post-transcriptional levels. The discovery of miRNAs as post-transcriptional regulators prompted us to look into miRNA-mediated mechanisms in the control of spatiotemporal gene expression during spermatogenesis. Although it has been shown that murine testes express abundant miRNAs (9, 14, 19, 23, 24), their physiological roles, especially functions of individual testicular miRNAs, remain largely unknown. In the present study, we focused on miR-449 because it was identified as the most up-regulated testicular miRNA upon meiotic initiation during postnatal testicular development and in adult testes through our microarray analyses (supplemental Fig. S1 and Tables S3 and S4). Interestingly, the miR-449 cluster appears to be under the regulation of both CREMτ, a critical master regulator in developing male germ cells (43–45), and SOX5, a major postmeiotic transcriptional factor (34, 46, 47). Given that both transcriptional factors are required for developing male germ cells to progress from mitotic to meiotic and the subsequent postmeiotic/haploid development, it is likely that miR-449 mainly functions in spermatocytes and haploid spermatids, which is consistent with our finding that miR-449 is predominantly and exclusively expressed in spermatocytes and spermatids in the adult testes (Fig. 1). Despite the fact that the miR-449 cluster KO mice show no discernible phenotype, an important role of miR-449 in male germ cell development cannot be entirely excluded at this time because our data suggest that miR-34b/c can compensate for the absence of miR-449 based upon the following lines of evidence. 1) miR-449 and miR-34b/c display similar expression profiles during testicular development and in adult spermatogenesis, and they are localized to the exact same spermatogenic cell types, namely spermatocytes and spermatids (Fig. 1) (14). 2) miR-449 and miR-34b/c possess identical an “seed sequence,” a core element usually located between the second and seventh nucleotides essential for base pairing with target mRNAs, and thus can theoretically target a similar set of mRNAs. Our reporter assays demonstrate that they both target genes that belong to the E2F-pRb pathway (Fig. 4). 3) miR-449 and miR-34b/c can induce similar mRNA transcriptomic changes when overexpressed in cultured cells in vitro (Fig. 5). 4) miR-34b/c levels are significantly up-regulated in miR-449-null testes, although the mechanism behind this up-regulation remains unknown (Fig. 4). The ideal way of proving the compensatory effects of miR-34b/c in the absence of miR-449 is to generate miR-34b/c-miR-449 double knock-out mice, which is underway in our laboratories.
It is not surprising to observe a high degree of overlap in both expression profiles and potential mRNA targets between miR-449 and miR-34b/c because it has been commonly accepted that one miRNA can target numerous mRNAs and one particular mRNA can be targeted by multiple miRNAs (48). This interwoven relationship between miRNAs and their mRNA targets results from the fact that many miRNAs possess similar or even identical “seed sequences” through which miRNAs bind their target sequences in the 3′-UTRs of mRNAs, and a 3′-UTR of a particular mRNA usually contains multiple sequence motifs that can be targeted in theory by multiple miRNAs with differential seed sequences. Consequently, this “multi-to-multi” relationship may represent a “fail-safe” mechanism in which in the case that one miRNA is missing other miRNAs with similar/identical seed sequences or those with different seed sequences but that can bind adjacent regions in the same 3′-UTR can compensate for the lost function of that particular miRNA. This hypothesis may also provide an answer to the disproportional number of miRNAs compared with that of mRNAs (∼1000 miRNAs versus ∼25,000 mRNAs) in the mouse genome. Many single miRNA or miRNA cluster knock-out mouse lines have been reported (49), but very few of them display obvious phenotypes under non-stress conditions. When under stress, some of these miRNA KO mice behave or react differently compared with their WT littermates, and abnormalities start to appear (50, 51). These findings suggest that miRNAs function mostly as a “fine tuning” mechanism responsible for stress responses by maintaining appropriate homeostasis of gene expression at post-transcriptional levels. It would be interesting to see whether miR-449 KO males would display any pathological changes if subjected to stress conditions in our future studies.
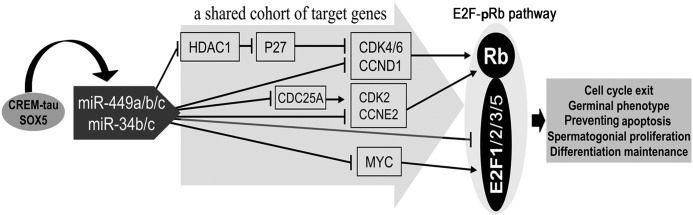
Previous studies have demonstrated that miR-34b/c target multiple genes belonging to the E2F-pRb pathway, including Ccnd1, Bcl2, E2f2, E2f3, and Myc (15, 30, 41). Intriguingly, in previous studies, miR-449 has been shown to target a wealth of cell cycle regulators that are also part of the E2F-pRb pathway, including Cdk25a, Cdk4/6, Ccne2, and E2f5 (13, 26–28), and the selected five miR-34b/c targets (Ccnd1, Bcl2, E2f2, E2f3, and Myc) were also validated as miR-449 targets by the present study (Figs. 4 and 5). Based upon these data, we here propose a model for the miR-449-miR-34b/c pathway to act to control male germ cell development during murine testicular development and spermatogenesis (Fig. 6). In this model, CREMτ and SOX5 activate the expression of miR-449 upon meiotic initiation. miR-449 in collaboration with miR-34b/c then suppresses the expression of factors that belong to the E2F-pRb pathway, including CDK4/6, CCND1, CDK25A, MYC, and E2F1/2/3/5. These changes further cause reduced levels of E2F proteins and an increase in hypophosphorylated pRb protein levels, which will further reduce E2F activities and thus affect male germ cell development during both meiotic and haploid phases of spermatogenesis.
FIGURE 6.
Proposed model for miR-449/miR-34 to act through E2F-pRb pathway in control of male germ cell development during murine testicular development and spermatogenesis. In this model, CREMτ and SOX5 activate the expression of miR-449 upon meiotic initiation. miR-449 in collaboration with miR-34b/c then suppresses the expression of factors that belong to the E2F-pRb pathway, including CDK4/6, CCND1, CDK25A, MYC, and E2F1/2/3/5. These changes further cause reduced levels of E2F proteins and an increase in hypophosphorylated pRb protein level, which will further reduce E2F activities and thus affect male germ cell development during both the meiotic and haploid phases of spermatogenesis.
Previous studies have demonstrated that the E2F-pRb pathway plays a critical role in the regulation of male germ cell development in the testis (52, 53). The mitotic phase of spermatogenesis requires the active E2F-pRb pathway as evidenced by the fact that E2F1 depletion causes highly reduced spermatogonial proliferation and marked testicular atrophy (54). However, upon the initiation of meiosis, activities of the E2F-pRb pathway appear to be down-regulated probably because this suppression allows male germ cells to exit from the mitotic cell cycle and to enter the meiotic program as suggested by several earlier studies (52, 55–57). In addition, this suppression may enhance the germinal phenotype by inhibiting mitotic proliferating activity and promoting meiotic gene expression (53). This notion is also supported by previous studies showing that miR-34c is capable of inducing a shift of the global mRNA pattern in HeLa cells toward germinal lineage and a shift of chicken embryonic stem cells toward germ cell fate (14). Moreover, lowered E2F1 activity appears to correlate with protective effects that prevent meiotic male germ cells (i.e. spermatocytes) from undergoing apoptosis because earlier studies have shown that overexpression of E2F1 leads to massive apoptosis of spermatocytes (10, 58). Spermatocytes represent the developing male germ cells that are most prone to apoptosis because these meiotic cells have to undergo crossover/homologous recombination, which is known to require double strand breaks. The double strand breaks must be repaired after meiotic recombination is finished, and any chromosomal mishaps during the meiotic phase can trigger the meiotic checkpoint mechanism, leading to spermatocyte apoptosis through p53-dependent and -independent pathways (39, 59–61). It is highly likely that the drastic up-regulation of miR-449 and miR-34b/c in the early meiotic phase represents a mechanism of suppressing E2F activity during meiotic and postmeiotic male germ cell development. Of course, the ultimate evidence has to come from cells or mice with a simultaneous inactivation of both miR-449 and miR-34b/c, an investigation already underway in our laboratories. Taken together, our data suggest that the miR-449 cluster is most abundantly expressed in developing male germ cells in the meiotic and haploid phases of spermatogenesis and that the miR-449 cluster and miR-34b/c function redundantly to suppress the activity of E2F-pRb pathway during the meiotic phase of spermatogenesis.
This work was supported, in whole or in part, by National Institutes of Health Grants HD050281 and HD060858 (to W. Y.). This work was also supported by National Natural Science Foundation of China Grants 30771139 and 30900776, Shanghai Natural Science Foundation Grant 09ZR1416200, Science and Technology Commission of Shanghai Municipality Grant 10DZ2270600, and Shanghai Leading Academic Discipline Project Grant S30201 (to C. X.).

This article contains supplemental Figs. S1–S4 and Tables S1–S4.
- miRNA
- microRNA
- miR-449
- microRNA-449
- miR-34
- microRNA-34
- qPCR
- quantitative real time PCR
- CREMτ
- cAMP-responsive element modulator τ
- E2F
- E2F transcription factor
- pRb
- retinoblastoma protein
- P
- postnatal day
- pre-miR-449
- miR-449 precursor.
REFERENCES
- 1. Yan W. (2009) Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol. Cell. Endocrinol. 306, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Idler R. K., Yan W. (2012) Control of messenger RNA fate by RNA binding proteins: an emphasis on mammalian spermatogenesis. J. Androl., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Papaioannou M. D., Nef S. (2010) MicroRNAs in the testis: building up male fertility. J. Androl. 31, 26–33 [DOI] [PubMed] [Google Scholar]
- 4. Kimble J. (2011) Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb. Perspect. Biol. 3, a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Czech B., Hannon G. J. (2011) Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wheeler G., Ntounia-Fousara S., Granda B., Rathjen T., Dalmay T. (2006) Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 580, 2195–2200 [DOI] [PubMed] [Google Scholar]
- 7. Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Kato I. (2006) The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 34, 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V., Lindblad-Toh K., Lander E. S., Kellis M. (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A. O., Landthaler M., Lin C., Socci N. D., Hermida L., Fulci V., Chiaretti S., Foà R., Schliwka J., Fuchs U., Novosel A., Müller R. U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D. B., Choksi R., De Vita G., Frezzetti D., Trompeter H. I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C. E., Nagle J. W., Ju J., Papavasiliou F. N., Benzing T., Lichter P., Tam W., Brownstein M. J., Bosio A., Borkhardt A., Russo J. J., Sander C., Zavolan M., Tuschl T. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marcet B., Chevalier B., Luxardi G., Coraux C., Zaragosi L. E., Cibois M., Robbe-Sermesant K., Jolly T., Cardinaud B., Moreilhon C., Giovannini-Chami L., Nawrocki-Raby B., Birembaut P., Waldmann R., Kodjabachian L., Barbry P. (2011) Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 13, 693–699 [DOI] [PubMed] [Google Scholar]
- 11. Greco S., De Simone M., Colussi C., Zaccagnini G., Fasanaro P., Pescatori M., Cardani R., Perbellini R., Isaia E., Sale P., Meola G., Capogrossi M. C., Gaetano C., Martelli F. (2009) Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23, 3335–3346 [DOI] [PubMed] [Google Scholar]
- 12. Redshaw N., Wheeler G., Hajihosseini M. K., Dalmay T. (2009) MicroRNA-449 is a putative regulator of choroid plexus development and function. Brain Res. 1250, 20–26 [DOI] [PubMed] [Google Scholar]
- 13. Yang X., Feng M., Jiang X., Wu Z., Li Z., Aau M., Yu Q. (2009) miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 23, 2388–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M. H., Durand P., Samarut J., Pain B., Rouault J. P. (2010) Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bommer G. T., Gerin I., Feng Y., Kaczorowski A. J., Kuick R., Love R. E., Zhai Y., Giordano T. J., Qin Z. S., Moore B. B., MacDougald O. A., Cho K. R., Fearon E. R. (2007) p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 16. Hermeking H. (2007) p53 enters the microRNA world. Cancer Cell 12, 414–418 [DOI] [PubMed] [Google Scholar]
- 17. Yan N., Lu Y., Sun H., Tao D., Zhang S., Liu W., Ma Y. (2007) A microarray for microRNA profiling in mouse testis tissues. Reproduction 134, 73–79 [DOI] [PubMed] [Google Scholar]
- 18. Bao J., Wu Q., Song R., Jie Z., Zheng H., Xu C., Yan W. (2011) RANBP17 is localized to the XY body of spermatocytes and interacts with SPEM1 on the manchette of elongating spermatids. Mol. Cell. Endocrinol. 333, 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu J., Bao J., Wang L., Hu Y., Xu C. (2011) MicroRNA-184 downregulates nuclear receptor corepressor 2 in mouse spermatogenesis. BMC Dev. Biol. 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Y., Zheng M., Shi Q., Zhang L., Zhen W., Chen W., Zhang Y. (2008) An epididymis-specific secretory protein HongrES1 critically regulates sperm capacitation and male fertility. PLoS One 3, e4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobsen A., Wen J., Marks D. S., Krogh A. (2010) Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 20, 1010–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu Z., Goodyear S. M., Rao S., Wu X., Tobias J. W., Avarbock M. R., Brinster R. L. (2011) MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 12740–12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buchold G. M., Coarfa C., Kim J., Milosavljevic A., Gunaratne P. H., Matzuk M. M. (2010) Analysis of microRNA expression in the prepubertal testis. PLoS One 5, e15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ro S., Park C., Sanders K. M., McCarrey J. R., Yan W. (2007) Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 311, 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lizé M., Herr C., Klimke A., Bals R., Dobbelstein M. (2010) MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle 9, 4579–4583 [DOI] [PubMed] [Google Scholar]
- 26. Noonan E. J., Place R. F., Pookot D., Basak S., Whitson J. M., Hirata H., Giardina C., Dahiya R. (2009) miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 28, 1714–1724 [DOI] [PubMed] [Google Scholar]
- 27. Bou Kheir T., Futoma-Kazmierczak E., Jacobsen A., Krogh A., Bardram L., Hother C., Grønbæk K., Federspiel B., Lund A. H., Friis-Hansen L. (2011) miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol. Cancer 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noonan E. J., Place R. F., Basak S., Pookot D., Li L. C. (2010) miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget 1, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Shaughnessy P. J., Hu L., Baker P. J. (2008) Effect of germ cell depletion on levels of specific mRNA transcripts in mouse Sertoli cells and Leydig cells. Reproduction 135, 839–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hermeking H. (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199 [DOI] [PubMed] [Google Scholar]
- 31. Lizé M., Pilarski S., Dobbelstein M. (2010) E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 17, 452–458 [DOI] [PubMed] [Google Scholar]
- 32. Mendell J. T. (2008) miRiad roles for the miR-17–92 cluster in development and disease. Cell 133, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novotny G. W., Sonne S. B., Nielsen J. E., Jonstrup S. P., Hansen M. A., Skakkebaek N. E., Rajpert-De Meyts E., Kjems J., Leffers H. (2007) Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17–92 cluster. Cell Death Differ. 14, 879–882 [DOI] [PubMed] [Google Scholar]
- 34. Connor F., Cary P. D., Read C. M., Preston N. S., Driscoll P. C., Denny P., Crane-Robinson C., Ashworth A. (1994) DNA binding and bending properties of the post-meiotically expressed Sry-related protein Sox-5. Nucleic Acids Res. 22, 3339–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 [DOI] [PubMed] [Google Scholar]
- 36. Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., Jaenisch R., Sharp P. A., Jacks T. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berezikov E. (2011) Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet 12, 846–860 [DOI] [PubMed] [Google Scholar]
- 38. Feng M., Yu Q. (2010) miR-449 regulates CDK-Rb-E2F1 through an auto-regulatory feedback circuit. Cell Cycle 9, 213–214 [DOI] [PubMed] [Google Scholar]
- 39. Schwartz D., Goldfinger N., Kam Z., Rotter V. (1999) p53 controls low DNA damage-dependent premeiotic checkpoint and facilitates DNA repair during spermatogenesis. Cell Growth Differ. 10, 665–675 [PubMed] [Google Scholar]
- 40. Li N., Fu H., Tie Y., Hu Z., Kong W., Wu Y., Zheng X. (2009) miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 275, 44–53 [DOI] [PubMed] [Google Scholar]
- 41. Li Y., Guessous F., Zhang Y., Dipierro C., Kefas B., Johnson E., Marcinkiewicz L., Jiang J., Yang Y., Schmittgen T. D., Lopes B., Schiff D., Purow B., Abounader R. (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bueno M. J., Malumbres M. (2011) MicroRNAs and the cell cycle. Biochim. Biophys. Acta 1812, 592–601 [DOI] [PubMed] [Google Scholar]
- 43. Macho B., Brancorsini S., Fimia G. M., Setou M., Hirokawa N., Sassone-Corsi P. (2002) CREM-dependent transcription in male germ cells controlled by a kinesin. Science 298, 2388–2390 [DOI] [PubMed] [Google Scholar]
- 44. De Cesare D., Fimia G. M., Brancorsini S., Parvinen M., Sassone-Corsi P. (2003) Transcriptional control in male germ cells: general factor TFIIA participates in CREM-dependent gene activation. Mol. Endocrinol. 17, 2554–2565 [DOI] [PubMed] [Google Scholar]
- 45. Monaco L., Kotaja N., Fienga G., Hogeveen K., Kolthur U. S., Kimmins S., Brancorsini S., Macho B., Sassone-Corsi P. (2004) Specialized rules of gene transcription in male germ cells: the CREM paradigm. Int. J. Androl. 27, 322–327 [DOI] [PubMed] [Google Scholar]
- 46. Denny P., Swift S., Connor F., Ashworth A. (1992) An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 11, 3705–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kiselak E. A., Shen X., Song J., Gude D. R., Wang J., Brody S. L., Strauss J. F., 3rd, Zhang Z. (2010) Transcriptional regulation of an axonemal central apparatus gene, sperm-associated antigen 6, by a SRY-related high mobility group transcription factor, S-SOX5. J. Biol. Chem. 285, 30496–30505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peter M. E. (2010) Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29, 2161–2164 [DOI] [PubMed] [Google Scholar]
- 49. Park C. Y., Choi Y. S., McManus M. T. (2010) Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 19, R169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leung A. K., Sharp P. A. (2010) MicroRNA functions in stress responses. Mol. Cell 40, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W. (2009) A microRNA imparts robustness against environmental fluctuation during development. Cell 137, 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singh S., Johnson J., Chellappan S. (2010) Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochim. Biophys. Acta 1799, 788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lizé M., Klimke A., Dobbelstein M. (2011) MicroRNA-449 in cell fate determination. Cell Cycle 10, 2874–2882 [DOI] [PubMed] [Google Scholar]
- 54. Hoja M. R., Liu J. G., Mohammadieh M., Kvist U., Yuan L. (2004) E2F1 deficiency impairs murine spermatogenesis and augments testicular degeneration in SCP3-nullizygous mice. Cell Death Differ. 11, 354–356 [DOI] [PubMed] [Google Scholar]
- 55. Nguyen D. X., McCance D. J. (2005) Role of the retinoblastoma tumor suppressor protein in cellular differentiation. J. Cell. Biochem. 94, 870–879 [DOI] [PubMed] [Google Scholar]
- 56. Spiller C. M., Wilhelm D., Koopman P. (2010) Retinoblastoma 1 protein modulates XY germ cell entry into G1/G0 arrest during fetal development in mice. Biol. Reprod. 82, 433–443 [DOI] [PubMed] [Google Scholar]
- 57. Nalam R. L., Andreu-Vieyra C., Braun R. E., Akiyama H., Matzuk M. M. (2009) Retinoblastoma protein plays multiple essential roles in the terminal differentiation of Sertoli cells. Mol. Endocrinol. 23, 1900–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holmberg C., Helin K., Sehested M., Karlström O. (1998) E2F-1-induced p53-independent apoptosis in transgenic mice. Oncogene 17, 143–155 [DOI] [PubMed] [Google Scholar]
- 59. Lu W. J., Chapo J., Roig I., Abrams J. M. (2010) Meiotic recombination provokes functional activation of the p53 regulatory network. Science 328, 1278–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hamer G., Roepers-Gajadien H. L., van Duyn-Goedhart A., Gademan I. S., Kal H. B., van Buul P. P., de Rooij D. G. (2003) DNA double-strand breaks and γ-H2AX signaling in the testis. Biol. Reprod. 68, 628–634 [DOI] [PubMed] [Google Scholar]
- 61. Yuan L., Liu J. G., Hoja M. R., Lightfoot D. A., Höög C. (2001) The checkpoint monitoring chromosomal pairing in male meiotic cells is p53-independent. Cell Death Differ. 8, 316–317 [DOI] [PubMed] [Google Scholar]