Background: Because of their biochemical properties, newly synthesized ribosomal proteins are prone to aggregation.
Results: Yar1 directly interacts with free Rps3, accompanies it from the cytoplasm to the nucleus and maintains its solubility.
Conclusion: Yar1 acts as an anti-aggregation factor for Rps3.
Significance: Ribosomal proteins require protection from aggregation.
Keywords: Chaperone Chaperonin, Nuclear Transport, Protein Aggregation, Ribosome Assembly, Ribosomes, Yeast, Ribosomal Protein, Rps3, Yar1
Abstract
2000 ribosomes have to be synthesized in yeast every minute. Therefore the fast production of ribosomal proteins, their efficient delivery to the nucleus and correct incorporation into ribosomal subunits are prerequisites for optimal growth rates. Here, we report that the ankyrin repeat protein Yar1 directly interacts with the small ribosomal subunit protein Rps3 and accompanies newly synthesized Rps3 from the cytoplasm into the nucleus where Rps3 is assembled into pre-ribosomal subunits. A yar1 deletion strain displays a similar phenotype as an rps3 mutant strain, showing an accumulation of 20S pre-rRNA and a 40S export defect. The combination of an rps3 mutation with a yar1 deletion leads to an enhancement of these phenotypes, while increased expression of RPS3 suppresses the defects of a yar1 deletion strain. We further show that Yar1 protects Rps3 from aggregation in vitro and increases its solubility in vivo. Our data suggest that Yar1 is a specific chaperone for Rps3, which serves to keep Rps3 soluble until its incorporation into the pre-ribosome.
Introduction
The synthesis of ribosomes is one of the major activities of a eukaryotic cell involving the action of almost 200 trans-acting factors that participate in the formation of a large 60S and a small 40S subunit (1). The challenge of this process is to correctly assemble one (in the case of 40S) to three (60S) ribosomal RNAs (rRNAs)2 and many different ribosomal proteins to form a complexly structured molecular machine, which is capable of accurately translating the genetic code into the amino acid sequence of proteins.
In the biogenesis pathway, a common precursor particle for the small and large ribosomal subunits is formed in the nucleolus, the 90S particle, containing the 35S pre-rRNA or processed versions thereof, ribosomal proteins and a large number of non-ribosomal factors. This precursor undergoes a complex series of protein assembly and disassembly as well as rRNA processing (see Fig. 5A) and modification events. During these maturation steps, an rRNA cleavage event separates the common precursor into a pre-40S and pre-60S particle. These particles independently undergo further maturation events, which do not only take place in the nucleolus and the nucleoplasm, but also following nuclear export of pre-ribosomal particles, within the cytoplasm. For recent reviews on ribosome biogenesis see Refs. 2–6.
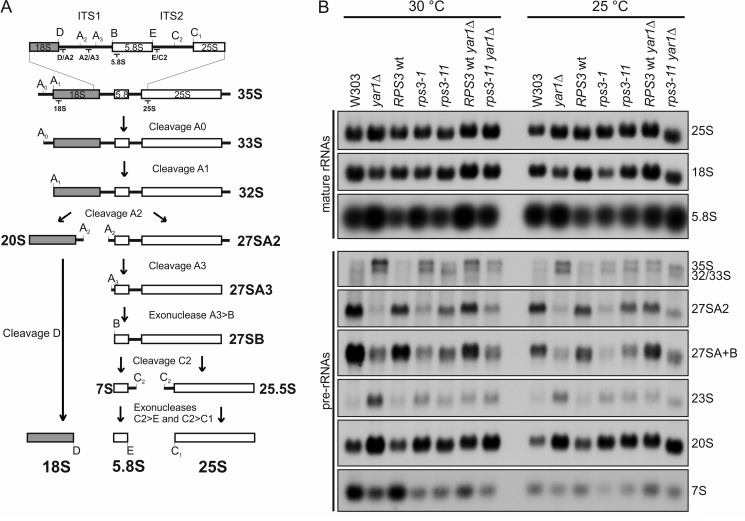
FIGURE 5.
yar1 and rps3 mutants accumulate 20S pre-rRNA. A, simplified rRNA processing pathway in yeast. Only the major pathway for generation of the 5′-end of the 5.8S rRNA is shown. The rRNA cleavage sites and the binding sites of the probes used for Northern blotting are indicated. ITS1 and 2: Internal transcribed spacers 1 and 2. In the course of pre-rRNA processing, the 35S pre-rRNA undergoes a series of endonucleolytic processing events at sites A0, A1, and A2 that lead to the separation of the 20S and 27SA2 pre-rRNAs. Endo- and exonucleolytic processing steps of the 27SA2 pre-rRNA finally yield the mature 25S and 5.8S rRNAs contained in 60S subunits. In the cytoplasm, the final processing step in 40S maturation takes place when the 20S pre-rRNA is converted into the 18S rRNA by endonucleolytic cleavage at processing site D. The aberrant 23S RNA, which is generated by premature cleavage of the 35S pre-rRNA at site A3 is not shown. B, steady-state levels of pre-rRNA and mature rRNA in yar1 and rps3 mutants. Cells were grown at 25 °C or 30 °C to an A600 of 0.6. RNA was isolated, separated by agarose gel electrophoresis, and transferred to a nylon membrane. Pre-rRNA processing intermediates were detected by Northern blotting using the following probes: “A2/A3” for detection of 35S, 33S/32S, 27SA2, and 23S RNAs, “E/C2” for detection of 27SA+B (27SA2, 27SA3 and 27SB) and 7S pre-rRNAs, “D/A2” for detection of the 20S pre-rRNA. Sequences of the probes are given in “Experimental Procedures”; binding sites of the probes are indicated in A.
In addition to trans-acting factors, ribosomal proteins themselves also participate in ribosome biogenesis (7, 8). The exact roles of ribosomal proteins in this process still remain to be resolved, however, it is likely that the timely association of ribosomal proteins to the emerging ribosomal subunits is necessary to maintain and presumably also to form the correct tertiary structure of the rRNA.
Most ribosomal proteins join pre-ribosomal particles early in the ribosome biogenesis pathway, which necessitates transport from their translation site in the cytoplasm to their assembly site in the nucleus. While research on ribosome biogenesis has mostly focused on the maturation steps of pre-ribosomal particles, less is known about the path of newly translated ribosomal proteins to their incorporation site. Nuclear import of ribosomal proteins has been proposed to be mainly mediated by the importin Kap123 (9). Beside their function as import receptors, importins have been reported to exert a stabilizing function on positively charged import substrates such as histones and ribosomal proteins (10). Furthermore, the yeast Hsp70/Hsp40 chaperone system SSB-RAC, known to be engaged in co-translational folding of proteins and the nascent polypeptide-associated complex NAC were proposed to function in preventing ribosomal proteins and ribosome assembly intermediates from aggregation (11, 12).
We are studying the yeast 40S ribosomal subunit protein Rps3 as a model to investigate the path of ribosomal proteins from their translation site in the cytoplasm to their assembly site. Like most ribosomal proteins, Rps3 joins pre-ribosomal particles in the nucleus. Failure of Rps3 assembly results in late 40S maturation defects such as 40S export defects and the accumulation of 20S pre-rRNA (7). Initially, Rps3 is only weakly associated to pre-40S particles. It only becomes stably incorporated during a structural re-arrangement of helix 33 of the 18S rRNA. This leads to the formation of the characteristic protrusion of 40S subunits termed the “beak structure” (13). Non-ribosomal binding partners of Rps3 include the pre-40S components Ltv1 and Enp1, as well as the ankyrin repeat protein Yar1 (13, 14).
Here, we report that Yar1 directly interacts with Rps3 in vitro and in vivo in a ribosome-free complex. Yar1 localizes to the cytoplasm and the nucleus and is exported in an Xpo1-dependent manner. Although Yar1 is non-essential, it becomes particularly important when Rps3 is not fully functional. Its absence results in 20S pre-rRNA processing and 40S export defects. We further show that in vivo and in vitro, Yar1 increases the solubility of Rps3. Our data suggest that Yar1 is a specific chaperone for Rps3, which accompanies Rps3 from the cytoplasm into the nucleus and maintains its solubility until incorporation into evolving ribosomal subunits.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains used in this study are listed in supplemental Table S1. Deletion disruption and C-terminal tagging at the genomic locus were performed as described previously (15–17). All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in supplemental Tables S2 and S3.
Screen for rps3 Mutants that Are Synthetically Lethal with yar1Δ
To generate rps3 mutants that are synthetically lethal with yar1Δ, we developed a protocol that utilizes the principles of the red-white sectoring genome-wide synthetic lethal screen and allows to specifically screen for synthetic lethality between two genes (18). A library of mutated rps3 alleles was generated by random PCR mutagenesis of the RPS3 open reading frame using TaqDNA polymerase (NEB) with reduced concentration of dATP (120 μm instead of 200 μm) and cloning into a pRS315 vector between the unmutagenized RPS3 promoter and terminator sequences. Plasmids harboring mutagenized RPS3 were transformed into a yar1Δ rps3Δ ade2 ade3Δ screening strain containing the plasmids pRS316-RPS3-GFP (URA3-marker) and pHT4467ΔCEN-ADE3-HIS3-YAR1. Transformants were selected on SDC-leu plates and replica plated on 5-FOA containing plates. Growth on 5-FOA plates (and hence loss of the pRS316-RPS3 plasmid) is only possible for mutants containing rps3 alleles that support viability. On 5-FOA plates, most colonies exhibited a white or red-white sectoring phenotype, indicating the ability to lose the YAR1-plasmid (containing also the ADE3 marker). A red color on 5-FOA plates indicated the inability to lose the YAR1 containing plasmid and hence synthetic lethality. rps3 plasmids conferring this phenotype were isolated and re-transformed into the RPS3/YAR1 shuffle strain to confirm synthetic lethality.
Fluorescence in Situ Hybridization and Microscopy
Fluorescence in situ hybridization was carried out as described previously (19), using a Cy3-labeled ITS1 specific probe (5′-Cy3-ATG CTC TTG CCA AAA CAA AAA AAT CCA TTT TCA AAA TTA TTA AAT TTC TT-3′) for detection of ITS1-containing pre-rRNAs. Cells were examined by fluorescence microscopy on a Zeiss Axioskop microscope. Live yeast cells were imaged by fluorescence microscopy using either a Zeiss Axioskop microscope or an Olympus BX54 microscope. Leptomycin B was provided by Alexis Biochemicals.
Sucrose Gradient Analysis
Cells were grown at 25 °C in 100 ml of YPD medium to logarithmic growth phase (A600 of ∼0.6). 100 μg/ml cycloheximide was added to the cultures and after incubation for 5 min on ice, cells were pelleted and resuspended in lysis buffer (10 mm Tris/HCl, pH 7.5, 100 mm NaCl, 30 mm MgCl2, 100 μg/ml cycloheximide). After cell lysis with glass beads, 5 A260 units of the cell extracts were loaded onto 5–45% sucrose gradients and centrifuged at 180,000 × g for 2 h 45 min at 4 °C. Gradients were analyzed using a UA-6 system (Teledyne ISCO) with continuous monitoring at A254 nm.
RNA Isolation and Northern Blotting
Total RNA preparations were performed from 20 A600 units using the mechanical disruption protocol of the RNeasy minikit (Qiagen). 3 μg of RNA per sample were separated on 1.5% MOPS-agarose gels as described in the manual for the RNeasy minikit. The RNA was transferred overnight onto a Hybond N nylon membrane (Amersham Biosciences) and then cross-linked to the membrane by UV. Hybridization was performed overnight at 42 °C in 500 mm NaPO4 buffer, pH 7.2, 7% SDS, 1 mm EDTA using 5′-32P-labeled oligonucleotides with the following sequences: D/A2, 5′-GAC TCT CCA TCT CTT GTC TTC TTG-3′; A2/A3, 5′-TGT TAC CTC TGG GCC C-3′, E/C2, 5′-GGC CAG CAA TTT CAA GTT A-3′; 25 S, 5′-CTC CGC TTA TTG ATA TGC-3′; 18 S, 5′-CAT GGC TTA ATC TTT GAG AC-3′; 5.8 S, 5′-GCG TTC TTC ATC GAT GC-3′. The membranes were washed three times for 20 min at 42 °C in 40 mm NaPO4 buffer, pH 7.2, 1% SDS, and radioactivity was detected by exposing x-ray films. Membranes were regenerated by washing in 1% SDS.
Purification of Recombinant Proteins
The expression of His6-Yar1, His6-Rps3, FLAG-Rps3, as well as the co-expression of His6-Yar1 and FLAG-Rps3 was performed in a BL21 (DE3) Rosetta STAR Escherichia coli strain using the pETDuet-1 Vector (Novagen). Cells were cultured in LB-medium at 37 °C to an A600 of 0.2 to 0.3, shifted to 16 °C and induced with 0.3 mm IPTG for 20 h. Cells were harvested, and pellets were resuspended in lysis-buffer (150 mm NaCl, 50 mm Tris/HCl pH 7.4, 10 mm imidazole, 1× HP protease inhibitor (Sigma)) and lysed either in a French Pressure Cell Press (SLM Instruments) or by sonication. Cell debris and insoluble proteins were removed by centrifugation for 30 min at 40,000 × g and 4 °C. For His6 tag purifications, the supernatant was incubated with Ni-NTA-agarose beads (Qiagen) for 1 h at 4 °C on a turning wheel to bind the proteins. After washing three times in lysis buffer, bound protein was eluted with 300 mm imidazole.
For FLAG tag purification, the supernatant was incubated with anti-FLAG M2-agarose (Sigma) and after washing, the elution was performed using FLAG peptide according to manufacturer's instructions (Sigma). For investigation of the aggregation behavior of Rps3, purified FLAG-Rps3 was incubated in the absence or presence of a ∼20-fold excess of recombinant His6-Yar1 for 30 min at 4 °C on a turning wheel. Subsequently, protein aggregates were separated from soluble proteins by centrifugation at 200,000 × g for 1 h. Equal amounts of pellets and TCA-precipitated supernatants were loaded onto a 14% SDS-polyacrylamide gel and the amounts of soluble FLAG-Rps3 were analyzed by Western blotting.
Size exclusion chromatography was performed using an ÄKTA-FPLC system (GE Healthcare) with a Superdex 200 HiLoad 16/600 column (GE Healthcare) in 150 mm NaCl, 50 mm Tris/HCl (pH 7.4). Purified proteins and gel filtration fractions were analyzed on 14% SDS-polyacrylamide gels.
Overexpression of Rps3 and Yar1 in Yeast Cells and Solubility Test
FLAG-Rps3 was overexpressed in the yar1Δ strain from a plasmid containing the copper-inducible CUP1-promoter. The protein was either expressed alone or together with Yar1, which was constitutively expressed from a plasmid containing an ADH1-promoter. The strains were grown to logarithmic growth phase (A600 of ∼0.5). Prior to and 30 min after induction with 0.5 mm CuSO4, 100 ml of the cultures were harvested. Cells were lysed by mechanical disruption with glass beads in 50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1× FY protease inhibitor (Sigma), 0.5 mm PMSF, and 1 mm DTT. After centrifugation of the lysates for 5 min at 1,000 × g, the supernatants were subjected to centrifugation at 200,000 × g for 1 h. Equal amounts of pellets and TCA-precipitated supernatants were loaded onto 14% SDS-polyacrylamide gels, and the amounts of FLAG-Rps3 in both fractions were analyzed by Western blotting.
Tandem Affinity Purification (TAP) and Mass Spectrometry
TAP purifications of TAP-tagged bait proteins were performed in a buffer containing 50 mm Tris/HCl, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2, and 0.075% Nonidet P-40 as described previously (17). Tobacco etch virus (TEV) protease was preincubated with RiboLock RNase inhibitor (Fermentas) and for cleavage, dithiothreitol was added to a final concentration of 1 mm to the buffer. For analysis of the protein composition of the purified material, the eluates were TCA-precipitated and dissolved in SDS sample buffer. Samples from Yar1-TAP purifications were separated on 14% SDS-polyacrylamide gels, which allowed good separation of Yar1 (22 kDa) and Rps3 (26.5 kDa). Samples from TAP-purifications of pre-ribosomal particles were separated on NuPAGE SDS 4–12% gradient polyacrylamide gels (Invitrogen) and stained with colloidal Coomassie (Sigma). Protein spots were excised from gels and tryptically digested according to (20). Protein identification by LC-MS/MS was performed as described (21).
Western Blotting
Western blot analysis was performed using the following antibodies: polyclonal anti-Yar1 antibody (1:5,000) generated against full-length recombinant Yar1 in rabbits (Eurogentec); anti-Rps3 antibody (1:30,000 provided by Matthias Seedorf); anti-Rps8 antibody (1:5,000, provided by Giorgio Dieci); anti-GAPDH (glyceraldehydes-3-phosphate dehydrogenase) antibody (1:40,000, Cell Signaling Technology); anti-FLAG antibody (1:4,000, Sigma); anti-calmodulin-binding protein (CBP) antibody (1:10,000, Upstate (Millipore); secondary anti-rabbit horseradish peroxidase-conjugated antibody (1:15,000) (Sigma); secondary anti-mouse horseradish peroxidase-conjugated antibody (1:15,000) (Sigma). Proteins were visualized using an enhanced chemiluminescence detection kit (ECL; GE Healthcare).
RESULTS
Yar1 Forms a Stable Complex with Non-ribosome-bound Rps3
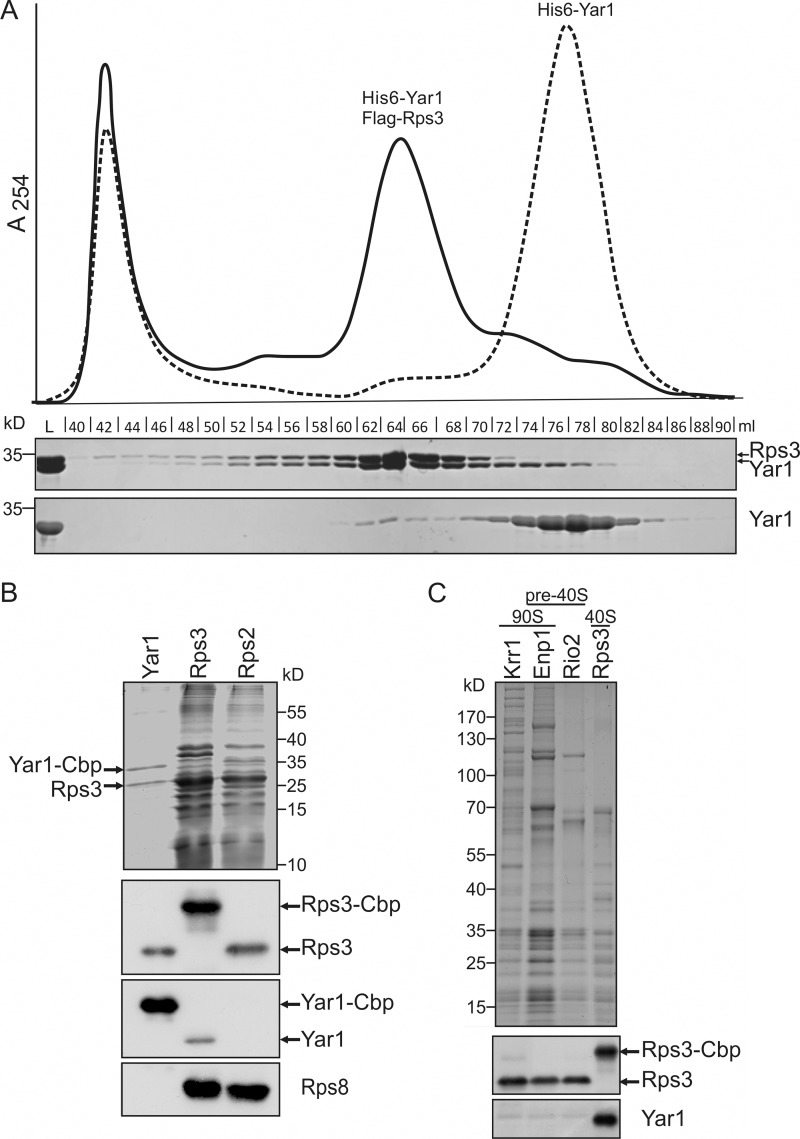
Yeast two-hybrid analyzes have suggested an interaction between Rps3 and the ankyrin repeat protein Yar1 (14). To further characterize the interaction between Yar1 and Rps3, we co-expressed both proteins in E. coli and affinity purified Yar1 via an N-terminal His6 tag. Close to stoichiometric amounts of Rps3 were recovered, confirming direct interaction of the proteins (Fig. 1A). Furthermore, both proteins co-migrated in one peak in size exclusion chromatography, verifying complex formation (Fig. 1A).
FIGURE 1.
Yar1 directly interacts with Rps3 in vitro and in vivo. A, Yar1 and Rps3 interact in vitro. His6-Yar1 and FLAG-Rps3 were co-expressed in E. coli and affinity purified via the His6 tag on Yar1. The purified material was subjected to gel filtration chromatography on a Superdex 200 column. Load (L) and collected fractions were analyzed by SDS-PAGE and Coomassie staining. As a control, purified His6-Yar1 (dashed line in the chromatogram and lower gel) was analyzed. Note that a second stoichiometric band (corresponding to Rps3) co-purified with Yar1. Rps3 eluted together with the major pool of Yar1 from the size exclusion column in a peak that occurred earlier than the Yar1 peak, confirming a larger size due to complex formation. B, Yar1 and Rps3 form a complex in vivo. Yar1-TAP, Rps3-TAP, and Rps2-TAP were purified from yeast cells. Samples were analyzed by SDS-PAGE and Coomassie staining. The indicated proteins were identified by mass spectrometry. The faint band at >55 kDa in the Yar1-TAP purification contains Hsp60 and Hsp70 proteins as well as Dbp2, which are most likely contaminations in the purification. To test for the presence of Yar1 and Rps3 in the purification, samples were analyzed by Western blotting using specific anti-Yar1 and anti-Rps3 antibodies. The 40S subunit ribosomal protein Rps8 was detected as a control and was only present in the 40S subunit-purifications (Rps3-TAP and Rps2-TAP) but not in the Yar1-TAP complex. Cbp, calmodulin-binding protein. C, Yar1 does not bind to pre-ribosomal particles. The 90S component Krr1, the 90S and pre-40S-factor Enp1, the pre-40S component Rio2 and Rps3 were TAP-purified. Samples were adjusted to equal levels of Rps3 and analyzed by SDS-PAGE followed by Coomassie staining and Western blotting using anti-Yar1 and anti-Rps3 antibodies.
To investigate the in vivo interaction between Yar1 and Rps3, we affinity purified Yar1 fused to a C-terminal TAP tag from yeast cells (Fig. 1B). A second almost stoichiometric band co-purified with Yar1 and was identified by mass spectrometry and Western blotting as Rps3. We conclude that Rps3 is the main binding partner of Yar1. As no other bands from ribosomal proteins were visible on the Coomassie-stained gel and no Rps8 was detected by Western blotting, Yar1 interacts only with free and not ribosome bound Rps3. Consistent with these findings, Yar1 was missing in an Rps2-TAP-purification, which purifies 40S ribosomal subunits containing Rps3 (Fig. 1B). In contrast, Rps3-TAP purification yielded not only 40S ribosomal subunits but as expected also Yar1 protein (Fig. 1B). To determine whether small amounts of Yar1 bound to Rps3 are found in pre-ribosomal subunits, we purified Rps3-containing 90S and pre-40S particles and tested for the presence of Yar1 by Western blotting (Fig. 1C). Yar1 was only present when Rps3 was used as bait and was not detected in any of the Rps3 containing 90S or pre-40S particles. All these data confirm that Yar1 forms a complex with free Rps3, suggesting that the interaction between Yar1 and Rps3 takes place before association of Rps3 with pre-ribosomal particles.
Yar1 Accompanies Rps3 from the Cytoplasm into the Nucleus
Interaction of Yar1 with Rps3 may occur in the cytoplasm after translation of Rps3 or in the nucleus before association of Rps3 with pre-ribosomal particles. Alternatively, Yar1 might accompany Rps3 from the cytoplasm to the nucleus and deliver it to 90S particles. To investigate these possibilities, we analyzed the cellular localization of Yar1. C-terminally GFP-tagged Yar1 was detected predominately in the cytoplasm, although a faint nuclear staining was also visible (Fig. 2A). To address whether Yar1 shuttles between the nucleus and the cytoplasm, we investigated the effect of export inhibition on Yar1 localization. For this purpose, a strain was used carrying a point mutation in XPO1/CRM1, which encodes the general exportin responsible for the nuclear export of NES-containing proteins (22, 23). Nuclear export can be blocked in this strain by treatment with the inhibitor leptomycin B (LMB) (24). An increased nuclear localization of Yar1-GFP was observed in the mutant strain after LMB treatment (Fig. 2B), demonstrating that Yar1 is a shuttling protein, which is most likely exported by Xpo1.
FIGURE 2.
Yar1 is a shuttling protein. A, Yar1 has a cytoplasmic steady-state localization. A strain containing a chromosomal YAR1-GFP fusion was grown to mid-log phase and the localization of the fusion protein was inspected by fluorescence microscopy. Note that Yar1-GFP is predominantly localized in the cytoplasm, while only a faint nuclear staining is visible. B, Yar1 transiently enters the nucleus. A leptomycin sensitive crm1 strain deleted for YAR1 and containing YAR1-GFP on a centromeric plasmid was grown at 30 °C in SDC-medium to logarithmic growth phase. Cells were inspected by fluorescence microscopy with or without treatment with 200 ng/ml leptomycin B (LMB) for 60 or 120 min. The crm1 strain transformed with plasmids expressing Rps3-GFP and Rpl25-GFP served as positive control for export inhibition of pre-40S and pre-60S subunits. C, Rps3 contains a monopartite classical NLS. Multiple Sequence Alignment of the 15 N-terminal amino acids of Rps3 from Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Homo sapiens (Hs). D, N-terminal 15 amino acids of Rps3 target a 3xyEGFP reporter to the nucleus. Amino acids 1–15 of Rps3 were fused to a triple yEGFP reporter and the fusion protein was expressed from a plasmid under the control of the ADH1 promoter in cells expressing the nucleolar marker protein Nop58-RedStar2. As controls, 3xyEGFP and SV40NLS-3xyEGFP were also localized.
Considering that Yar1 is present in the cytoplasm and nucleus and that most of the Yar1 population appears to be bound to Rps3 (Fig. 1B), it is feasible that Rps3 interacts with Yar1 in both compartments and that the two proteins are imported into the nucleus as a complex. Nuclear import of proteins is usually mediated by interaction of an import factor with a nuclear localization signal (NLS) in the cargo protein. We could not find an NLS within the amino acid sequence of Yar1, however, Rps3 contains a putative monopartite classical NLS in the N terminus (Fig. 2C). A fragment of Rps3 comprising the first 15 amino acids and including the putative NLS was sufficient to target C-terminally fused GFP into the nucleus, indicating that Rps3 contains a functional NLS (Fig. 2D). We propose that Yar1 associates with Rps3 in the cytoplasm and is transported into the nucleus in complex with Rps3. To investigate the function of Yar1 along this path, we next analyzed the phenotypes of specific mutants.
YAR1 Genetically Interacts with RPS3
Although Yar1 is a non-essential protein, yar1 deletion strains are delayed in growth, especially at low temperatures (Ref. 25 and Fig. 3A). This could mean that the interaction between Yar1 and Rps3 is required for optimal cell growth, possibly by ensuring sufficient supply of Rps3 to the ribosome biogenesis pathway. It has been previously reported that overexpression of RPS3 from a galactose-inducible promoter suppresses the growth defects of a yar1Δ strain (14). Consistent with these findings, we found that even the presence of RPS3 on a centromeric plasmid under the control of its endogenous promoter was sufficient to compensate the yar1 deletion phenotype (Fig. 3A). This suppression by plasmid-encoded RPS3 was also observed when the chromosomal copy of RPS3 was deleted (Fig. 3B; note that in an rps3Δ background with wild-type RPS3 on a plasmid, yar1 deletion affects growth only slightly).
FIGURE 3.
YAR1 and RPS3 genetically interact. A, increased RPS3 dosage suppresses the growth defect of yar1Δ cells. yar1Δ cells were transformed with plasmids containing RPS3, YAR1, and RPS15 and spotted in serial 10-fold dilution steps onto SDC-leu plates. Wild-type (WT) and yar1Δ cells transformed with empty plasmid served as controls. Plates were incubated at 25, 30, and 37 °C for 3 days. Suppression was observed with a plasmid carrying RPS3, but not with the negative control plasmid carrying a gene encoding another small subunit ribosomal protein, RPS15. B, growth phenotypes of viable rps3/yar1 and rps3 mutant strains. RPS3 and RPS3/YAR1 shuffle strains transformed with plasmids carrying the indicated wild-type and mutant rps3 alleles were shuffled on 5-FOA-containing plates and then spotted in 10-fold serial dilution steps onto YPD plates. Plates were incubated for 2 days at the indicated temperatures.
To further investigate whether a partial loss of function of Rps3 could increase the need for Yar1, we screened for rps3 alleles showing synthetic lethality with the yar1Δ deletion mutant (see “Experimental Procedures” for a detailed description of the procedure). Indeed, an rps3 mutant (rps3-1) was isolated that showed a synthetic lethal phenotype in combination with yar1Δ (supplemental Fig. S1A). Hence, in a strain carrying the rps3-1 allele, the function of Yar1 is essential. Further growth assays of the rps3-1 mutant (containing wild-type YAR1) showed that the strain was slow growing at all temperatures (Fig. 3B). The mutated rps3-1 allele contained three point mutations, resulting in T46A, R64K, and E135K amino acid exchanges. To create viable rps3 yar1 mutant strains for further phenotypic studies, we separated the three point mutations of rps3-1 and tested the growth phenotypes of the resulting mutants. Indeed, the E135K strain, hereafter termed rps3-11, was viable although slow growing in combination with yar1 deletion, in particular at low temperatures. The rps3-11 mutation alone, however, only caused very mild growth defects (Fig. 3B and supplemental Fig. S1B). In contrast, neither the T46A, nor the T46A/R64K mutations enhanced the growth defects of a yar1Δ strain (supplemental Fig. S1B).
yar1 and rps3 Mutation Leads to 40S Biogenesis Defects
To determine the physiological reason behind the growth defects observed in the mutants, we subjected all viable mutants showing growth defects (Fig. 3, A and B) to further phenotypic analyzes.
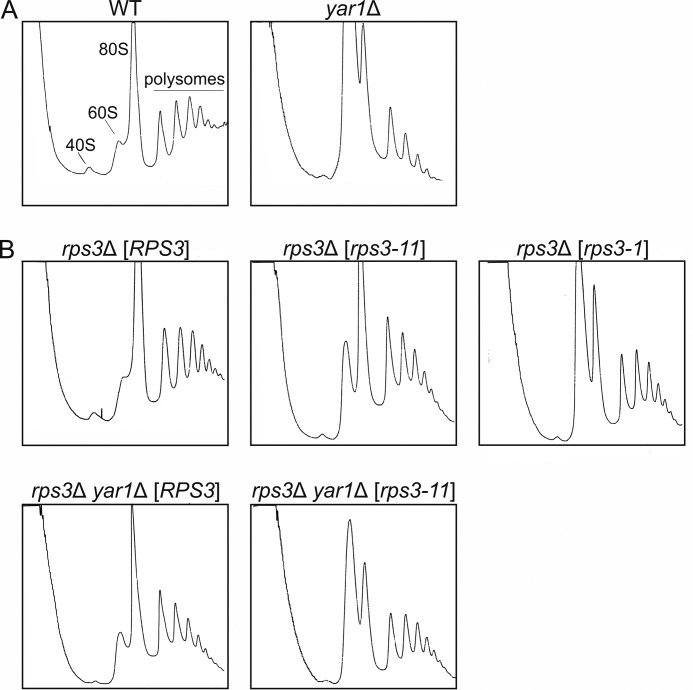
First, we recorded polysome profiles to analyze the levels of free ribosomal subunits and polysomes in yar1 and rps3 mutants (Fig. 4). The experiments were carried out with cells grown at 25 °C as the synthetic enhancement of rps3-11 with yar1Δ was strongest at 25 °C (Fig. 3B). In accordance with previous data from the Lycan laboratory, yar1Δ cells showed a reduced 40S peak and a drastically increased free 60S peak, both characteristic of 40S synthesis defects (14). Consequently, less polysomes were present in the mutant (Fig. 4A). In accordance with suppressing the growth defect of a yar1Δ mutant, expression of RPS3 from a centromeric plasmid almost completely rescued the reduction in 40S synthesis observed in the yar1Δ mutant (Fig. 4B). However, when wild-type RPS3 was exchanged with the rps3-11 allele, a significant enhancement of the phenotypes was observed in the yar1Δ strain, with a very small 40S peak, a very high 60S peak and low levels of polysomes. Similar defects were also observed in the rps3-1 mutant in the presence of wild-type YAR1 (Fig. 4B).
FIGURE 4.
40S synthesis defects in yar1 deletion-cells are enhanced by rps3 mutations. Wild-type and yar1Δ cells, containing either the chromosomal copy of RPS3 (A) or an rps3 deletion complemented by a plasmid carrying either the RPS3, rps3-1 or rps3-11 alleles (B) were grown at 25 °C to an A600 of 0.6. Five A260 units of the cell extracts were loaded onto 5–45% sucrose gradients and fractionated at 200,000 × g for 2.45 h. Polysome profiles were obtained by measuring the UV absorbance at 254 nm.
To characterize the ribosome biogenesis defects of yar1 and rps3 mutants in more detail, we next analyzed the levels of various rRNA precursors by Northern blotting (Fig. 5). In the yar1Δ strain grown at 30 and 25 °C, an accumulation of 20S pre-rRNA was observed, which is the direct precursor of the mature 18S rRNA (Fig. 5, A and B). Furthermore, 35S pre-rRNA levels were increased. This is characteristic for an inhibition of early pre-rRNA cleavages at A0, A1, and A2 frequently observed as a feedback reaction to later rRNA processing defects (26). The inhibition of these early processing steps resulted in accumulation of the aberrant 23S RNA. In addition, the delayed processing of 35S pre-rRNA led to a reduction of the levels of all other pre-rRNAs formed from the 35S pre-rRNA, including 27S and 7S pre-rRNAs. Again, while the rps3-11 strain and the yar1Δ strain with plasmid encoded RPS3 showed no significant defects, the combination resulted in an enhancement of the phenotypes observed, including the accumulation of 20S pre-rRNA. Similar rRNA processing defects were also observed in the rps3-1 strain with a YAR1 wild-type background.
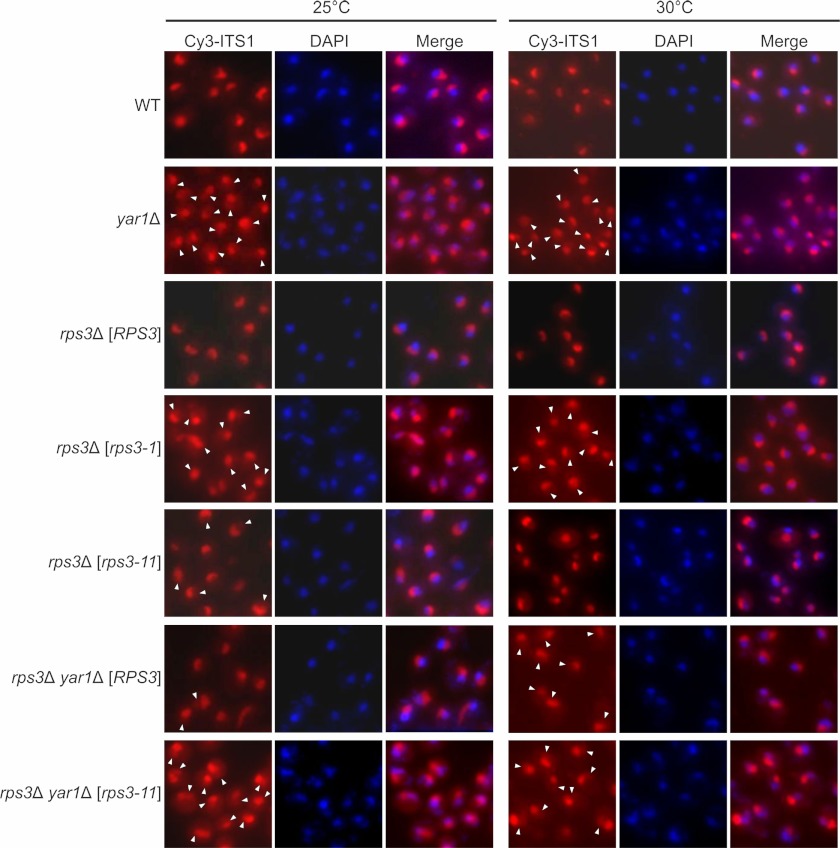
The processing of the 20S pre-rRNA into mature 18S rRNA occurs in the cytoplasm after pre-40S export. Consequently, an accumulation of 20S pre-rRNA could be explained either by the inhibition of a cytoplasmic maturation step or by a 40S export defect. To distinguish whether 20S pre-rRNA accumulates in the cytoplasm or the nucleus of yar1 and rps3 mutants, we performed in situ hybridization using a fluorescently labeled oligonucleotide complementary to the 5′ region of ITS1 (Fig. 6). This probe detects all 40S subunit rRNA precursors, but no mature 18S rRNA. Because of the relatively low abundance of pre-40S precursors in the nucleoplasm and cytoplasm, wild-type cells display an exclusively nucleolar ITS1-signal. The yar1 deletion strain, as well as the rps3-1 mutant, showed nucleoplasmic accumulation of 5′-ITS1 containing rRNA, indicative of a nuclear export defect. Furthermore, a faint cytoplasmic staining was observed particularly at 25 °C, suggesting a slight delay in cytoplasmic 20S processing. The rps3-11 mutant showed only a very mild export defect. However, in a yar1 deletion background the export defect of the rps3-11 mutant was enhanced (Fig. 6). Together, these data show that the absence of Yar1, a partial loss of function of Rps3, and a combination of yar1 and rps3 mutations all lead to 40S export defects.
FIGURE 6.
yar1 and rps3 mutants show 40S export defects. Wild-type (WT) and yar1Δ cells, either containing the chromosomal copy of RPS3 (upper two rows) or an rps3 deletion complemented by a plasmid carrying either the RPS3, rps3-1 or rps3-11 alleles (lower five rows) were grown at 25 °C or 30 °C to an A600 of 0.5. Cells were fixed with formaldehyde, spheroplasted and subjected to fluorescence in situ hybridization (FISH) using a Cy3-labeled probe complementary to a sequence in the D/A2 segment of ITS1. To visualize the nucleoplasm, cells were stained with DAPI. Arrowheads indicate the nucleoplasm in cells showing nucleoplasmic pre-rRNA accumulation.
Yar1 Protects free Rps3 from Aggregation
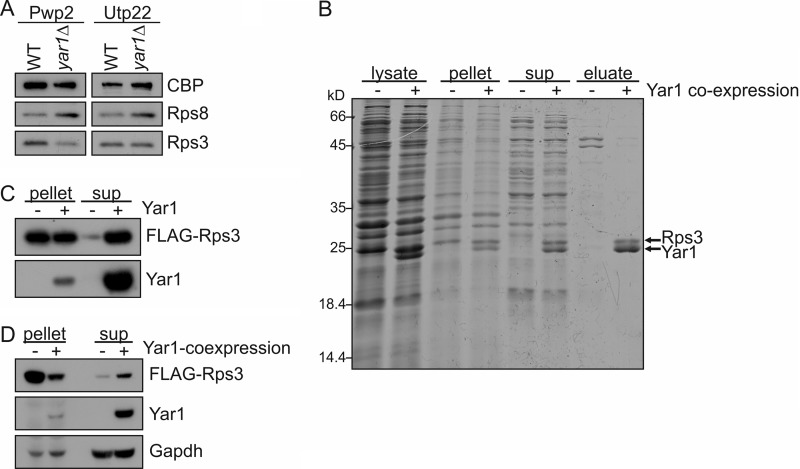
Although Yar1 is not associated with pre-ribosomal particles, the deletion of Yar1 results in 40S maturation defects. This indicates that the effect of Yar1 on 40S maturation must be exerted through Rps3. 40S export defects are also observed upon Rps3 depletion (7). This suggests that in the yar1Δ strain less Rps3 reaches its assembly site on nucleolar pre-ribosomes. Consistently, 90S particles purified from a yar1Δ strain contained reduced Rps3 levels (Fig. 7A). This effect was especially strong in early 90S particles purified via Pwp2-TAP, while a less pronounced but reproducible reduction was observed in later 90S particles purified via Utp22-TAP. No such reduction of Rps3 was observed in 40S particles (data not shown), suggesting that only 90S particles containing Rps3 further maturate into pre-40S particles.
FIGURE 7.
Yar1 keeps Rps3 soluble. A, 90S particles from a yar1Δ strain contain less Rps3. Pwp2-TAP and Utp22-TAP were purified from yeast cells. Samples were analyzed by SDS-PAGE and Western blotting using anti-CBP, anti Rps8 and anti-Rps3 antibodies. Note that in both purifications, Rps3 levels are reduced when compared with Rps8, which was used as a loading control for pre-ribosomal particles. B, Rps3 expressed in E. coli is only soluble upon co-expression of Yar1. Rps3 was either expressed alone as a His6 tag fusion (−) or co-expressed as a FLAG tag fusion with His6-Yar1 (+) in E. coli. Cells were lysed by sonication and lysates were centrifuged at 40,000 × g to pellet insoluble material. The supernatant was used for affinity purification of His6-Rps3 and His6-Yar1. Samples from the lysate, the 40,000 × g pellet, the 40,000 × g supernatant (sup), and the eluate from the affinity purification were analyzed by SDS-PAGE and Coomassie staining. C, Yar1 protects purified Rps3 from precipitation. FLAG-Rps3 was expressed in E. coli, affinity purified, and incubated for 30 min at 4 °C in the presence (+) or absence (−) of purified His6-Yar1. Thereafter, samples were subjected to centrifugation at 200,000 × g for 1 h, and equal amounts of pellet and supernatant fractions were analyzed by SDS-PAGE and Western blotting. D, overexpression of Yar1 increases the solubility of Rps3 in yeast cells. (−), expression of plasmid encoded FLAG-Rps3 under the control of the CUP1-promoter was induced in a yar1Δ strain for 30 min. (+), in addition, plasmid encoded Yar1 was overexpressed from an ADH1-promoter. After mechanical disruption of the cells, lysates were centrifuged at 200,000 × g for 1 h, and equal amounts of pellet and supernatant fractions were analyzed by SDS-PAGE and Western blotting.
Reduced assembly of Rps3 could be due to reduced efficiency of nuclear import of Rps3 or the degradation or precipitation of free Rps3. Notably, we observed that Rps3 was largely insoluble when expressed in E. coli, whereas a high proportion of the protein became soluble when Yar1 was co-expressed, indicating that Yar1 protects Rps3 from precipitation (Fig. 7B). Despite the very low solubility of Rps3 expressed in the absence of Yar1, we attempted to purify FLAG-Rps3 expressed in E. coli. Although some FLAG-Rps3 was recovered in this purification, centrifugation at 200,000 g resulted in precipitation of most of the protein, suggesting aggregate formation (Fig. 7C). In contrast, a high proportion of FLAG-Rps3 remained in the 200,000 × g supernatant when purified His6-Yar1 was added prior to centrifugation (Fig. 7C). This further supports the idea that Yar1 maintains the solubility of Rps3. To investigate whether Yar1 also counteracts aggregation of Rps3 in yeast cells, we overexpressed Rps3 and investigated whether co-expression of Yar1 affects the solubility of Rps3. For this purpose, FLAG-Rps3 was expressed from the copper-inducible CUP1 promoter in a yar1Δ strain either in the absence or presence of a plasmid constitutively expressing Yar1 from an ADH1 promoter. After 30 min of induction of FLAG-Rps3 expression, cell lysates were centrifuged at 200,000 g and supernatant fractions (containing soluble protein) as well as pellet fractions (containing aggregated FLAG-Rps3 and FLAG-Rps3 incorporated into ribosomal subunits) were analyzed by Western blotting. Indeed, the amount of soluble FLAG-Rps3 was significantly increased when Yar1 was overexpressed (Fig. 7D). We suggest that by keeping freshly synthesized Rps3 soluble, Yar1 acts as a chaperone for Rps3.
DISCUSSION
The constant supply of ribosomal proteins is crucial for a growing cell to maintain a maximal rate of ribosome synthesis. Hence, it is conceivable that mechanisms exist which ensure that ribosomal proteins are not only synthesized in high amounts, but also remain soluble and are efficiently targeted to the ribosome. In this study, we discovered an anti-aggregation function of the non-ribosomal protein Yar1, which it exhibits exclusively on the small ribosomal subunit protein Rps3. Hence we suggest that Yar1 functions as a specific chaperone for Rps3.
Because of their extensive interactions with ribosomal RNA, ribosomal proteins usually contain a high proportion of positive charges, which are known to cause aggregation in the presence of polyanions such as RNA (10). Yar1 is composed of two ankyrin repeats. Ankyrin repeats are helix-turn-helix motifs of 33 amino acid residues that exhibit an L-shaped topology and exclusively function in mediating protein-protein interactions (27). Since Yar1 is a small protein (22 kDa) that does not contain any further domains, it is likely that the main function of Yar1 is to bind Rps3, thereby preventing its aggregation. Protection from aggregation with RNA could be achieved by shielding the positive charges of the basic Rps3 protein (pI 10.2). It is tempting to speculate that Yar1 could act as an RNA mimic for Rps3 considering the high content of negative charges (pI 4.2) found in Yar1.
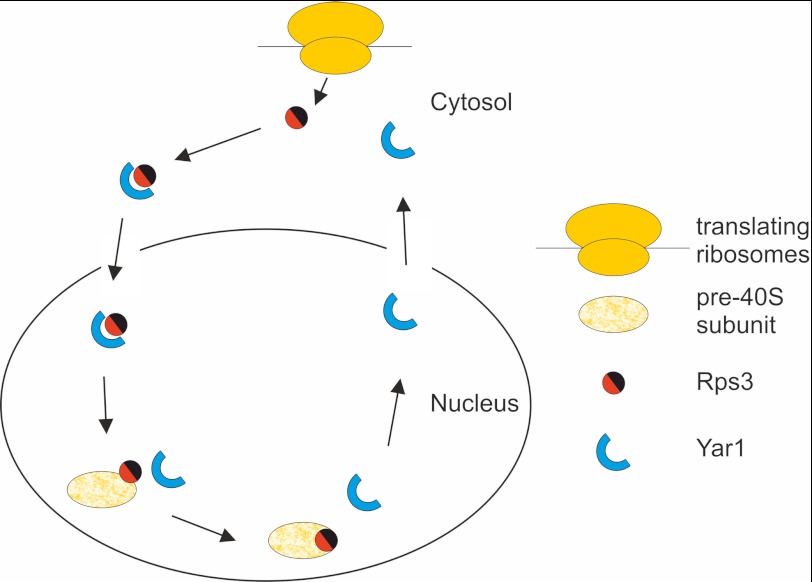
Newly translated Rps3 travels from the cytoplasm through the nuclear pores and into the nucleus where it assembles with pre-ribosomal particles. Therefore, it needs to be protected from aggregation along this entire path. Consistently, we found that Yar1 is localized both in the cytoplasm and the nucleus. Yar1 shows a predominantly cytoplasmic steady-state localization, however accumulation is observed in the nucleus after inhibition of the export receptor Xpo1. This is in contrast to previous data from the Lycan laboratory, where nuclear accumulation of Yar1-GFP was not observed upon leptomycin B treatment of an LMB-sensitive xpo1 mutant (28). An explanation for this discrepancy may be that Seiser et al. used a wild-type strain containing Yar1-GFP on a plasmid, probably resulting in competition between the chromosomal wild-type copy and plasmid encoded GFP-tagged Yar1. According to our data, Yar1 is a shuttling protein that binds Rps3 in the cytoplasm and is presumably imported into the nucleus in complex with Rps3, possibly via the N-terminal NLS of Rps3. The low amount of Yar1 detected in the nucleus under steady-state conditions indicates that it is quickly exported into the cytoplasm after dissociation from Rps3, where it can encounter a new Rps3 molecule (Fig. 8).
FIGURE 8.
Model for the function of Yar1. Newly translated Rps3 is bound by Yar1, which shields the positive charges of Rps3 (symbolized in red). After translocation of Yar1 and Rps3 into the nucleus the Yar1-Rps3 complex is disassembled and Rps3 joins pre-ribosomal particles. After its dissociation from Rps3, Yar1 is exported back into the cytoplasm.
Apparently, the requirement for Yar1 is indirectly proportional to the cellular concentration of Rps3: When Rps3 levels are high, the function of Yar1 is not required for optimal growth. This is probably because sufficient soluble Rps3 is present in the cell to reach pre-ribosomal particles even in the absence of a chaperone. When wild-type levels of Rps3 are expressed, the absence of Yar1 reduces the amount of soluble Rps3, resulting in 40S export defects and a reduced production of mature 40S subunits, eventually leading to reduced growth rates. When Rps3 is not fully functional (as in the case of the rps3-1 mutant), the absence of Yar1 is lethal. This may be due to an insufficient amount of Rps3 reaching pre-ribosomal particles in order to ensure synthesis of the critical number of 40S subunits necessary for growth.
Recent reports have highlighted the importance of the general chaperone network in assisting ribosome biogenesis (reviewed in Ref. 12). Ribosomal proteins and ribosome biogenesis factors have been found in aggregates in strains deleted for the Hsp70 chaperone SSB (11). Furthermore, Zuo1, which is involved in stimulation of SSB, and its homologue Jjj1 have been shown to bind to pre-ribosomal particles and participate in ribosome biogenesis (29–32). However, the exact role of the general chaperone network in ribosome biogenesis remains unclear, and as to date, no direct substrates have been described. Additionally, importins have been shown to protect the human ribosomal proteins S7, S3a, L4, L6, and L18a from aggregation with RNA and were suggested to function as general chaperones for ribosomal proteins (10).
The existence of a specific anti-aggregation factor for Rps3 suggests that for some proteins, the general chaperone network of the cell is insufficient for production of the required amounts of soluble protein. The need for additional, more specific factors makes particular sense for ribosomal proteins, which are not only highly expressed but beyond that also prone to aggregation. For these reasons, it is likely that Rps3 is not the only ribosomal protein with a specific chaperone. Chaperone-like functions have also been suggested for Rrb1, a non-ribosomal binding partner of the ribosomal protein Rpl3, Sqt1, an assembly factor for the ribosomal protein Rpl10, as well as Rpf2 and Rrs1, which are involved in the recruitment of 5S rRNA and the ribosomal proteins Rpl5 and Rpl11 into pre-60S subunits (33–36). It remains open to future investigations to address whether similar aggregation-preventing mechanisms also exist for other ribosomal proteins.
Acknowledgments
We thank Lisa Kappel for helpful suggestions, Marén Gnädig for generation of the Yar1-TAP and Yar1-GFP strain, Ed Hurt for providing plasmids, and Matthias Seedorf and Giorgio Dieci for providing antibodies.
The research was funded by Grant T404-B12 from the Austrian Science Fund (FWF) (to B. P.), Grant PP00P3_123341 from the Swiss National Science Foundation (to D. K.), and Grant P21991 from the FWF (to H. B.).

This article contains supplemental Tables S1–S3 and Fig. S1.
- rRNA
- ribosomal RNA
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- 5-FOA
- 5-fluoroorotic acid
- ITS
- internal transcribed spacer
- LMB
- leptomycin B
- NLS
- nuclear localization sequence
- pre-rRNA
- precursor rRNA
- SDC
- synthetic dextrose complete
- CBP
- calmodulin-binding protein.
REFERENCES
- 1. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2. Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kressler D., Hurt E., Bassler J. (2010) Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 4. Strunk B. S., Karbstein K. (2009) Powering through ribosome assembly. RNA 15, 2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karbstein K. (2011) Inside the 40S ribosome assembly machinery. Curr. Opin. Chem. Biol. 15, 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panse V. G., Johnson A. W. (2010) Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferreira-Cerca S., Pöll G., Gleizes P. E., Tschochner H., Milkereit P. (2005) Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 20, 263–275 [DOI] [PubMed] [Google Scholar]
- 8. Pöll G., Braun T., Jakovljevic J., Neueder A., Jakob S., Woolford J. L., Jr., Tschochner H., Milkereit P. (2009) rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One 4, e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rout M. P., Blobel G., Aitchison J. D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715–725 [DOI] [PubMed] [Google Scholar]
- 10. Jäkel S., Mingot J. M., Schwarzmaier P., Hartmann E., Görlich D. (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., Deuerling E. (2010) A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karbstein K. (2010) Chaperoning ribosome assembly. J. Cell Biol. 189, 11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]
- 14. Loar J. W., Seiser R. M., Sundberg A. E., Sagerson H. J., Ilias N., Zobel-Thropp P., Craig E. A., Lycan D. E. (2004) Genetic and biochemical interactions among Yar1, Ltv1, and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 168, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 16. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 17. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 18. Bender A., Pringle J. R. (1991) Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grosshans H., Hurt E., Simos G. (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 14, 830–840 [PMC free article] [PubMed] [Google Scholar]
- 20. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 21. Birner-Gruenberger R., Susani-Etzerodt H., Kollroser M., Rechberger G. N., Hermetter A. (2008) Lipolytic and esterolytic activity-based profiling of murine liver. Proteomics 8, 3645–3656 [DOI] [PubMed] [Google Scholar]
- 22. Neville M., Stutz F., Lee L., Davis L. I., Rosbash M. (1997) The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7, 767–775 [DOI] [PubMed] [Google Scholar]
- 23. Stade K., Ford C. S., Guthrie C., Weis K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 24. Neville M., Rosbash M. (1999) The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18, 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lycan D. E., Stafford K. A., Bollinger W., Breeden L. L. (1996) A new Saccharomyces cerevisiae ankyrin repeat-encoding gene required for a normal rate of cell proliferation. Gene 171, 33–40 [DOI] [PubMed] [Google Scholar]
- 26. Venema J., Tollervey D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311 [DOI] [PubMed] [Google Scholar]
- 27. Li J., Mahajan A., Tsai M. D. (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178 [DOI] [PubMed] [Google Scholar]
- 28. Seiser R. M., Sundberg A. E., Wollam B. J., Zobel-Thropp P., Baldwin K., Spector M. D., Lycan D. E. (2006) Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Demoinet E., Jacquier A., Lutfalla G., Fromont-Racine M. (2007) The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA 13, 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer A. E., Hung N. J., Yang P., Johnson A. W., Craig E. A. (2007) The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer A. E., Hoover L. A., Craig E. A. (2010) The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60S subunit factor Arx1. J. Biol. Chem. 285, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Albanèse V., Reissmann S., Frydman J. (2010) A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189, 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schaper S., Fromont-Racine M., Linder P., de la Cruz J., Namane A., Yaniv M. (2001) A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr. Biol. 11, 1885–1890 [DOI] [PubMed] [Google Scholar]
- 34. Iouk T. L., Aitchison J. D., Maguire S., Wozniak R. W. (2001) Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol. 21, 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eisinger D. P., Dick F. A., Denke E., Trumpower B. L. (1997) SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol. Cell. Biol. 17, 5146–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M. P., Hiley S. L., Hughes T., Woolford J. L., Jr. (2007) Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]