Background: The dystrophin complex stabilizes the cell membrane by linking the cytoskeletal network to the extracellular matrix.
Results: α-Catulin interacts directly with dystrobrevin, a component of the dystrophin complex, in muscle.
Conclusion: The interaction of α-catulin with dystrobrevin contributes to the integrity of the dystrophin complex in muscle.
Significance: A molecular interaction potentially important for muscle pathogenesis is identified.
Keywords: Caenorhabditis elegans Genetics, Cytoskeleton, Dystrophin, Muscle, Muscular Dystrophy
Abstract
The dystrophin complex is a multimolecular membrane-associated protein complex whose defects underlie many forms of muscular dystrophy. The dystrophin complex is postulated to function as a structural element that stabilizes the cell membrane by linking the contractile apparatus to the extracellular matrix. A better understanding of how this complex is organized and localized will improve our knowledge of the pathogenic mechanisms of diseases that involve the dystrophin complex. In a Caenorhabditis elegans genetic study, we demonstrate that CTN-1/α-catulin, a cytoskeletal protein, physically interacts with DYB-1/α-dystrobrevin (a component of the dystrophin complex) and that this interaction is critical for the localization of the dystrophin complex near dense bodies, structures analogous to mammalian costameres. We further show that in mouse α-catulin is localized at the sarcolemma and neuromuscular junctions and interacts with α-dystrobrevin and that the level of α-catulin is reduced in α-dystrobrevin-deficient mouse muscle. Intriguingly, in the skeletal muscle of mdx mice lacking dystrophin, we discover that the expression of α-catulin is increased, suggesting a compensatory role of α-catulin in dystrophic muscle. Together, our study demonstrates that the interaction between α-catulin and α-dystrobrevin is evolutionarily conserved in C. elegans and mammalian muscles and strongly suggests that this interaction contributes to the integrity of the dystrophin complex.
Introduction
Since the identification of dystrophin as the cause of Duchenne muscular dystrophy, a number of transmembrane proteins and peripheral membrane proteins have been found to interact with dystrophin to form a macromolecular protein complex, collectively called the dystrophin-associated protein complex (DAPC)3 (1–3). Defects in components of the DAPC are associated with a variety of pathological conditions, including muscular dystrophy, cardiomyopathy, myoclonic dystonia, and vasospasm. Interestingly, specific components of the DAPC differ in various tissues or cell types, and different forms of the DAPC co-exist within the same cell (1). Regardless of such differences in specific components, the DAPC interacts with the extracellular matrix and cytoskeletal elements. This interaction provides structural stability to the cells by forming a link between the cytoskeleton and the extracellular matrix and allows the DAPC to serve as a scaffolding complex for signaling molecules, such as ion channels.
In skeletal muscle, the DAPC is enriched at costameres, although it is present throughout the sarcolemma (4). Costameres are subsarcolemmal protein assemblies that align with the Z lines of myofibrils and physically couple force-generating sarcomeres with the sarcolemma (5). Costameres are considered to be a muscle-specific form of focal adhesions because most of the cytoskeletal components are shared. The DAPC is thought to anchor the sarcolemma to costameres and stabilize the sarcolemma against physical forces transduced through costameres during muscle contraction or stretching (6). Consistent with this idea, defects of the DAPC in humans and mice lead to a disorganized costameric lattice and disruption of sarcolemmal integrity (7, 8). Certain cortical cytoskeletal proteins that interact with the DAPC show increased expression levels in the absence of dystrophin (9). Such increased expression of the cytoskeletal proteins has been postulated to be a compensatory response to fortify the weakened costameric lattice. Therefore, gaining knowledge on how these cytoskeletal proteins interact with the DAPC and other proteins at specific domains of the plasma membrane is critical for improving our understanding of muscular dystrophy pathogenesis.
The genome of the nematode Caenorhabditis elegans encodes most of the DAPC components (10). Similar to the mammalian DAPC, the C. elegans dystrophin homolog, DYS-1, forms a multimeric protein complex with other homologs of DAPC components at the muscle membrane. For instance, a physical interaction between DYS-1 and DYB-1 (an α/β-dystrobrevin homolog) was demonstrated, and STN-1, an α/β-syntrophin homolog, was also shown to bind DYB-1 (11). In addition, a mutation in dys-1 disrupts normal localization of SGCA-1, an α-sarcoglycan homolog (12). Importantly, defects in genes encoding components of the DAPC cause a unique locomotory phenotype, known as the head-bending phenotype (13). The phenotype is characterized by an exaggerated bending of the anterior part of the body when mechanical stimuli are applied to the posterior part of the body, as opposed to a normal body curvature in wild-type animals (12). Recent electrophysiological evidence shows that the head-bending phenotype is likely to be associated with a defect in action potential during periods of elevated synaptic activity (14). In C. elegans, the DAPC localizes near dense bodies, structures that are analogous to costameres (12). Both dense bodies and costameres are muscle-specific forms of integrin-mediated attachments between the extracellular matrix and the actin cytoskeleton (15). Although the structural role of the DAPC in C. elegans has not been explored, there is convincing evidence that the DAPC plays a role in localizing signaling molecules (10). For example, the DAPC localizes SLO-1, the calcium-activated potassium BK channel, at dense bodies (12). A compromise in the integrity of the DAPC causes a disruption of normal SLO-1 localization in muscle. A defect in action potential in DAPC mutants was indeed attributed, at least in part, to a defect in SLO-1 localization (14).
In a C. elegans forward genetic study that identifies mutants exhibiting abnormal localization of SLO-1 in muscle, we previously reported that mutations in α-catulin/ctn-1, encoding a putative cytoskeletal protein, cause a disruption of DAPC localization near dense bodies (16). Consistent with the idea that SLO-1 localization is controlled by the DAPC, we observed that mutations in α-catulin/ctn-1 disrupt SLO-1 localization as well. However, the mechanism by which CTN-1 interacts with the DAPC has not been clearly defined.
In this study we show that CTN-1 interacts with the dystrophin complex through DYB-1/dystrobrevin in C. elegans. We further demonstrate that CTN-1 and DYB-1 are dependent on each other for their localization near dense bodies. Disruption of the interaction between CTN-1 and DYB-1, or the lack of either of these proteins, compromises the localization of the dystrophin complex near dense bodies in C. elegans. We also found that the interaction between α-catulin and α-dystrobrevin is conserved in mouse skeletal muscle. Intriguingly, we uncovered that in mdx mice, a mouse model of Duchenne muscular dystrophy, the sarcolemmal expression level of α-catulin is considerably increased, indicating a compensatory role of α-catulin in the damaged sarcolemma.
EXPERIMENTAL PROCEDURES
C. elegans Strains
Strains were maintained on nematode growth medium agar plates at 20 °C as described previously (17). The following C. elegans strains were used for this study: N2, dyb-1(cx36), dys-1(eg33), ctn-1(eg1167), cimIs12[GFP::sgca-1, rol-6(d)], cimIs9 [GFP::ctn-1, Pttx-3mRFP], and cimIs11 [GFP::dyb-1, Pttx-3mRFP].
Antibodies
Generation of monoclonal antibodies against α-catulin has been described previously (18). Monoclonal antibodies 3C4 and 5G8, recognizing two different epitopes in α-catulin, were used in all of the experiments with the exception of Fig. 5E and supplemental Fig. S2 (rabbit pAb; Epitomics). The following additional commercial antibodies were used: anti-β-catenin (rabbit pAb; Sigma), anti-α-catenin (mouse mAb; BD Transduction Laboratories), anti-vinculin (rabbit pAb; Sigma), anti-α-dystrobrevin (goat and rabbit; Santa Cruz Biotechnology), α-bungarotoxin-Alexa Fluor 488 (Invitrogen), anti-GFP antibody (clone N86/8; UC Davis/NIH NeuroMab facility), anti-dystrophin (MANDRA1 clone 7A10; Developmental Hybridoma Bank) antibody, and anti-α-tubulin antibody (AA4.3; Developmental Hybridoma Bank).
FIGURE 5.
α-Catulin forms a complex with α-dystrobrevin in mouse muscles. A, α-dystrobrevin co-precipitates with α-catulin in mouse muscle extract. Mouse muscle extract was immunoprecipitated with either control antibody (unimmunized rabbit IgG) or anti-α-dystrobrevin antibody (H300). Immunoprecipitated proteins were analyzed by Western blotting (WB) with anti-α-catulin antibodies. The same membrane was stripped and reprobed with α-dystrobrevin antibody. Arrows indicate the specific protein detected. Asterisks indicate isoforms of dystrobrevin. Arrowhead indicates nonspecific protein. B, α-catulin co-precipitates with α-dystrobrevin in mouse muscle extract. Mouse muscle extract was immunoprecipitated with anti-α-catulin antibody (clone 5G8) and probed with α-dystrobrevin (upper). The same blot was stripped and reprobed with α-catulin antibody (lower). Arrows indicate the specific protein band detected. Total, extract input; Control, control nonspecific antibody. C, frozen cross-section of mouse muscle was stained with anti-α-catulin antibody (clone 5G8) and anti-α-dystrobrevin antibody (H300) and examined under a confocal microscope. Shown are images obtained by two different laser lines at a single Z-plane. M.W., molecular mass.
Animal Use
Mice are taken care of and used in accordance of the procedures approved by the Institutional Animal Care and Use Committee of the Rosalind Franklin University, Ghent University, and Harvard University.
Immunoprecipitation and Identification of Proteins by Liquid Chromatography–Tandem Mass Spectrometry
Monoclonal antibodies 3C4 and 5G8 were covalently coupled to M-280 tosylactivated Dynabeads® (Dynal Biotech) according to the manufacturer's protocol. For co-immunoprecipitation of α-catulin and associated proteins, lysates of HEK293T cells were prepared in PBS containing 0.5% Nonidet P-40 and a protease inhibitor mixture (Roche Diagnostics). Lysates (4 mg of protein each) were incubated overnight with washed and preequilibrated antibody-coupled beads. The beads were washed five times in 5-fold diluted lysis buffer followed by boiling for 5 min in Laemmli buffer. Samples were subjected to SDS-PAGE followed by silver staining (Silver Stain Plus kit; Bio-Rad). After excision of the bands and in-gel digestion with trypsin, the samples were subjected to LC-MS/MS, as described previously (19). Interactions of α-catulin with α-dystrobrevin and other proteins in HEK293T cells were further validated by standard co-immunoprecipitation and Western blot analyses (see below).
DNA Constructs and Transgenic Worms
To generate a CTN-1 expression vector construct that allows us to determine the localization of CTN-1, we inserted a GFP sequence at the translation initiation site of ctn-1 cDNA, and then the ctn-1 promoter sequence (∼4 kb) was inserted before the GFP sequence. Deletion constructs of CTN-1 were generated in a similar manner, where cDNAs of CTN-1Δ1–279 and CTN-1Δ551–784 were fused in-frame to a GFP sequence under the control of the ctn-1 promoter. CTN-1Δ382–416 lacking the coiled-coil domain was constructed using overlapping extension PCR using full-length CTN-1 cDNA fused with GFP under the control of the ctn-1 promoter as the template. To generate the Pdyb-1GFP::dyb-1 construct, the 3-kb dyb-1 promoter sequence was inserted upstream of the GFP coding sequence, and the open reading frame of dyb-1 cDNA was inserted in-frame at the C terminus of GFP. The GFP-tagged SGCA-1 construct was described previously. The entire sgca-1 coding sequence and ∼2 kb upstream of the translation initiation codon was first cloned, and a GFP sequence was inserted in-frame right after the signal sequence of SGCA-1.
Transgenic strains carrying extrachromosomal arrays were made by injecting DNA constructs (2.5 ng/μl) along with a co-injection marker DNA [pRF4(rol-6(d)) or ttx-3::mRFP, 30 ng/μl] into the gonads of hermaphrodite animals at a total DNA concentration of 100 ng/μl. Integration of extrachromosomal arrays to the genome was performed by the UV/trimethyl psolaren method (20).
Yeast Two-hybrid Screen
The yeast two-hybrid screen was performed using the Matchmaker GAL4-based yeast two-hybrid system (Clontech). ctn-1 cDNA encoding the CTN-1 fragment (1–377 amino acids) was used as the bait and was cloned into the pGBKT7 vector in-frame with the GAL4 DNA binding domain, followed by transformation into the Y187 yeast strain. A C. elegans cDNA library was constructed in the pGADT7 vector such that the proteins encoded by the inserts were fused to the 3′ end of the GAL4 activation domain. The library was then transformed into the Y2H yeast strain. The Y187 strain expressing the bait was mated with the Y2H strain expressing the library, and the diploids that showed positive interactions were selected on quadruple dropout plates (−histidine, −alanine, −leucine, and −tryptophan) containing 40 μg/ml X-α-Gal. Approximately 8.7 million independent clones were screened for positive interactions. cDNA inserts from positive clones were sequenced and identified.
For yeast two-hybrid assays used for identifying interacting domains of CTN-1 and DYB-1, ctn-1 cDNAs were cloned into the pGBKT7 vector in-frame with the GAL4 DNA binding domain, and dyb-1 cDNAs were cloned into the PGADT7 vector in-frame with the GAL4 activation domain. The ctn-1 constructs were transformed into the Y187 yeast strain, and the dyb-1 constructs were transformed into the Y2H Gold strain. Mating was performed by inoculating and growing appropriate yeast colonies in YPD medium, followed by selection of diploids that showed positive two-hybrid interaction on quadruple dropout plates (−histidine, −alanine, −leucine, and −tryptophan) containing 40 μg/ml X-α-Gal. Empty vectors served as negative controls, and constructs were tested for autoactivation.
Body Curvature Analysis
Body curvature analysis was described previously (21). A single animal was transferred onto an unseeded nematode growth medium agar plate, and its movement was recorded at 20 frames/s. We limited image acquisition within 15–60 s after transfer because the head-bending phenotype is prominent when animals are stimulated to move forward rapidly. A script written on ImgePro Plus (Media Cybernetics) automatically recognizes the animal in each image contained in an acquired image stack and assigns 13 points spaced equally from the tip of the nose to the tail along the midline of the body and produces the pixel coordinates of the 13 points. First supplementary angles were calculated from the coordinates of the first three points with MATLAB software. First angle data were obtained when the head swing of an animal reached the maximal extension to the dorsoventral side.
Western Blot Analysis and Co-immunoprecipitation
Worms at the young adult stage were collected and directly lysed by heating at 95 °C for 10 min in SDS sample loading buffer. Total protein extracts, after removal of debris by centrifugation, were resolved onto a 7.5% SDS-polyacrylamide gel and transferred onto PVDF membranes. Western blot analysis was performed using anti-GFP antibody, anti-α-tubulin antibody, and HRP-conjugated goat anti-mouse IgG (Millipore). To quantify the level of specific proteins in Western blot analyses, the band intensities were normalized with tubulin (worm extracts) or GAPDH (mouse muscle extracts) levels. The Western blots from three independent extracts were quantified using the ImageJ program. Statistics analyses were performed using Prism (GraphPad Software).
Muscles (soleus, extensor digitorum longus, gastrocnemius, and tibialis anterior) from 4-week-old mice (C57BL/10ScSnJ) were dissected out and frozen. The frozen muscles were thawed and sonicated in IP buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.25% deoxycholate, 10% glycerol). The lysate was centrifuged for 10 min at 20,000 × g, and the supernatant was used for immunoprecipitation. The extracts were incubated with primary antibodies and protein A/G-agarose beads (Santa Cruz Biotechnology) at 4 °C overnight. The beads retaining the immune complexes were washed three times with IP buffer and heated in SDS sample loading buffer for 5 min. The supernatants resulting from centrifugation at 20,000 × g for 5 min were loaded onto 7.5% SDS-polyacrylamide gel and were subjected to Western blot analysis.
Immunofluorescent Staining of Mouse Muscle Tissue
Gastrocnemius or tibialis anterior muscles from 4-week-old wild-type (C57BL/10ScSnJ) mdx (C57BL/10ScSn-mdx/J) and α-DB−/− mice (22) were dissected, frozen in liquid nitrogen-isopentane, and cut into 6- or 9-μm cryostat sections. Sections were fixed with cold methanol, permeabilized with 0.5% Triton X-100 diluted in PBS for 30 min, and blocked with 10% goat serum diluted in PBS for 30 min. To prevent nonspecific binding with mouse primary antibodies, we incubated sections with 0.1 mg/ml AffiniPure Fab anti-mouse IgG (H+L) fragment (Jackson ImmunoResearch) for 1 h. Primary antibodies were diluted in 10% goat serum in PBS, and the tissues were incubated for 1 h. Secondary goat antibodies coupled to Alexa Fluor 488 were used in 10% goat serum in PBS (1:500 dilution). We made sure that under our staining conditions no staining was observed in the omission of primary antibodies or with irrelevant primary antibodies. The sections were mounted in anti-fade ProlongGold (Invitrogen), were imaged under a wide-field fluorescence microscope (Zeiss AxioObserver Z1) with identical exposure time and gain settings and were processed identically.
Confocal Microscopy
Worms were mounted on a 4% agar pad containing 25 mm NaN3, and fluorescence images were acquired using a Zeiss LSM510 confocal microscope with 63× objective (oil immersion, NA 1.4).
RESULTS
α-Catulin Forms Complex with Dystrobrevin
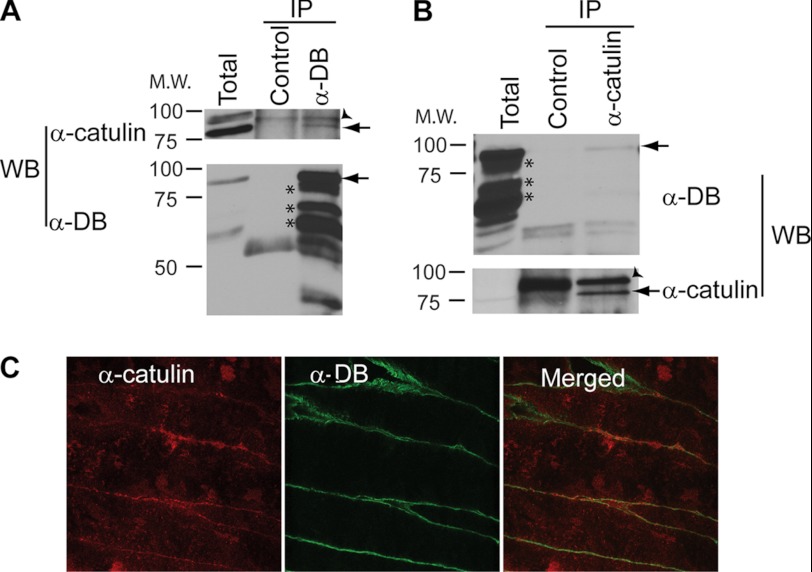
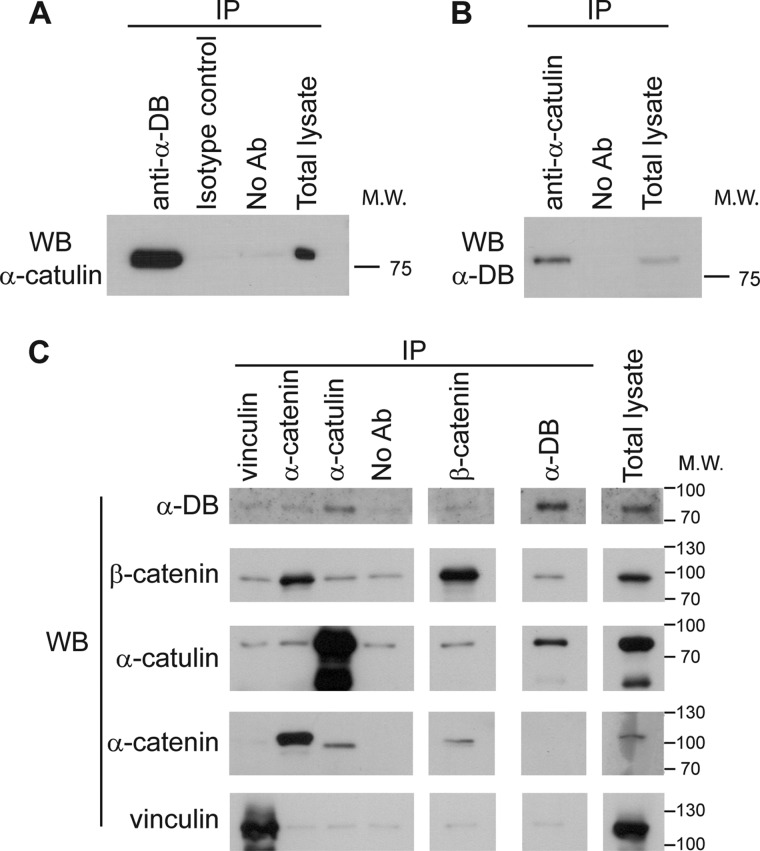
To gain further insight into the molecular function of α-catulin, we employed two independent approaches to identify proteins that interact with α-catulin: a proteomics approach (i.e. co-immunoprecipitation followed by mass spectrometry) and a yeast two-hybrid screen. In the proteomics approach, we immunoprecipitated α-catulin from HEK293T cell lysate with two monoclonal antibodies (3C4 and 5G8) specific for α-catulin and identified proteins that co-precipitated with α-catulin by mass spectrometry. One of the co-purified proteins was identified as α-dystrobrevin on the basis of six different peptides (data not shown). Complex formation between α-catulin and α-dystrobrevin in HEK293T cells was further confirmed by co-immunoprecipitation and Western blot analysis. When α-dystrobrevin was immunoprecipitated, the co-precipitated α-catulin was detected by Western blotting using an anti-α-catulin antibody, and conversely, when α-catulin was immunoprecipitated, the co-precipitated α-dystrobrevin was detected by Western blotting using an anti-α-dystrobrevin antibody (Fig. 1, A and B). We also examined the specificity of this complex formation by immunoprecipitation experiments in which we used antibodies against either vinculin or α-catenin, which are homologs of α-catulin, or against β-catenin, an interaction partner of these homologs. None of these proteins was co-immunoprecipitated with α-dystrobrevin, demonstrating that α-dystrobrevin exists in a specific complex with α-catulin (Fig. 1C).
FIGURE 1.
α-Catulin forms complex with α-dystrobrevin in HEK293T cells. A, α-catulin co-precipitates with α-dystrobrevin. HEK293T cells were lysed, and IP was performed on HEK293T lysates using (lanes from left to right): anti-α-dystrobrevin antibody (α-DB), isotype control antibody or protein G-Sepharose without antibody added. These immunoprecipitates, as well as total cell lysate (right lane), were analyzed by Western blotting (WB) using an antibody against α-catulin (3C4). B, α-dystrobrevin co-precipitates with α-catulin. HEK293T lysates were immunoprecipitated with anti-α-catulin antibody (3C4) or without antibody added. Immunoprecipitates (left) and total cell lysate (right) were immunoblotted using an antibody against α-dystrobrevin. C, α-dystrobrevin specifically interacts with α-catulin but not with other α-catulin homologs. HEK293T lysates were subjected to IP with the indicated antibodies or with no antibody added (serving as control for nonspecific adherence to the protein G-Sepharose). The immunoprecipitates were analyzed by Western blotting using the same set of antibodies.
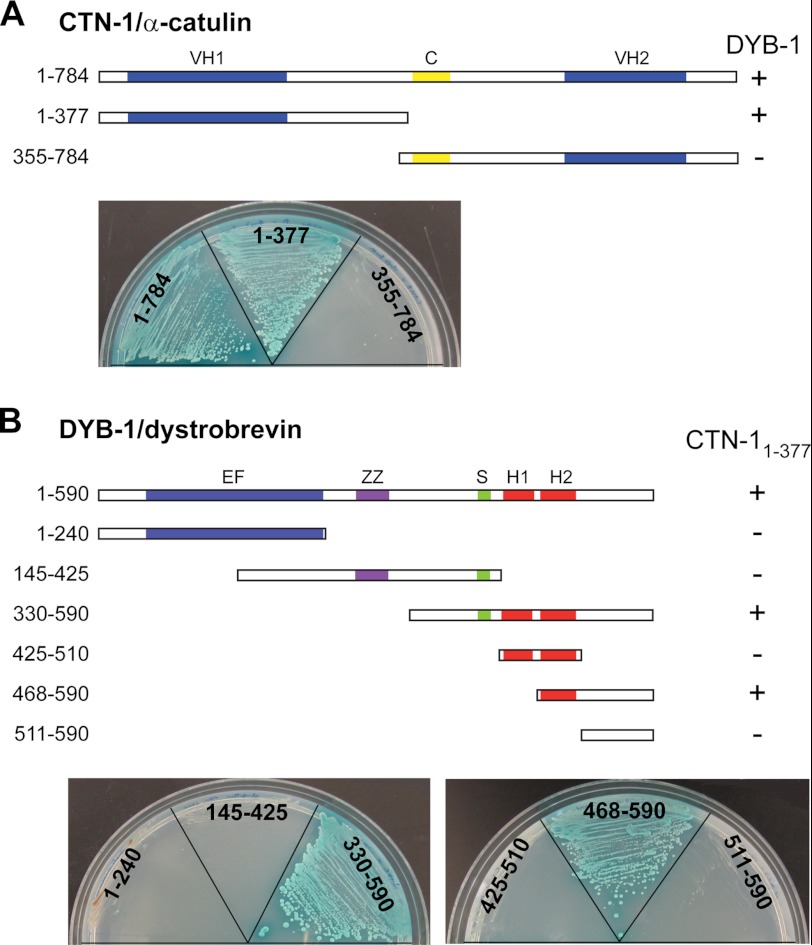
As a complementary approach to identify interaction partners of α-catulin, we performed a yeast two-hybrid screen of a C. elegans cDNA library. CTN-1 has three regions that have homology to α-catenin/vinculin, previously designated as the VH1 domain, the coiled-coil domain, and the VH2 domain (16, 23). This domain organization as well as the overall sequence homology is similar to that of mammalian α-catulin. Because our rescue experiments show that the VH1 domain is functionally the most important among the three domains (see below), we chose a fragment (1–377 amino acids) that includes the VH1 domain as the bait. In this screen, we isolated a fragment of DYB-1 (395–590 amino acids) as a CTN-1-interacting partner. To further confirm which domain of CTN-1 specifically interacts with DYB-1, we subcloned DYB-1 full-length cDNA into a yeast two-hybrid assay vector and performed an assay using full-length CTN-1 and two truncated CTN-1 forms, CTN-11–377 (VH1 domain) and CTN-1355–784 (coiled-coil domain and VH2 domain). We found that the full-length CTN-1 and CTN-11–377, but not CTN-1355–784, interact with DYB-1, indicating that the N-terminal VH1 domain of CTN-1 mediates the interaction with DYB-1 (Fig. 2A).
FIGURE 2.
Identification of interaction between DYB-1/dystrobrevin and CTN-1/α-catulin using yeast two-hybrid analysis. A, VH domain 1 (amino acids 1–377) of CTN-1 interacts with DYB-1, whereas the coiled-coil (C) and VH2 domains (amino acids 355–784) do not. The full-length and truncated CTN-1 cDNA sequences were fused with the DNA binding domain of GAL4 transcription factor, whereas the DYB-1 full-length cDNA sequence was fused with the activation domain. These constructs were transformed into two haploid yeast strains, and the resulting strains were mated with each other. Diploids showing a positive interaction were selected on quadruple dropout plates (−histidine, −alanine, −leucine, −tryptophan) containing X-α-Gal. + indicates the diploid yeast colonies that grow on selective plates and display blue color, whereas − indicates the diploid yeast that cannot grow on selective plates. B, various constructs of DYB-1 were tested for a positive interaction with the VH1 domain of CTN-1. The C-terminal region of DYB-1 was found to interact with CTN-1. EF, EF hand; ZZ, ZZ-type zinc finger domain; S, syntrophin binding domain; H1 and H2, coiled-coil motifs.
Next, we sought to determine which domain of DYB-1 interacts with CTN-1. DYB-1 is highly conserved from C. elegans to humans (24). Mutations in dyb-1 cause the head-bending phenotype, which is shared by other C. elegans DAPC mutants (25). Similar to mammalian α-dystrobrevin, DYB-1 is reported to interact with DYS-1 and STN-1, an α/β-syntrophin homolog (11). DYB-1/α-dystrobrevin has the following recognizable domains: an EF domain containing two EF hand motifs, a zinc-finger domain (ZZ), a syntrophin binding domain (S), and a coiled-coil domain containing two α-helices (H1 and H2) (Fig. 2B). Using six different truncated DYB-1 constructs that have a single domain or a combination of these domains, we performed a yeast two-hybrid assay to determine specific DYB-1 domains or regions that are critical for the interaction with CTN-11–377. We found that the C-terminal region of DYB-1 that includes the second H2 helix is required for the interaction with CTN-11–377 (Fig. 2B). Taken together, these results indicate that the VH1 domain of CTN-1/α-catulin directly interacts with the C-terminal region of DYB-1/α-dystrobrevin.
VH1 Domain of CTN-1 Is Critical for CTN-1 Localization and Function
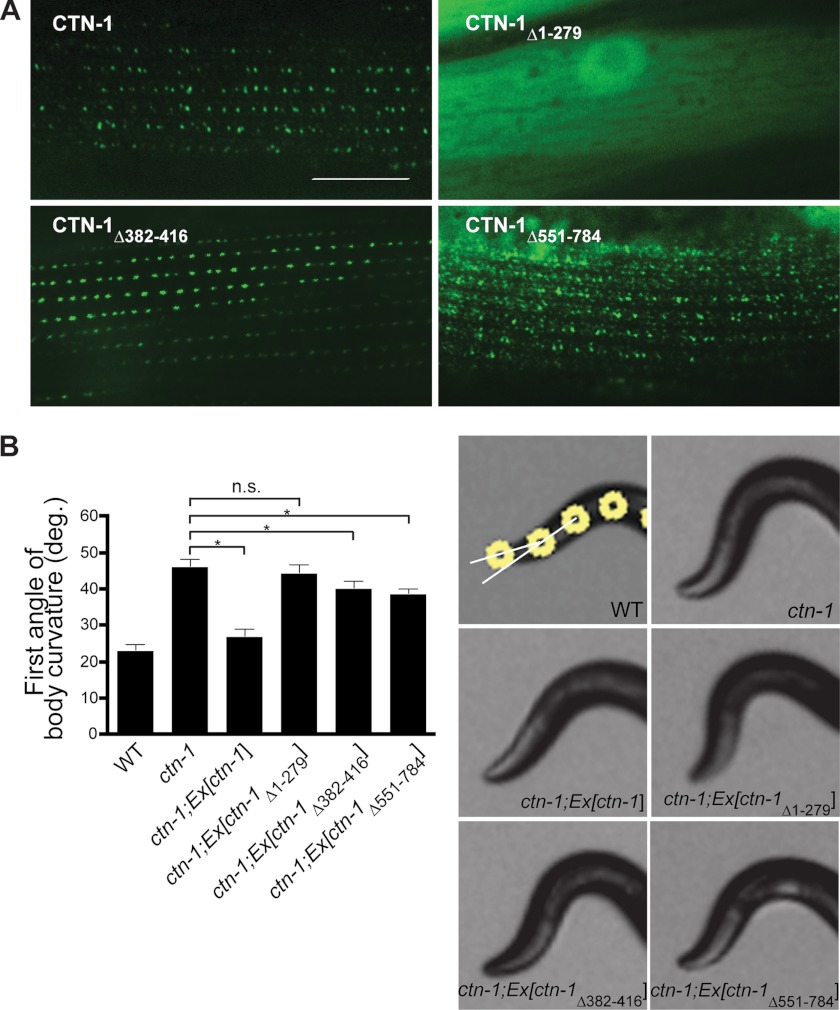
Next, we examined the functional contribution of each of the three domains of CTN-1 in vivo. We generated C. elegans transgenic animals in ctn-1(eg1167)-null mutants by introducing several ctn-1 constructs designed to express GFP-tagged full-length CTN-1, CTN-1Δ1–279 lacking the VH1 domain, CTN-1Δ382–416 lacking the coiled-coil domain, or CTN-1Δ551–784 lacking the C-terminal region. In these transgenic animals, we investigated the localization pattern of CTN-1 in muscle. We previously reported that CTN-1 and SLO-1 appear as puncta near dense bodies (16). Consistent with this report, the full-length CTN-1 exhibits a punctate pattern at the muscle membrane (Fig. 3A). As reported previously (12, 16), the major difference between SLO-1 (or CTN-1) and typical dense body markers, such as vinculin/DEB-1, is that SLO-1 and CTN-1 puncta are round in shape, whereas dense bodies show a dash-like pattern. CTN-1Δ1–279 lacking the VH1 domain is localized diffusely in the cytosolic compartment (Fig. 3A). A weak dash-like pattern was also occasionally observed at the membrane of some muscle cells (data not shown). Because the dash-like pattern was observed only in muscle cells that express high levels of the truncated CTN-1, it appears that this pattern could be an artifact resulting from overexpression. On the other hand, CTN-1Δ551–784 lacking the C-terminal region shows a punctate pattern, which is much more diffused than that of the full-length CTN-1. This result indicates that although the C-terminal region is not critical as the N-terminal region, it has a certain role in exact localization of CTN-1 near dense bodies. Finally, CTN-1Δ382–416 lacking the coiled-coil domain shows a punctate pattern similar to that of the full-length CTN-1, albeit with some differences in puncta intensity and size. Together, these results indicate that although the VH2 domain and coiled-coiled domain may have some roles in CTN-1 localization, it is the VH1 domain that is critical for binding to DYB-1 and for the localization of CTN-1 near dense bodies.
FIGURE 3.
VH1 domain of CTN-1 is critical for CTN-1 localization and function. A, representative GFP images from the muscle cells of C. elegans transgenic animals expressing GFP-tagged full-length CTN-1 or deleted forms of CTN-1. CTN-1Δ1–279 lacking the VH1 domain is localized primarily in the cytosolic compartment. CTN-1Δ551–784 lacking the C-terminal region shows a diffused punctate pattern, whereas the expression pattern of the CTN-1Δ382–416 construct that lacks the coiled-coil domain is similar to that of the full-length construct. Scale bar is 10 μm. B, first curvature angle obtained from wild-type (WT), ctn-1(eg1167) mutant, and several transgenic ctn-1 rescue strains. Error bars, S.E. n.s., not significant (p > 0.05). *, p < 0.01, unpaired Student's t test, n >5. Representative photos show the anterior portion of wild-type, ctn-1, and ctn-1 transgenic rescue strains. White circles indicate the four most anterior of the 13 midline points for a wild-type animal (21). The first curvature angles are angles between two white lines resulting from the first three dots.
To determine whether the localization defects we observed with these constructs affect the head-bending phenotype, we performed rescue experiments in ctn-1(lf) mutants with transgenes expressing the CTN-1 constructs. Consistent with our localization data, we found that the CTN-1 full-length construct, but not the VH1 deletion construct, rescues the head-bending phenotype of ctn-1 mutants (Fig. 3B). Deletion constructs lacking either the VH2 or the coiled-coil domain partially rescue the phenotype, although they do not rescue it to the level that the full-length construct does. Together, these results indicate that the VH1 domain of CTN-1 plays a critical role in muscle function, as seen by its importance in localizing CTN-1 to the correct regions of the muscle membrane, and in the rescue of the head-bending phenotype.
Localization of Both CTN-1 and DYB-1 in C. elegans Muscle Is Interdependent
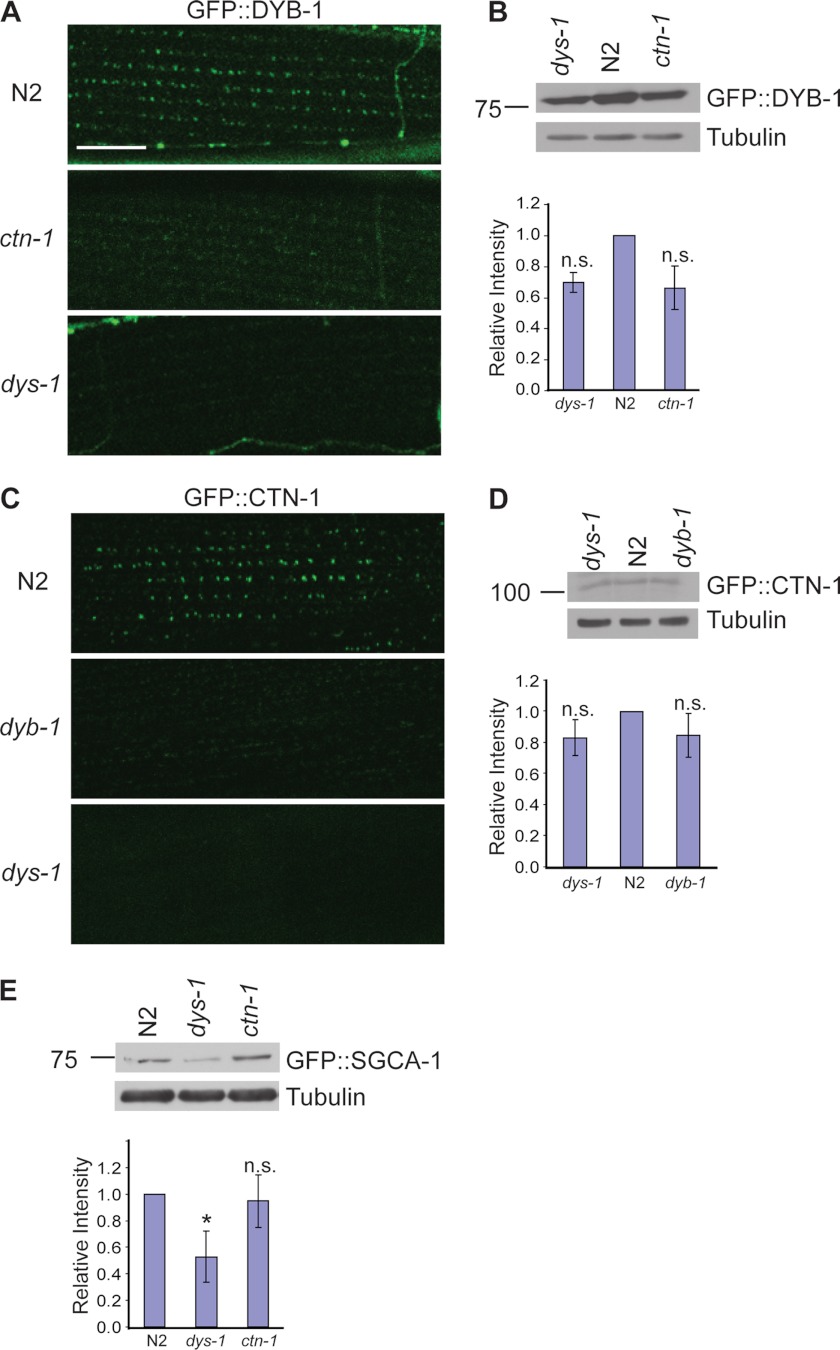
Previously, we showed that a ctn-1 mutation disrupts the localization of the DAPC; however, we were not able to identify the molecular link between CTN-1 and the DAPC (16). We now find that CTN-1 physically interacts with DYB-1, a component of the DAPC in muscle. This new finding raises the possibility that the interaction between CTN-1 and DYB-1 may mediate the proper localization and/or stability of the entire DAPC complex. Thus, we investigated whether the localization of DYB-1 is altered at the muscle membrane of dys-1 and ctn-1 mutant animals. We generated integrated transgenic lines expressing DYB-1::GFP under the control of the dyb-1 promoter and chose the line displaying the lowest expression level of DYB-1 (cimIs11). This transgene rescues the head-bending phenotype of dyb-1 mutants, indicating that DYB-1::GFP in transgenic animals reflects DYB-1 localization in vivo. Consistent with previous reports on the localization pattern of SGCA-1 and SLO-1 (12, 16), DYB-1::GFP exhibits a similar punctate pattern at the muscle membrane of wild-type animals. However, this localization pattern of DYB-1::GFP was almost completely abolished in dys-1 or ctn-1 mutants (Fig. 4A). The loss of the puncta in dys-1 or ctn-1 mutants could be due to defects in DYB-1 localization, or due to a reduction in DYB-1 stability. In mammalian skeletal muscles, failure of the DAPC to assemble at the sarcolemma reduces the stability and amount of DAPC components (26–28). We examined the DYB-1::GFP protein levels by Western blot analysis. We found that DYB-1::GFP levels were not significantly reduced either in the dys-1 mutants or in the ctn-1 mutants compared with that of wild-type (Fig. 4B). These data indicate that the loss of the puncta in dys-1 and ctn-1 mutants is largely due to the defects in DYB-1 localization.
FIGURE 4.
Proper localizations of both α-dystrobrevin and α-catulin in muscle are interdependent. A, C. elegans transgenic animals expressing integrated GFP-tagged DYB-1 under the control of the dyb-1 promoter were examined under a confocal microscope. DYB-1::GFP exhibits a distinctive punctate pattern in wild-type animals. However, this pattern is lost in both ctn-1(eg1167) and dys-1(eg33) mutant animals. Scale bar is 10 μm. B, protein levels of DYB-1::GFP in both ctn-1(eg1167) and dys-1(eg33) mutants are not significantly changed when examined by Western blot analysis using anti-GFP antibodies. Upper, representative immunoblot. Lower, quantification of relative band intensity normalized to the level of tubulin. Error bars, S.D. The change of the relative level was not significant (two-tailed Student's t test, p > 0.05). C, C. elegans transgenic animals expressing integrated GFP-tagged CTN-1 under the control of the ctn-1 promoter were examined under a confocal microscope. In wild-type animals, CTN-1::GFP exhibits a punctate pattern similar to that of DYB-1::GFP. This pattern is lost in both dyb-1(cx36) and dys-1(eg33) mutant animals. D, protein levels of CTN-1::GFP in both dyb-1(cx36) and dys-1(eg33) mutants are only slightly decreased compared with wild type when examined by Western blot analysis using anti-GFP antibodies. Upper, representative immunoblot. Lower, quantification of the relative band intensity normalized to the level of tubulin. Error bars, S.D. The change of the relative level was not significant (two-tailed Student's t test, p > 0.05). E, protein levels of SGCA-1::GFP were compared in wild-type, dys-1(eg33), and ctn-1(eg1167) animals by Western blot analysis. Compared with wild-type and ctn-1(eg1167) animals, dys-1(eg33) mutants show a significant decrease in SGCA-1::GFP expression. Upper, representative immunoblot. Lower, quantification of the relative band intensity normalized to the level of tubulin. Error bars, S.D. *, p < 0.05 (two-tailed Student's t test).
Next, we examined the localization pattern of CTN-1 at the muscle membrane. Similarly, we generated an integrated transgenic line expressing CTN-1::GFP under the control of the ctn-1 promoter (cimIs9). This transgene rescues the head-bending phenotype, indicating that CTN-1 localization in these transgenic animals reflects its normal in vivo localization. In wild-type animals CTN-1::GFP shows a punctate pattern as we reported previously (16). However, in both dys-1 and dyb-1 mutants, the punctate pattern of CTN-1::GFP disappeared (Fig. 4C). We also examined the protein levels by Western blot analysis and found that the level of CTN-1::GFP protein was not significantly changed in either the dys-1 mutant or the dyb-1 mutant (Fig. 4D). This also indicates that loss of CTN-1::GFP puncta in dys-1 and dyb-1 mutants is largely due to defects in CTN-1 localization. Taken together, our data show that the proper localizations of both DYB-1 and CTN-1 at the muscle membrane are dependent on the presence of both of these proteins.
In mouse skeletal muscle, α-dystrobrevin can be localized at the sarcolemma in the absence of a direct link to dystrophin (29). Furthermore, in α-dystrobrevin-deficient mice, other DAPC components, including dystrophin and components of the sarcoglycan complex, are still localized at the sarcolemma (22). In contrast, in dystrophin-deficient mdx muscle, all of the DAPC components are destabilized and disappear from the sarcolemma (26). These observations raise the possibility that α-dystrobrevin is not critical for the assembly of the DAPC but is involved in the localization of the DAPC at specific domains within the sarcolemma, whereas dystrophin is a key molecule that assembles the DAPC. According to this hypothesis, in dyb-1 and ctn-1 mutants, the DAPC would be dispersed throughout the muscle membrane, but would not be clustered to a region close to dense bodies. If this is the case, we expect to observe a significant reduction in the protein levels of core DAPC components in dys-1 mutants, but not in ctn-1 mutants. To test this possibility, we compared the protein levels of SGCA-1, a core component of the DAPC, in dys-1 mutants with that of ctn-1 mutants (Fig. 4E). Indeed, we found that the SGCA-1 level was significantly reduced in dys-1 mutants but was not significantly affected in ctn-1 mutants. These results strongly suggest that CTN-1 and DYB-1 do not affect the formation and assembly of the core DAPC and that instead they have a role in organizing the DAPC within the subcellular regions near dense bodies.
In Mouse Skeletal Muscle, α-Catulin and α-Dystrobrevin Exist in Same Protein Complex and Co-localize in Muscle
Based on the findings from our C. elegans genetic study, we explored the possible roles of α-catulin and α-dystrobrevin in mammalian skeletal muscle. In mammals, the single α-dystrobrevin gene produces multiple isoforms generated by alternative use of the promoter or by post-transcriptional regulation (30). Among these isoforms, α-dystrobrevin-1 (α-DB1) is the only isoform with a C-terminal extension that is homologous to the CTN-1 binding sequence of C. elegans DYB-1 (31, 32). Although α-DB1, the largest isoform, is clearly expressed in mouse skeletal muscle, it was not clear whether α-catulin is also present in skeletal muscle. Using two monoclonal antibodies specific for α-catulin, we were able to detect its expression in mouse skeletal muscle (Fig. 5A). Because we have shown in our studies that α-catulin plays a crucial role in the localization of DAPC in striated body wall muscles in C. elegans, we examined whether α-catulin interacts with α-dystrobrevin in mouse skeletal muscle. We performed reciprocal co-immunoprecipitation and Western blotting experiments using mouse muscle extracts. Immunoprecipitation with an anti-α-dystrobrevin antibody brought down α-catulin along with several different isoforms of α-dystrobrevin in wild-type muscle extracts (Fig. 5A), but not in α-dystrobrevin-null (α-DB−/−) muscle extracts (supplemental Fig. S1). Conversely, immunoprecipitation with an anti-α-catulin antibody co-precipitated α-catulin along with α-DB1, but not with other isoforms of α-dystrobrevin (Fig. 5B). These results indicate that α-catulin and α-DB1 exist in the same protein complex in mouse skeletal muscle. Further evidence that α-catulin may interact with α-dystrobrevin in mouse muscle was obtained by immunostaining of muscle. Both α-catulin and α-dystrobrevin are stained at the muscle membrane, and the two proteins are partially co-localized, although α-catulin shows variable staining in other intracellular areas (Fig. 5C).
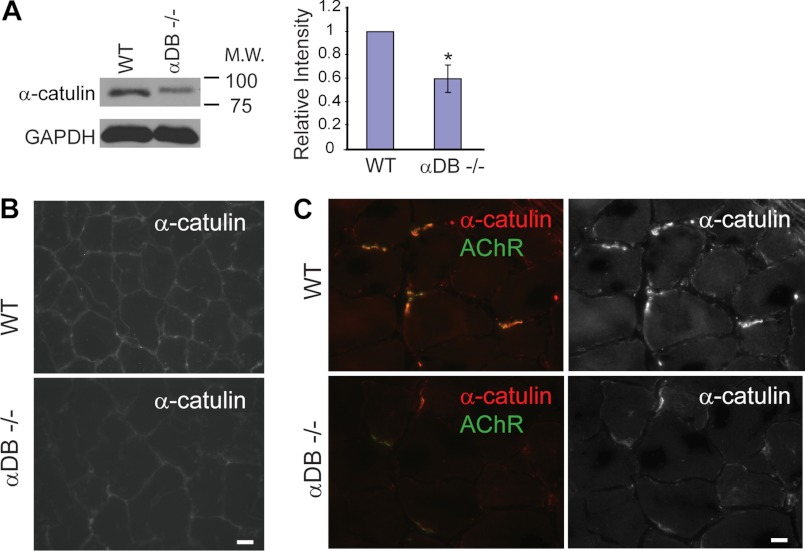
Based on physical interaction and partial co-localization of α-catulin and α-dystrobrevin, we further examined whether the expression of α-catulin is altered in α-DB−/− muscle. In Western blot analyses from muscle extracts, we found that the protein level of α-catulin is significantly reduced in α-DB−/− mice compared with control mice (Fig. 6A). Furthermore, α-catulin staining showed a reduction throughout the sarcolemma in α-DB−/− muscle (Fig. 6B). Notably, in α-DB−/− mice we observed a great reduction of α-catulin, along with acetylcholine receptors, at the neuromuscular junctions, where α-catulin staining is stronger than other areas (Fig. 6C). These results are consistent with our C. elegans results that the localization of CTN-1 is dependent on the presence of DYB-1.
FIGURE 6.
α-Catulin levels are reduced at the sarcolemma and neuromuscular junctions of α-dystrobrevin-deficient mouse muscle. A, α-catulin level is reduced in α-dystrobrevin-null (α-DB−/−) hindlimb muscle extract. Left, representative immunoblot with anti-α-catulin antibody (clone 5G8). Right, quantification of relative intensity normalized to the level of GAPDH. Error bars, S.D. *, p < 0.05 (two-tailed Student's t test). B, α-catulin is reduced at the sarcolemma of α-DB−/− muscle (tibialis anterior). Scale bar, 10 μm. C, α-catulin is reduced at the neuromuscular junctions of α-DB−/− muscle (tibialis anterior). AChR, the neuromuscular junctions were identified by the presence of acetylcholine receptors (α-bungarotoxin).
Expression of α-Catulin Is Increased When DAPC Is Compromised in Mouse Skeletal Muscle
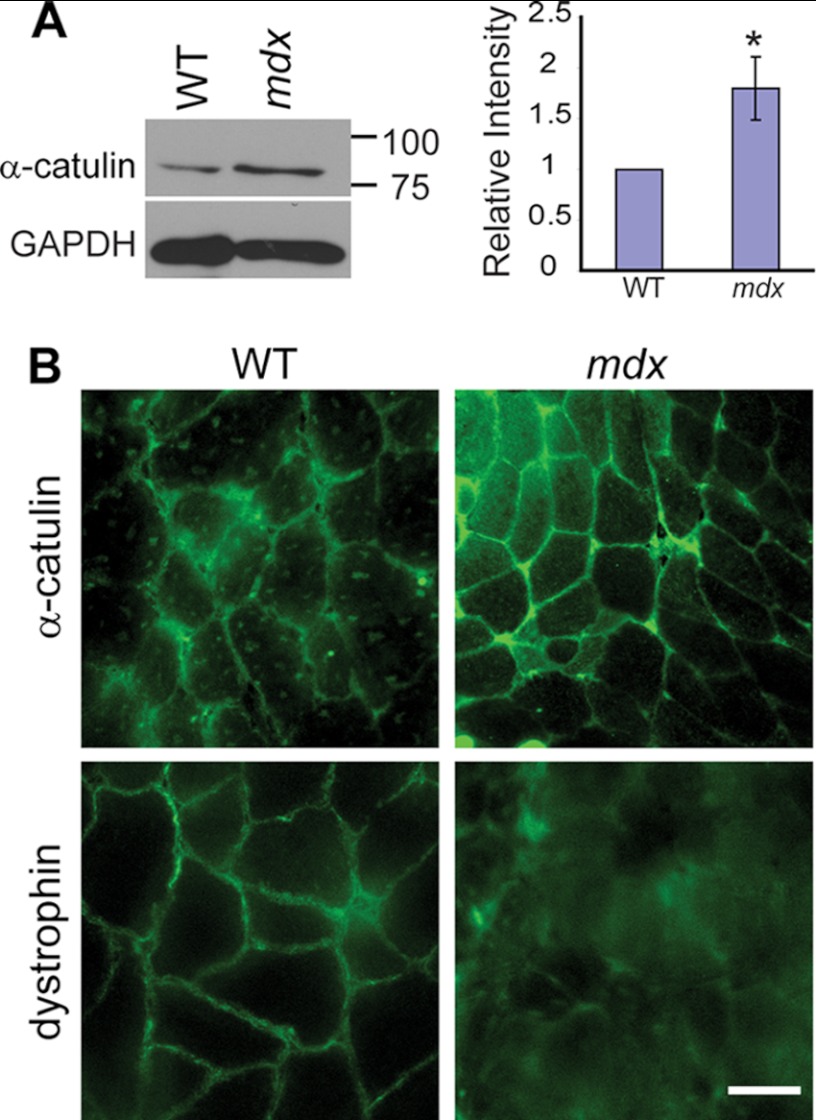
After establishing the interaction between α-catulin and α-dystrobrevin, we further explored the relationship between the DAPC and α-catulin in mouse skeletal muscle. In C. elegans muscle, we showed that DAPC and α-catulin are dependent on each other for their proper localization at the subdomain of the muscle membrane. In mammalian skeletal muscles, the DAPC is known to interact with many cytoskeletal proteins. Furthermore, the expression of these cytoskeletal proteins is increased when the DAPC is compromised. This compensatory increase is thought to play a role in protecting the weakened sarcolemma (9, 33, 34). Therefore, we compared the expression levels of α-catulin in gastrocnemius muscles from 4-week-old wild-type mice and mdx mice, which lack functional dystrophin. We chose 4-week-old mice for our experiments because mdx mice at that age have not yet undergone extensive degeneration and regeneration of skeletal muscle. In a Western blot analysis, we found that the amount of α-catulin protein in mdx muscle was significantly increased compared with wild-type muscle (Fig. 7A). These results indicate that, as a compensatory mechanism, dystrophic skeletal muscle increases the expression or stability of α-catulin. Next, we examined whether this increased α-catulin is limited to the sarcolemma or is diffused throughout the cytoplasmic compartment. Immunostaining of frozen muscle sections shows that α-catulin was increased throughout the sarcolemma of mdx mice, although α-catulin was more prominent in the sarcolemma of small muscle fibers that are likely to represent occasional regenerating muscle fibers (Fig. 7B and supplemental Fig. S2). Although we cannot completely exclude that other inflammatory cells or regenerating skeletal muscle cells contribute to an increased α-catulin level, these results indicate that, in mdx mice, α-catulin is specifically elevated at the sarcolemma. We conclude that the increased levels of α-catulin in mdx muscle are the result of a compensatory response to the disrupted DAPC integrity.
FIGURE 7.
Increased levels of α-catulin in mdx mouse muscles. A, α-catulin level is increased in mdx hindlimb muscle extracts. Upper, representative Western blot of muscle extracts from 4-week-old control or mdx mice using anti-α-catulin antibody (clone 5G8). Lower, quantification of relative band intensity normalized to the level of GAPDH. Error bars, S.D. *, p < 0.05 (two-tailed Student's t test). B, frozen sections of gastrocnemius muscles from control or mdx mouse were stained with either anti-α-catulin antibody (clone 5G8) or anti-dystrophin. The stained sections were examined under a wide-field fluorescent microscope. Scale bar, 30 μm.
DISCUSSION
In this study, we uncovered that CTN-1/α-catulin interacts with DYB-1/dystrobrevin in C. elegans and also in mouse muscles. Our data show that disruption of the interaction between CTN-1 and DYB-1 abolishes normal localization of the DAPC in C. elegans muscle. Importantly, the interaction between α-catulin and α-dystrobrevin is conserved in mammalian muscle, as shown by the fact that in mouse muscle α-catulin exists in the same protein complex as α-DB1, and the two proteins co-localize by immunofluorescence microscopy. Intriguingly, we observed a compensatory increase of α-catulin expression levels in the skeletal muscle of dystrophin-deficient mdx mice, in which the DAPC is disassembled and the mechanical link between the costamere and the sarcolemma is lost.
Despite extensive proteome analysis based on biochemical purification of the DAPC in mammalian skeletal muscles, α-catulin has not been identified as a protein that interacts with the DAPC. α-Dystrobrevin in mammals produces multiple isoforms generated by alternative promoter use or by post-transcriptional regulation (30). Most of the α-dystrobrevin isoforms are distributed throughout the sarcolemma and the neuromuscular junction. With the exception of an isoform lacking the syntrophin and dystrophin binding sites, all of the α-dystrobrevin isoforms interact with the DAPC at the sarcolemma (31, 32). Among these many different isoforms, α-DB1 is the sole α-DB isoform that co-precipitates with α-catulin. Hence, the amount of α-catulin in the purified mammalian dystrophin complex could be relatively low. This may partially explain the failure to identify α-catulin using a proteomics approach. In contrast, C. elegans expresses a single form of dystrobrevin that interacts with α-catulin. We identified ctn-1 as an important player in DAPC localization and function by means of a forward genetic screen in C. elegans (16), which allowed us to circumvent problems associated with biochemical purification such as abundance and solubility of proteins and stability of the interaction. This genetic screen can be used further to identify additional novel genes that have roles in the localization and function of the DAPC.
We previously reported that CTN-1 localizes SLO-1 channels near dense bodies through the DAPC in muscles (16). We showed that mutation in ctn-1 disrupts normal localization of SGCA-1, one of the DAPC components, and that mutations in various components of DAPC disrupt normal localization of SLO-1 in C. elegans striated muscle. In the present study, we found that CTN-1 interacts directly with α-dystrobrevin, one component of the DAPC, thereby providing the link between CTN-1 and DAPC in muscles. Our data are at odds with another result reported by Chen et al. (35). The observation that the localization of DYB-1 is not altered in the ctn-1 mutants led these authors to conclude that CTN-1 localizes SLO-1 near dense bodies independently of the dystrophin complex. We interpret that this discrepancy likely stems from the differences in the ctn-1 alleles used. The eg1167 allele used in our study has an early nonsense codon mutation at amino acid 144 that eliminates all of the functional domains, including the VH1 domain, which is necessary for binding to DYB-1. However, the zw1 allele used by the other group has a nonsense mutation at amino acid 398, so it is likely to still express a functional VH1 domain. We previously observed that another allele, cim6, which was isolated from the same genetic screen as was eg1167, retains the VH1 domain and is not a null allele, as the cim6 allele clearly causes the head-bending phenotype, but does not result in an abnormality in locomotory speed (16). Therefore, although our study does not necessarily exclude the possibility that CTN-1 interacts with SLO-1 directly, it indicates that CTN-1 requires the dystrophin complex for SLO-1 localization in muscle.
Lyssand et al. reported that α-catulin interacts with dystrobrevin in a human embryonic kidney cell line (36). They speculated that the C terminus of dystrobrevin mediates the interaction with α-catulin because α-catulin is only co-purified with the isoform that retains the extra C terminus. We confirmed this prediction and found that the C terminus of DYB-1 is indeed required for its binding to CTN-1. However, our data from yeast two-hybrid analysis indicate that the α-helix H2 proximal to the C-terminal region is also necessary for this binding. Another important difference between the report from Lyssand et al. and our results is that the earlier report did not find a correlation between α-dystrobrevin and α-catulin in mouse skeletal muscle. In contrast, we found that α-catulin is expressed at the sarcolemma of skeletal muscle and that its expression is reduced in α-DB−/− muscle. This discrepancy may have resulted from the specificity of the antibodies used in the previous study. The anti-α-catulin antibodies used in the present study were made by our group and have been validated in other studies (18). Although Lyssand et al. (36) showed that α-catulin interacts with α- and β-dystrobrevin 1 in tissue culture cells, it was not clear from those studies whether lack of α-catulin disrupts the integrity and localization of the DAPC. Given that disrupted integrity of the DAPC is extensively documented as a cause of muscular dystrophy, addressing this question is important for understanding the pathogenesis of muscular dystrophy. Our present study demonstrates that proper membrane localization of CTN-1/α-catulin and DYB-1/α-dystrobrevin is interdependent in vivo and directly links α-catulin to the localization of the DAPC near dense bodies, structures that are homologous to mammalian costameres.
The role of α-dystrobrevin in muscle degeneration has not been clear thus far. Although early biochemical studies established α-dystrobrevin as a component of the DAPC (37), genetic defects in α-dystrobrevin have not been linked to human muscular dystrophy. Furthermore, disruption of α-dystrobrevin in mice does not result in disintegration of the DAPC in muscle and does not cause massive skeletal and cardiac muscle degeneration (22). These data led to the idea that dystrobrevin contributes to muscle stability as a mediator of signaling function, rather than structural function, of the DAPC (22). Recent findings from vertebrate comparative genome analysis of α-dystrobrevin show that α-dystrobrevin generates numerous isoforms by alternative splicing (30). These diverse isoforms can control specific interactions with different signaling proteins by shuffling specific domains responsible for the interactions. In addition to interactions with dystrophin, syntrophins, and the sarcoglycan complex, α-dystrobrevin interacts, through its different domains, with several additional cytoskeletal proteins, including syncoilin (38), synemin (or desmuslin) (39), kinesin heavy chain (40), and DAMAGE (41). Such interactions with a variety of proteins correlate with the observation that the DAPC is broadly localized throughout the sarcolemma, although it is enriched at costameres. In C. elegans, dystrobrevin/DYB-1 and α-sarcoglycan/SGCA-1 are observed as puncta only near dense bodies and do not show broad expression in the muscle membrane. DYB-1 is also expressed as a single CTN-1-binding form, which is homologous to α-DB1. Together, these data indicate that the C. elegans DAPC is exclusively associated with a region close to dense bodies (12). Our current study identifies α-catulin as another protein that interacts with α-dystrobrevin in muscle. Importantly, mutations in ctn-1 cause defects in the localization of DYB-1 and SGCA-1/α-sarcoglycan near dense bodies without significantly altering the levels of DYB-1 and SGCA-1 proteins. Likewise, a mutation in dyb-1 also disrupts the localization of CTN-1 near dense bodies without reducing the amount of CTN-1 protein. Based on previous data and our new data, we propose that, although mutations in dyb-1 and ctn-1 do not significantly influence the formation and assembly of the DAPC, the interaction between DYB-1 and CTN-1 stabilizes or maintains the localization of the DAPC near dense bodies of C. elegans muscle. The absence of either protein disrupts the tight association of the dystrophin complex with unidentified cytoskeletal elements located near dense bodies, thereby compromising normal localization of the DAPC. By extension, the interaction between α-catulin and α-dystrobrevin is likely to play a role in maintaining or stabilizing the mammalian DAPC near costameres, which are structures homologous to C. elegans dense bodies. In this case, the DAPC form with α-DB1 and α-catulin is expected to be concentrated at costameres, and other forms of the DAPC that are not associated with α-catulin may be localized throughout the sarcolemma.
Several cytoskeletal proteins show compensatory increases when dystrophin is mutated. These proteins include utrophin, γ-actin, and syncoilin (9, 33, 34, 42). Such compensatory increases have been postulated to fortify the weakened sarcolemma through recruitment of parallel mechanical linkages. Overexpression of utrophin protects mdx muscle from the disorganization of costameric actin. Similarly, overexpression of γ-actin improves the dystrophic phenotype of mdx muscle (43). In this context, the increase of sarcolemmal α-catulin levels in mdx muscle may be a compensatory mechanism that has a role in protection from sarcolemmal damage. Interestingly, such increases of α-catulin were not observed in α-DB−/− muscle, where other core components of the dystrophin complex remain intact and the dystrophic phenotype is mild. Thus, we speculate that a weakened or damaged sarcolemma due to a complete disruption of the dystrophin complex induces the expression of α-catulin. In C. elegans we observed a reduction of CTN-1 localization at the muscle membrane of dys-1 mutants. We conjecture that the compensatory mechanism may be an example of a complex gene-regulatory network present in higher organisms, but not in nematodes. Alternatively, a lack of a sarcolemmal repair mechanism in C. elegans muscle may preclude such compensatory programs.
In the future, it will be important to understand the role of α-catulin in the pathogenesis of muscle diseases. Furthermore, the identification of additional α-catulin-interacting proteins will illuminate how the DAPC is organized near dense bodies/costameres and how α-catulin is retained at the sarcolemma in the absence of the DAPC. In summary, evolutionary conservation of the DAPC composition and of the interaction of α-catulin with dystrobrevin from C. elegans to mammals strongly suggests that α-catulin plays an essential role in the correct localization and functioning of the dystrophin complex in striated muscle.
Acknowledgments
Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources. Anti-dystrophin antibody and anti-α-tubulin antibody were obtained from Developmental Hybridoma Bank developed under auspices of the NICHD, National Institutes of Health.
This work was supported, in whole or in part, by the National Institute of Health Grants R01NS058814 (to H. K.), R01NS059853 (to J. R. S.), and R01AR058043 (to J. X. D.). This work was also supported by the Concerted Research Actions (GOA) of Ghent University and the Research Foundation–Flanders (FWO-Vlaanderen) (to F. v. R.).

This article contains supplemental Figs. S1 and S2.
- DAPC
- dystrophin-associated protein complex
- α-DB1
- α-dystrobrevin-1
- IP
- immunoprecipitation
- VH
- vinculin homology.
REFERENCES
- 1. Durbeej M., Campbell K. P. (2002) Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 12, 349–361 [DOI] [PubMed] [Google Scholar]
- 2. Dalkilic I., Kunkel L. M. (2003) Muscular dystrophies: genes to pathogenesis. Curr. Opin. Genet. Dev. 13, 231–238 [DOI] [PubMed] [Google Scholar]
- 3. Davies K. E., Nowak K. J. (2006) Molecular mechanisms of muscular dystrophies: old and new players. Nat. Rev. Mol. Cell. Biol. 7, 762–773 [DOI] [PubMed] [Google Scholar]
- 4. Porter G. A., Dmytrenko G. M., Winkelmann J. C., Bloch R. J. (1992) Dystrophin co-localizes with β-spectrin in distinct subsarcolemmal domains in mammalian skeletal muscle. J. Cell Biol. 117, 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pardo J. V., Siliciano J. D., Craig S. W. (1983) Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J. Cell Biol. 97, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ervasti J. M. (2003) Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 278, 13591–13594 [DOI] [PubMed] [Google Scholar]
- 7. Williams M. W., Bloch R. J. (1999) Extensive but coordinated reorganization of the membrane skeleton in myofibers of dystrophic (mdx) mice. J. Cell Biol. 144, 1259–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rybakova I. N., Patel J. R., Ervasti J. M. (2000) The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 150, 1209–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanft L. M., Rybakova I. N., Patel J. R., Rafael-Fortney J. A., Ervasti J. M. (2006) Cytoplasmic γ-actin contributes to a compensatory remodeling response in dystrophin-deficient muscle. Proc. Natl. Acad. Sci. U.S.A. 103, 5385–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grisoni K., Martin E., Gieseler K., Mariol M. C., Ségalat L. (2002) Genetic evidence for a dystrophin-glycoprotein complex (DGC) in Caenorhabditis elegans. Gene 294, 77–86 [DOI] [PubMed] [Google Scholar]
- 11. Grisoni K., Gieseler K., Mariol M. C., Martin E., Carre-Pierrat M., Moulder G., Barstead R., Ségalat L. (2003) The stn-1 syntrophin gene of C. elegans is functionally related to dystrophin and dystrobrevin. J. Mol. Biol. 332, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 12. Kim H., Pierce-Shimomura J. T., Oh H. J., Johnson B. E., Goodman M. B., McIntire S. L. (2009) The dystrophin complex controls bk channel localization and muscle activity in Caenorhabditis elegans. PLoS Genet. 5, e1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carre-Pierrat M., Grisoni K., Gieseler K., Mariol M. C., Martin E., Jospin M., Allard B., Ségalat L. (2006) The SLO-1 BK channel of Caenorhabditis elegans is critical for muscle function and is involved in dystrophin-dependent muscle dystrophy. J. Mol. Biol. 358, 387–395 [DOI] [PubMed] [Google Scholar]
- 14. Sancar F., Touroutine D., Gao S., Oh H. J., Gendrel M., Bessereau J. L., Kim H., Zhen M., Richmond J. E. (2011) The dystrophin-associated protein complex maintains muscle excitability by regulating Ca2+-dependent K+ (BK) channel localization. J. Biol. Chem. 286, 33501–33510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moerman D. G., Williams B. D. (January 16, 2006) WormBook, ed. The C. elegans Research Community. doi/ 10.1895/wormbook.1.7.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- 16. Abraham L. S., Oh H. J., Sancar F., Richmond J. E., Kim H. (2010) An α-catulin homologue controls neuromuscular function through localization of the dystrophin complex and BK channels in Caenorhabditis elegans. PLoS Genet. 6, e1001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiesner C., Winsauer G., Resch U., Hoeth M., Schmid J. A., van Hengel J., van Roy F., Binder B. R., de Martin R. (2008) α-Catulin, a Rho signalling component, can regulate NF-κB through binding to IKK-β, and confers resistance to apoptosis. Oncogene 27, 2159–2169 [DOI] [PubMed] [Google Scholar]
- 19. Ghesquière B., Goethals M., Van Damme J., Staes A., Timmerman E., Vandekerckhove J., Gevaert K. (2006) Improved tandem mass spectrometric characterization of 3-nitrotyrosine sites in peptides. Rapid Commun. Mass Spectrom. 20, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 20. Gengyo-Ando K., Mitani S. (2000) Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64–69 [DOI] [PubMed] [Google Scholar]
- 21. Pierce-Shimomura J. T., Chen B. L., Mun J. J., Ho R., Sarkis R., McIntire S. L. (2008) Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 20982–20987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grady R. M., Grange R. W., Lau K. S., Maimone M. M., Nichol M. C., Stull J. T., Sanes J. R. (1999) Role for α-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat. Cell Biol. 1, 215–220 [DOI] [PubMed] [Google Scholar]
- 23. Janssens B., Staes K., van Roy F. (1999) Human α-catulin, a novel α-catenin-like molecule with conserved genomic structure, but deviating alternative splicing. Biochim. Biophys. Acta 1447, 341–347 [DOI] [PubMed] [Google Scholar]
- 24. Gieseler K., Mariol M. C., Bessou C., Migaud M., Franks C. J., Holden-Dye L., Ségalat L. (2001) Molecular, genetic, and physiological characterisation of dystrobrevin-like (dyb-1) mutants of Caenorhabditis elegans. J. Mol. Biol. 307, 107–117 [DOI] [PubMed] [Google Scholar]
- 25. Gieseler K., Bessou C., Ségalat L. (1999) Dystrobrevin- and dystrophin-like mutants display similar phenotypes in the nematode Caenorhabditis elegans. Neurogenetics 2, 87–90 [DOI] [PubMed] [Google Scholar]
- 26. Ohlendieck K., Campbell K. P. (1991) Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 115, 1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Straub V., Duclos F., Venzke D. P., Lee J. C., Cutshall S., Leveille C. J., Campbell K. P. (1998) Molecular pathogenesis of muscle degeneration in the δ-sarcoglycan-deficient hamster. Am J. Pathol. 153, 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Durbeej M., Cohn R. D., Hrstka R. F., Moore S. A., Allamand V., Davidson B. L., Williamson R. A., Campbell K. P. (2000) Disruption of the β-sarcoglycan gene reveals pathogenetic complexity of limb-girdle muscular dystrophy type 2E. Mol. Cell 5, 141–151 [DOI] [PubMed] [Google Scholar]
- 29. Crawford G. E., Faulkner J. A., Crosbie R. H., Campbell K. P., Froehner S. C., Chamberlain J. S. (2000) Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 150, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Böhm S. V., Constantinou P., Tan S., Jin H., Roberts R. G. (2009) Profound human/mouse differences in α-dystrobrevin isoforms: a novel syntrophin-binding site and promoter missing in mouse and rat. BMC Biol. 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peters M. F., Sadoulet-Puccio H. M., Grady M. R., Kramarcy N. R., Kunkel L. M., Sanes J. R., Sealock R., Froehner S. C. (1998) Differential membrane localization and intermolecular associations of α-dystrobrevin isoforms in skeletal muscle. J. Cell Biol. 142, 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nawrotzki R., Loh N. Y., Ruegg M. A., Davies K. E., Blake D. J. (1998) Characterisation of α-dystrobrevin in muscle. J. Cell Sci. 111, 2595–2605 [DOI] [PubMed] [Google Scholar]
- 33. Brown S. C., Torelli S., Ugo I., De Biasia F., Howman E. V., Poon E., Britton J., Davies K. E., Muntoni F. (2005) Syncoilin up-regulation in muscle of patients with neuromuscular disease. Muscle Nerve 32, 715–725 [DOI] [PubMed] [Google Scholar]
- 34. Tinsley J. M., Potter A. C., Phelps S. R., Fisher R., Trickett J. I., Davies K. E. (1996) Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature 384, 349–353 [DOI] [PubMed] [Google Scholar]
- 35. Chen B., Liu P., Wang S. J., Ge Q., Zhan H., Mohler W. A., Wang Z. W. (2010) α-Catulin CTN-1 is required for BK channel subcellular localization in C. elegans body wall muscle cells. EMBO J. 29, 3184–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lyssand J. S., Whiting J. L., Lee K. S., Kastl R., Wacker J. L., Bruchas M. R., Miyatake M., Langeberg L. K., Chavkin C., Scott J. D., Gardner R. G., Adams M. E., Hague C. (2010) α-Dystrobrevin-1 recruits α-catulin to the α1D-adrenergic receptor/dystrophin-associated protein complex signalosome. Proc. Natl. Acad. Sci. U.S.A. 107, 21854–21859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshida M., Yamamoto H., Noguchi S., Mizuno Y., Hagiwara Y., Ozawa E. (1995) Dystrophin-associated protein A0 is a homologue of the Torpedo 87K protein. FEBS Lett. 367, 311–314 [DOI] [PubMed] [Google Scholar]
- 38. Poon E., Howman E. V., Newey S. E., Davies K. E. (2002) Association of syncoilin and desmin: linking intermediate filament proteins to the dystrophin-associated protein complex. J. Biol. Chem. 277, 3433–3439 [DOI] [PubMed] [Google Scholar]
- 39. Bhosle R. C., Michele D. E., Campbell K. P., Li Z., Robson R. M. (2006) Interactions of intermediate filament protein synemin with dystrophin and utrophin. Biochem. Biophys. Res. Commun. 346, 768–777 [DOI] [PubMed] [Google Scholar]
- 40. Ceccarini M., Torreri P., Lombardi D. G., Macchia G., Macioce P., Petrucci T. C. (2005) Molecular basis of dystrobrevin interaction with kinesin heavy chain: structural determinants of their binding. J. Mol. Biol. 354, 872–882 [DOI] [PubMed] [Google Scholar]
- 41. Albrecht D. E., Froehner S. C. (2004) DAMAGE, a novel α-dystrobrevin-associated MAGE protein in dystrophin complexes. J. Biol. Chem. 279, 7014–7023 [DOI] [PubMed] [Google Scholar]
- 42. Newey S. E., Gramolini A. O., Wu J., Holzfeind P., Jasmin B. J., Davies K. E., Blake D. J. (2001) A novel mechanism for modulating synaptic gene expression: differential localization of α-dystrobrevin transcripts in skeletal muscle. Mol. Cell. Neurosci. 17, 127–140 [DOI] [PubMed] [Google Scholar]
- 43. Baltgalvis K. A., Jaeger M. A., Fitzsimons D. P., Thayer S. A., Lowe D. A., Ervasti J. M. (2011) Transgenic overexpression of γ-cytoplasmic actin protects against eccentric contraction-induced force loss in mdx mice. Skeletal Muscle 1, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]