Background: A G90D point mutation in rhodopsin causes congenital stationary night blindness.
Results: The G90D mutation alters the chromophore-binding pocket, mechanical rigidity, and energetic stability of dark state rhodopsin.
Conclusion: Significant perturbations are promoted by the G90D mutation.
Significance: Characterizing the effect of point mutations in rhodopsin allows for a better understanding of how these mutations lead to dysfunction in retinal diseases.
Keywords: 7-Helix Receptor, Atomic Force Microscopy, G Protein-coupled Receptors (GPCR), Membrane Proteins, Mutant, Receptor Structure-Function, Retinal Degeneration, Rhodopsin, Spectroscopy
Abstract
Several point mutations in rhodopsin cause retinal diseases including congenital stationary night blindness and retinitis pigmentosa. The mechanism by which a single amino acid residue substitution leads to dysfunction is poorly understood at the molecular level. A G90D point mutation in rhodopsin causes constitutive activity and leads to congenital stationary night blindness. It is unclear which perturbations the mutation introduces and how they can cause the receptor to be constitutively active. To reveal insight into these mechanisms, we characterized the perturbations introduced into dark state G90D rhodopsin from a transgenic mouse model expressing exclusively the mutant rhodopsin in rod photoreceptor cells. UV-visible absorbance spectroscopy revealed hydroxylamine accessibility to the chromophore-binding pocket of dark state G90D rhodopsin, which is not detected in dark state wild-type rhodopsin but is detected in light-activated wild-type rhodopsin. Single-molecule force spectroscopy suggested that the structural changes introduced by the mutation are small. Dynamic single-molecule force spectroscopy revealed that, compared with dark state wild-type rhodopsin, the G90D mutation decreased energetic stability and increased mechanical rigidity of most structural regions in the dark state mutant receptor. The observed structural, energetic, and mechanical changes in dark state G90D rhodopsin provide insights into the nature of perturbations caused by a pathological point mutation. Moreover, these changed properties observed for dark state G90D rhodopsin are consistent with properties expected for an active state.
Introduction
Rhodopsin is the light receptor that initiates phototransduction in rod photoreceptor cells of the retina. Over 120 point mutations have been discovered in the rhodopsin gene, causing retinitis pigmentosa or congenital stationary night blindness (1). Not all amino acid substitutions are equivalent in their effects; they can range from having little to no discernible consequences on receptor structure and function to causing receptor instability, misfolding, or malfunction. The link between the structural perturbations promoted by mutation and the resulting defects causing disease is still unclear.
Atomic force microscopy (AFM)-based3 single-molecule force spectroscopy (SMFS) and dynamic SMFS (DFS) directly probe and localize the complex network of molecular interactions stabilizing the structure and guiding the function of membrane proteins under nearly native conditions (2). In SMFS, membrane proteins are mechanically unfolded at a constant pulling velocity to quantify the force required to overcome interactions that stabilize structural segments of the protein. These interactions can be quantified at piconewton sensitivity and structurally located at (sub)nanometer resolution. In DFS, SMFS is conducted at many different pulling velocities to quantify the kinetic, energetic, and mechanical properties of structural segments stabilizing a membrane protein (3). Because SMFS and DFS not only quantify but also structurally localize these properties, both methods are well suited to characterize the perturbations a point mutation introduces in rhodopsin and to delineate the mechanisms by which malfunction occurs.
Initial SMFS studies of vertebrate rhodopsin in native membranes revealed the stabilization of the receptor into distinct structural segments by inter- and intramolecular interactions (4, 5). Additionally, SMFS has revealed stabilizing effects on receptor structure promoted by zinc ions and post-translational modifications (6, 7). More recently, SMFS and DFS have been used to determine the impact of amino acid sequence differences among rhodopsins from different vertebrate species (8). These studies revealed that despite a difference of 23 amino acid residues in the sequences of bovine and mouse rhodopsin, the network of molecular interactions in the homologous receptors are largely conserved and that both receptors maintain similar functional properties (8). Thus, rhodopsin can accommodate certain amino acid substitutions with minimal structural and functional perturbations. In contrast, single amino acid substitutions in critical positions can significantly influence the stability or functional properties of the receptor (9). Accordingly, molecular dynamics simulations mimicking SMFS experiments on rhodopsin suggested that mutations causing retinitis pigmentosa can lead to receptor instability and misfolding by perturbing critical networks of molecular interactions (10).
In the current study, the effects of a glycine to aspartic acid mutation at position 90 (G90D) in rhodopsin, which causes congenital stationary night blindness (11), was investigated by UV-visible absorbance spectroscopy, SMFS, and DFS. Patients harboring this mutation have extensive night blindness at an early age (11). In contrast to the majority of rhodopsin mutants causing retinitis pigmentosa, night blindness in patients harboring the G90D point mutation in rhodopsin is not a result of retinal degeneration, but rather, the result of constitutive activity displayed by the mutant receptor (11, 12). The origin of the constitutive activity from G90D rhodopsin causing disease is unclear. In dark state wild-type rhodopsin, the receptor is locked into an inactive state by the covalently linked inverse agonist 11-cis-retinal. In one hypothesis, the origin of constitutive activity caused by the mutation is proposed to arise from dark state chromophore-bound rhodopsin, and in the other, it is proposed to arise from chromophore-free opsin (13, 14). Electrophysiological studies on photoreceptor cells in the retina of a mouse model of congenital stationary night blindness suggest that dark state G90D rhodopsin can be active in the absence of light (14). Here, we characterized the structural, energetic, and mechanical properties of dark state G90D rhodopsin from this mouse model to determine the perturbations caused by the mutation and to gain insights into the origin of constitutive activity.
EXPERIMENTAL PROCEDURES
Mouse Rod Outer Segment (ROS) Disc Membrane Preparation
ROS disc membranes were prepared from the eyes of mice as described (8). Wild-type rhodopsin was obtained from C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME). G90D rhodopsin was from G90D mice, which are homozygous for both the G90D rhodopsin transgene and wild-type rhodopsin knock-out (15). The mice were exposed to a 12-h light/dark cycle environment and were dark-adapted overnight prior to sample preparation. The mice were between 4 and 6 weeks old, and 20 to 24 mice were used for each preparation of ROS disc membranes. ROS disc membranes were stored at −80 °C in 67 mm potassium phosphate, 1 mm magnesium acetate, 0.1 mm EDTA, 18% (w/v) sucrose, pH 7.0. The membranes were thawed and then washed twice in Ringer's buffer (130 mm NaCl, 3.6 mm KCl, 2.4 mm MgCl2, 1.2 mm CaCl2, 10 mm HEPES, 0.02 mm EDTA, pH 7.4) prior to use. Unless otherwise stated, all of the experimental procedures were carried out under dim red light to prevent photobleaching. The centrifugation steps were performed at 4 °C.
SMFS and DFS
SMFS was performed on ROS disc membranes prepared from wild-type and G90D mice similar to procedures reported previously (8), except for the following modifications. 10 μl of washed ROS disc membranes was diluted in 200 μl of Ringer's buffer. 30 μl of the diluted ROS disc membranes was adsorbed on freshly cleaved mica for 20 min. Ringer's buffer was exchanged several times to remove loose debris prior to experiments. SMFS was performed on ROS disc membranes in Ringer's buffer at room temperature (≈28 °C). Experiments were conducted in the dark using an AFM equipped with an 850-nm laser detection system (NanoWizardII, JPK Instruments, Berlin, Germany) and NPS Si3N4 cantilevers (Bruker, Karlsruhe, Germany) having nominal spring constants of 0.06–0.08 N/m. Cantilevers were calibrated in Ringer's buffer using the thermal fluctuation method (16, 17). DFS was conducted at constant pulling velocities of 300, 700, 1500, 3000, 4500, and 6000 nm/s. For pulling velocities >1500 nm/s, a 16-bit data acquisition card (NI PCI-6221; National Instruments, Munich, Germany) was used to obtain higher sampling frequencies. F tests were conducted as described (8). Previous SMFS studies showed that ROS disc membranes adsorb onto mica, predominantly exposing the extracellular surface of rhodopsin to the AFM tip (4). Accordingly, force-distance (F-D) curves collected from ROS disc membranes represented mechanical unfolding of rhodopsin from the N-terminal end and exhibited characteristic unfolding patterns such as those observed in the current study (see Figs. 2 and 3).
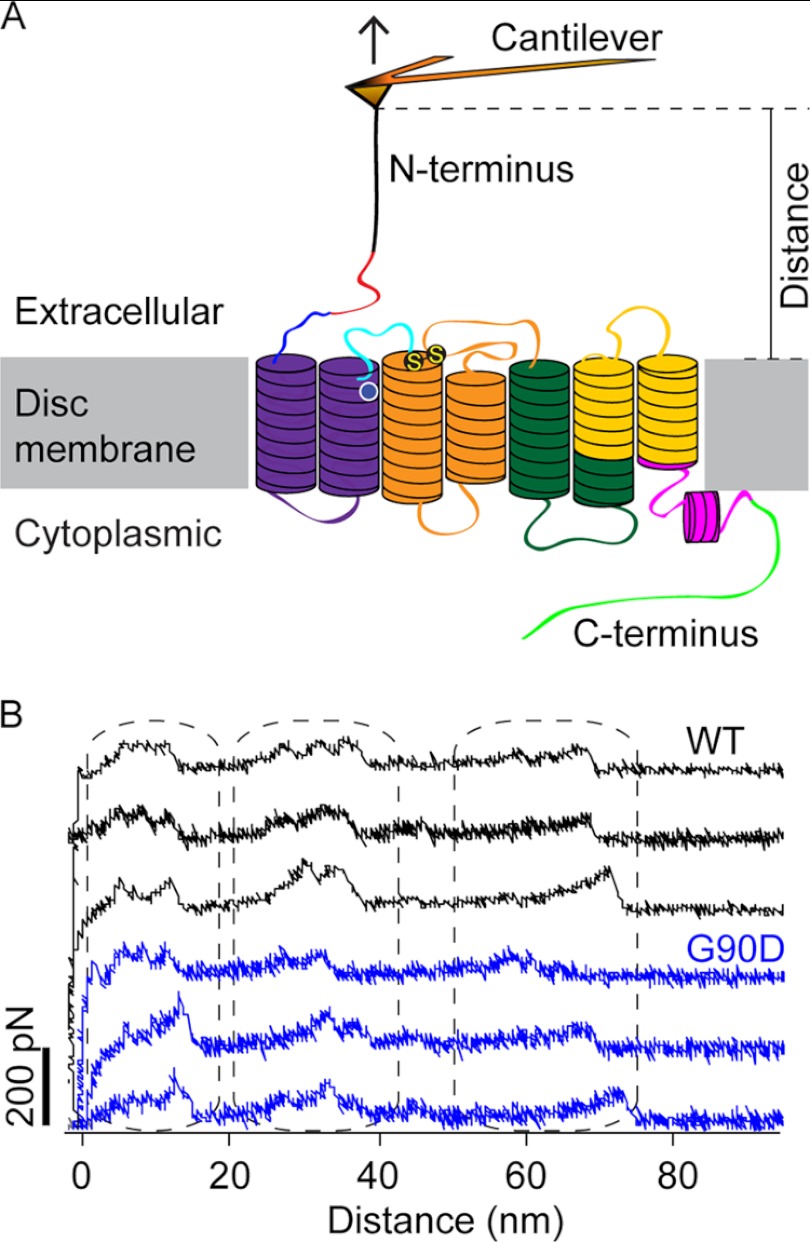
FIGURE 2.
SMFS of rhodopsin. A, illustration of SMFS on rhodopsin embedded in ROS disc membranes. In SMFS, the AFM cantilever tip is brought in contact with the N-terminal end of rhodopsin and then retracted from the membrane. This retraction applies mechanical stress and induces stepwise unfolding of rhodopsin. For each attempt, force and tip sample distances are recorded in F-D curves (as shown in B). The location of the G90D mutation is highlighted (white circle), and S-S denotes the conserved disulfide bond between residues Cys-110 and Cys-187. B, examples of F-D curves obtained from the mechanical unfolding of single wild-type (black) or G90D mutant (blue) rhodopsin. Each force peak of a F-D curve records the rupture of bonds and the subsequent unfolding of a stabilizing structural segment. The distance at which a force peak was detected localizes the molecular interaction that stabilized the structural segment.
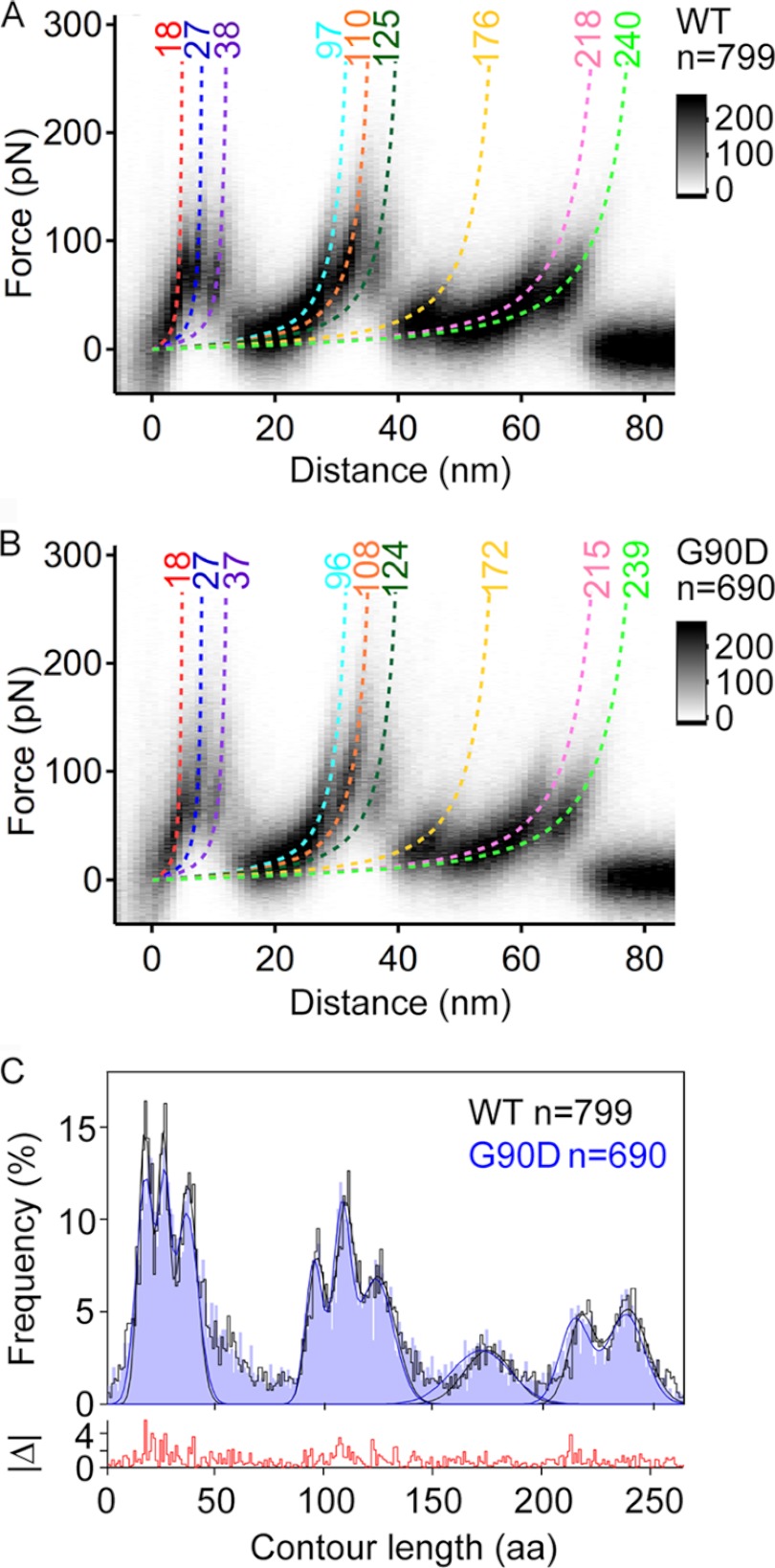
FIGURE 3.
Density maps of F-D curves and histogram of force peak positions. A and B, density maps generated from superimposition of all F-D curves collected for wild-type (A) and G90D mutant (B) rhodopsin. 799 F-D curves were superimposed for wild-type rhodopsin, and 690 F-D curves were superimposed for G90D rhodopsin. F-D curves were recorded at pulling velocities of 300, 700, 1500, 3000, 4500, and 6000 nm/s. The monochrome scale bars indicate relative intensities for each data set. Each force peak has been fitted with the wormlike chain (WLC) model (dashed curves) to reveal the contour length (in amino acids) of the stretched and unfolded polypeptide. This allowed for the localization of the stepwise unfolding of structural segments. The coloring of the wormlike chain curves corresponds to the structural segments they represent in Fig. 4. C, histograms showing the frequency of detecting a force peak at a certain contour length. A bin size of 1 amino acid (aa) was used. Gaussian curves are fitted to localize the most probable force peak positions (Table 1). The absolute frequency difference between both histograms (|Δ|) is shown at the bottom.
Estimating Parameters Describing Energy Barriers from DFS Data
Parameters describing the unfolding energy barrier of structural segments stabilizing rhodopsin (see Fig. 5) were estimated from DFS plots as described (3, 8). Briefly, the loading rate (r) dependence of the most probable unfolding force (F*) in DFS plots (see Fig. 6A) was analyzed using Equation 1 (18, 19),
where kB is the Boltzmann constant, and T is the absolute temperature. xu and ku were estimated by fitting DFS data (see Fig. 6A) to Equation 1 by nonlinear regression. The height of the free energy barrier was estimated with the Arrhenius equation,
where 1/τA is the Arrhenius frequency factor describing the diffuse relaxation time of a structural segment (20). τA was set 10−9 s (21, 22). The mechanical spring constant of a structural segment (κ) was estimated using Equation 3 (5, 23).
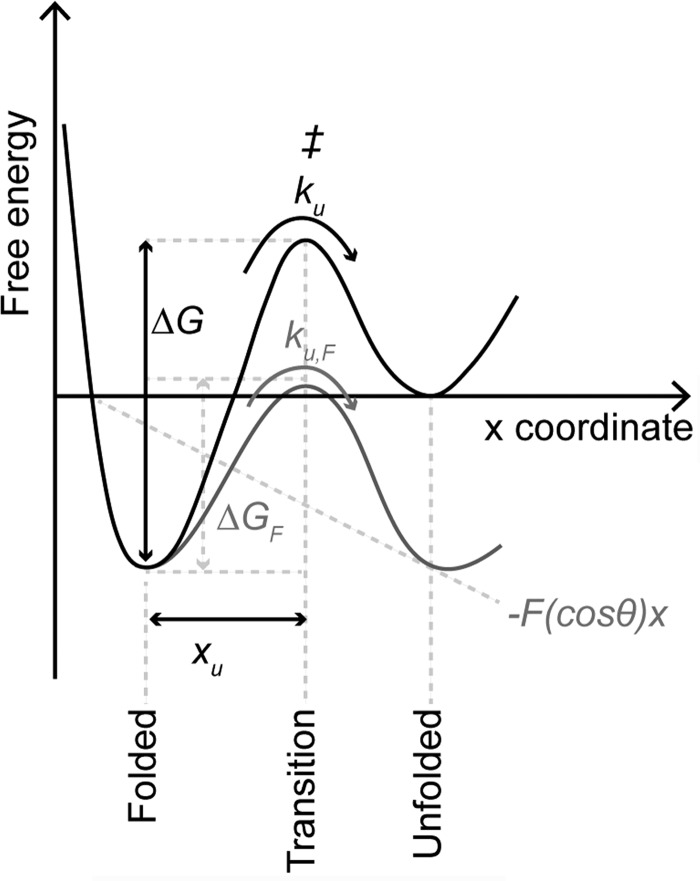
FIGURE 5.
Schematic of an unfolding energy barrier for a stable structural segment. The folded state of a structural segment is separated from the unfolded state by a free energy barrier. In the absence of an externally applied force (i.e., at equilibrium), the folded structure can unfold at certain transition rate (ku) by overcoming the transition state (‡). xu characterizes the distance from the folded state to the transition state. The height of the unfolding free energy barrier separating the folded state from the unfolded state is given by ΔG. According to the Bell-Evans model (18, 19, 47), an externally applied force (F) tilts the energy landscape by the mechanical energy (−F(cosθ)x) and lowers the free energy barrier (ΔG) to ΔGF. Lowering of the free energy barrier is described by the pulling direction x and the angle θ of the externally applied force (F).
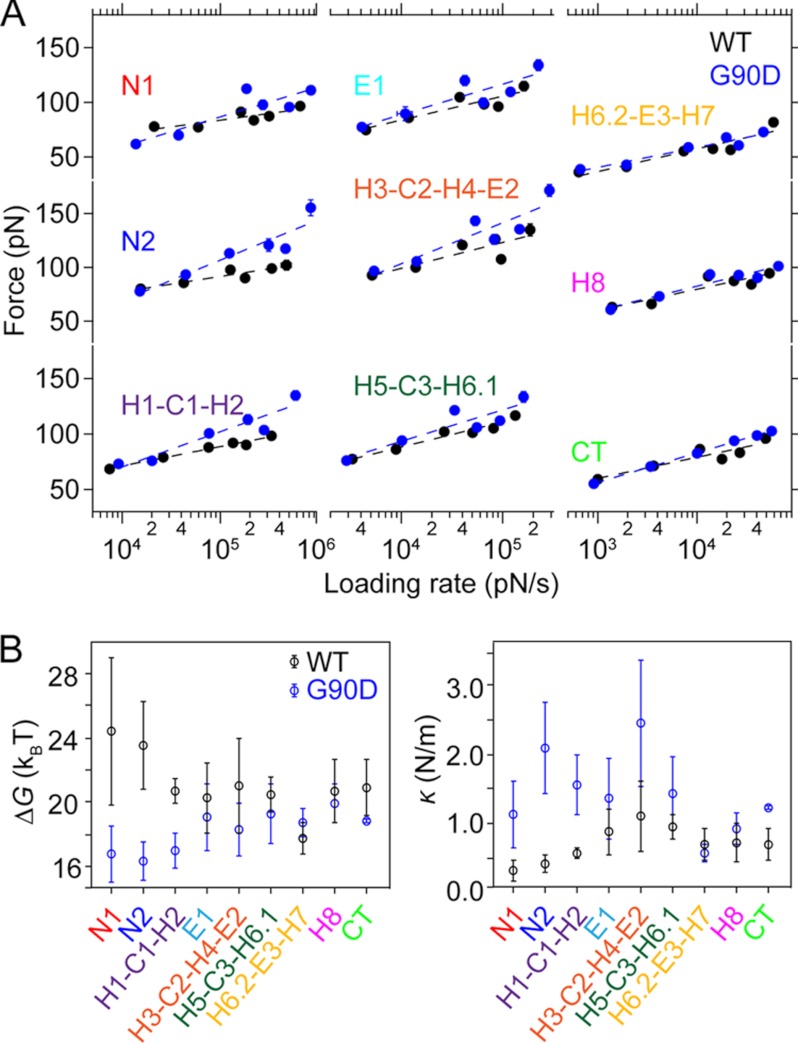
FIGURE 6.
Dynamic behavior of stable structural segments of wild-type and G90D mutant rhodopsin detected by DFS, and their energetic and mechanical properties. A, DFS plots for wild-type (black) and G90D mutant (blue) rhodopsin. Most probable force and most probable loading rates were determined from histograms in supplemental Figs. S2 and S3, respectively. If error bars (standard error) cannot be seen, then they are smaller than the data points. The data were fitted with the Bell-Evans model to estimate ku and xu (Fig. 5). B, the energetic stability (ΔG) and mechanical rigidity (κ) for each structural segment in wild-type (black) and G90D mutant (blue) rhodopsin are shown with standard deviations. The values were computed as described under “Experimental Procedures.”
UV-visible Absorbance Spectroscopy
Rhodopsin from dark-adapted mice was purified from eyes as described (8). Eyes from four or five mice were used for each purified preparation, which yielded on average 38 and 15 μg of purified wild-type and G90D rhodopsin, respectively. The absorbance spectrum of purified rhodopsin was obtained using a Lambda 35 UV-visible spectrophotometer (PerkinElmer Life Sciences). The absorbance maximum of purified wild-type and G90D rhodopsin was estimated by fitting the data in the visible region of the spectrum with a Gaussian function using GraphPad Prism version 5.0 for Mac OS X (San Diego, CA).
Hydroxylamine accessibility to the chromophore-binding pocket of rhodopsin was determined by UV-visible absorbance spectroscopy by monitoring the decrease in absorbance at 499 nm for wild-type and 485 nm for G90D rhodopsin. Hydroxylamine hydrochloride (Fisher Scientific) was dissolved in water at a stock concentration of 1 m with pH adjusted to 7.0. The assays were performed in 10 mm Bis-Tris propane, 500 mm NaCl, 2 mm n-dodecyl-β-d-maltoside at pH 7.5 using 0.8–2.4 μm purified rhodopsin. The temperature of the sample was maintained at 20 °C during experiments by a circulating water bath. The absorbance spectra were recorded in the absence of hydroxylamine (0 min) and at 0.5, 1, 3, 5, 10, 20, and 30 min following the addition of hydroxylamine to a final concentration of 20 mm. At the time points indicated, the range of wavelengths recorded for wild-type rhodopsin was between 494 and 502 nm, and that for G90D rhodopsin was between 481 and 489 nm. Absorbance spectra (200–800 nm) of both samples were recorded at 0 and 30 min and after a 1-min bleach at the end of the assay using a Fiber-Lite MI-150 Illuminator (Dolen-Jenner Industries, Boxborough, MA) equipped with a 480–520-nm band pass filter. The time-dependent decrease in maximum absorbance, monitored at 499 nm for wild-type rhodopsin and at 485 nm for G90D rhodopsin, was plotted as a function of time. Each experiment was repeated four times, and the average values were plotted with the standard deviations. Using GraphPad Prism, the data were approximated by an equation that describes a one-phase decay using nonlinear regression.
RESULTS
Transgenic mice expressing rhodopsin with a G90D point mutation were previously generated to serve as a model to study congenital stationary night blindness (15). In the current study, G90D rhodopsin homozygous mice on a null wild-type rhodopsin background were studied, which are referred to as G90D mice. Mutant G90D rhodopsin in the absence of wild-type rhodopsin can facilitate the proper formation of ROS (15). Thus, G90D mice provide a source of ROS disc membranes homogeneously expressing the mutant rhodopsin. We characterized dark state wild-type rhodopsin from wild-type mice and G90D rhodopsin from G90D mice by UV-visible absorbance spectroscopy, SMFS, and DFS to quantify and localize the perturbations occurring in the dark state receptor as a result of the G90D point mutation.
Characterization of Purified Wild-type and G90D Mutant Rhodopsin
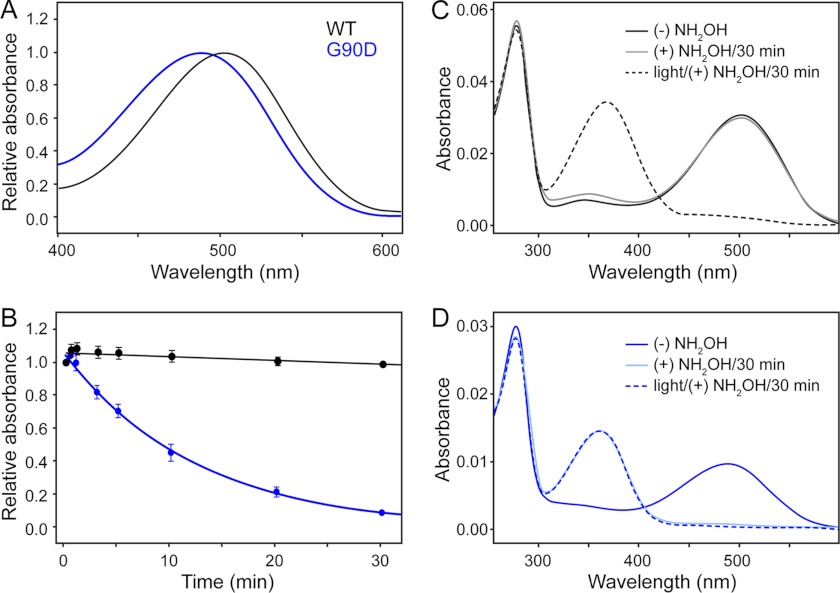
Structural perturbations of dark state G90D rhodopsin were first investigated on purified receptors. Wild-type and G90D rhodopsin were extracted and purified from ROS disc membranes in the retina of wild-type or G90D mice, respectively. We then characterized the absorbance maxima of purified rhodopsin. Rhodopsin covalently binds the chromophore 11-cis-retinal via a Schiff base linkage. The chromophore serves as an inverse agonist that in the dark locks the receptor in the inactive state and exhibits spectral properties sensitive to the Schiff base linkage and protein environment (24, 25). Purified G90D rhodopsin exhibited an absorbance maximum at a wavelength of 485 nm, which was blue-shifted compared with the 499-nm absorbance maximum of purified wild-type rhodopsin (Fig. 1A). The absorbance maximum of G90D rhodopsin from transgenic mice was similar to that determined from G90D rhodopsin obtained from heterologous expression systems (12, 26, 27). This blue shift in the absorbance spectrum indicates that interactions between 11-cis-retinal and amino acid side chains of transmembrane α-helices are altered (12).
FIGURE 1.
Effect of hydroxylamine on the absorbance spectra of wild-type (WT) and G90D mutant rhodopsin. A, UV-visible spectroscopy of purified wild-type (black) and G90D mutant (blue) rhodopsin. Wild-type rhodopsin exhibits an absorbance maximum at 498.8 ± 0.1 nm (n = 3), and G90D rhodopsin exhibits an absorbance maximum at 485.0 ± 0.3 nm (n = 3). n gives the number of experiments. The absorbance maximum was estimated by fitting the data with a Gaussian function. B, the effect of hydroxylamine on the absorbance maximum of wild-type (499 nm) and G90D mutant (485 nm) rhodopsin monitored as a function of time at 20 °C. The change in maximum absorbance was measured in the absence (0 min) and presence of 20 mm hydroxylamine at 0.5, 1, 3, 5, 10, 20, and 30 min, respectively. The half-life of the decrease in absorbance at 485 nm for G90D rhodopsin was 8.7 ± 1.2 min (n = 4). n gives the number of experiments. C and D, complete UV-visible absorbance spectra of purified wild-type (C) and G90D mutant (D) rhodopsin recorded simultaneously along with data points generated for B. The data show single representative experiments for wild-type and G90D mutant rhodopsin. The absorbance spectra of rhodopsin in the absence of hydroxylamine (solid black and blue lines), treatment with 20 mm hydroxylamine (NH2OH) (solid gray and light blue lines) for 30 min and after 1 min of bleaching in the presence of 20 mm hydroxylamine (dashed black and blue lines) are shown (see “Experimental Procedures”).
Next, we investigated the solvent accessibility of the Schiff base formed between 11-cis-retinal and Lys-296 in rhodopsin using a hydroxylamine reactivity assay (27). If accessible, hydroxylamine can hydrolyze the Schiff base to form 11-cis-retinal oxime, thereby reducing the absorbance maxima at 499 or 485 nm in purified wild-type and G90D rhodopsin, respectively. In wild-type rhodopsin, hydroxylamine has access to the Schiff base only after light activation (28). In the dark, no change is observed in absorbance at 499 nm of wild-type rhodopsin even upon extending the hydroxylamine exposure to 30 min (Fig. 1, B and C). In contrast, hydroxylamine reduced the absorbance at 485 nm of G90D rhodopsin in the dark (Fig. 1, B and D). This reactivity of hydroxylamine with dark state G90D rhodopsin indicates structural rearrangements of the transmembrane α-helices surrounding the chromophore-binding pocket similar to that found in light-activated wild-type rhodopsin.
Wild-type and G90D Mutant Rhodopsin Stabilize Similar Structural Segments
Results from UV-visible spectroscopy and the hydroxylamine reactivity assay suggest that dark state G90D rhodopsin exhibits structural alterations compared with dark state wild-type rhodopsin. These structural changes must be a result of altered intramolecular interactions promoted by the G90D point mutation. To quantify and localize the interactions in the mutant receptor, SMFS and DFS were applied. These approaches are particularly useful for rhodopsin because the receptor can be examined in the native membrane (i.e., ROS disc membranes) and in physiological buffer. Previous SMFS and DFS studies of vertebrate rhodopsin were conducted in a buffer optimized for AFM imaging (29). In the current study, SMFS was conducted in a Ringer's buffer, the ion composition and pH of which are similar to buffers used in suction electrode recording of mouse rod photoreceptor cells (30). Thus, rhodopsin was examined under physiologically relevant conditions.
Applying SMFS to dark state wild-type or G90D mutant rhodopsin in ROS disc membranes, the AFM tip was nonspecifically attached to the N-terminal end of rhodopsin (Fig. 2A). When withdrawing the AFM tip from the disc membrane, the N-terminal end was mechanically stretched to initiate the unfolding of a single rhodopsin molecule. During retraction of the AFM tip, we recorded the force required to stretch and unfold single rhodopsins over the pulling distance in F-D curves (Fig. 2B). Fully unfolded and stretched rhodopsin molecules exhibiting an intact disulfide bond between Cys-110 and Cys-187 exhibited F-D curves with an overall tip sample distance of ∼65–75 nm and three clusters of force peaks (outlined by dashed regions in Fig. 2B) (4, 8). Every force peak in a F-D curve represents the unfolding of a structural segment of rhodopsin. The magnitude of the force peak quantifies the pulling force that is required to overcome the molecular interactions stabilizing a particular structural segment of rhodopsin. The pattern of a F-D curve represents the sequential unfolding events of structural segments established by a single rhodopsin molecule. This force peak pattern can vary depending on the unfolding pathway an individual rhodopsin has taken (2).
The forces detected by SMFS depend on the loading rate (i.e., force applied versus time) at which the stability of the structural segments are probed (31). Thus, the force characterizing the strength of molecular interactions stabilizing a structural segment is dependent on the pulling velocity of the SMFS experiment. Probing these forces at different loading rates (i.e., pulling velocities) allows us to determine the energetic, kinetic, and mechanical parameters that describe the properties of the structural segments stabilizing rhodopsin (3, 8). To obtain these parameters, we conducted DFS of dark state wild-type and G90D rhodopsin embedded in native ROS disc membranes. Over 100 F-D curves were collected at each of the six pulling velocities (300, 700, 1500, 3000, 4500, and 6000 nm/s) tested (supplemental Fig. S1).
The comparison of F-D curves recorded at different pulling velocities showed that their force peaks occurred at similar positions and displayed similar patterns for wild-type and G90D rhodopsin (supplemental Fig. S1). This pulling velocity-independent position of force peaks is in agreement with previous observations on wild-type bovine and mouse rhodopsin (8). To further enhance the signal to noise ratio of single F-D curves, all of the F-D curves recorded for wild-type or G90D rhodopsin were overlaid to generate density maps (Fig. 3, A and B, and supplemental Fig. S1A). The density maps of both wild-type and G90D rhodopsin displayed nine major common unfolding events that are represented by force peaks of higher density.
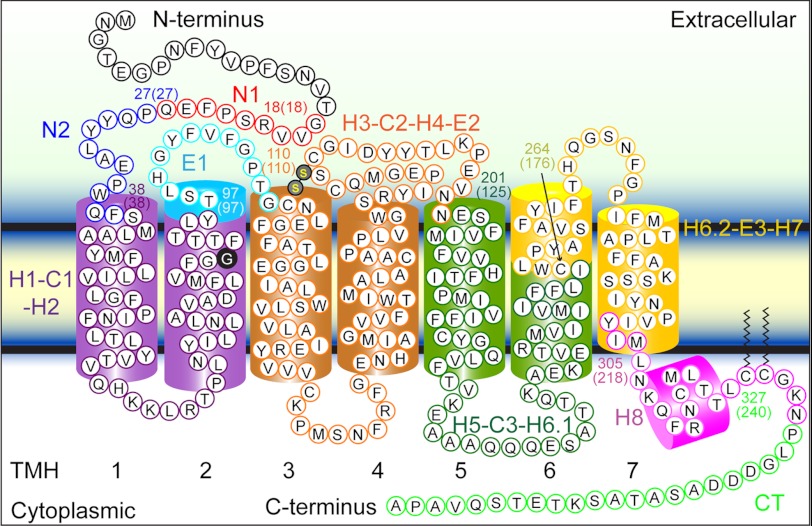
To localize the structural segments stabilizing wild-type or G90D rhodopsin, every force peak in each F-D curve was fitted with the wormlike chain model. The wormlike chain model estimates the contour length, thereby providing an estimate for the number of amino acid residues of the rhodopsin polypeptide chain unfolded and stretched above the membrane surface. One end of the unfolded polypeptide chain is tethered to the AFM tip, whereas the other end is held in place by the structural segment that remains stably embedded in the ROS disc membrane (Fig. 2A). Thus, the contour length of the unfolded peptide assigns the beginning of a stable structural segment (2).
Histograms of fitted contour lengths from every force peak of each F-D curve were generated for wild-type and G90D rhodopsin (Fig. 3C). Nine major peaks were discernable in both histograms, each corresponding to a common unfolding event detected in F-D curve overlays. Fitting a Gaussian function to each major peak in the histograms allowed the estimation of their contour lengths (Fig. 3C and Table 1). These contour lengths were used to assign the structural segments of wild-type and G90D rhodopsin (Fig. 4). Within the uncertainty of the measurements, the contour lengths estimated by the Gaussian function for each major unfolding event were similar between wild-type and G90D rhodopsin (Fig. 3C and Table 1). Thus, the location of structural segments stabilizing rhodopsin remained unchanged by the G90D mutation.
TABLE 1.
Most probable contour lengths of major unfolding force peaks detected for wild-type (WT) and G90D mutant mouse rhodopsin
The values were obtained by fitting peaks in the histogram of contour lengths in Fig. 3C with a Gaussian function.
| Structural segment | Contour length ± S.D. (amino acids) |
|
|---|---|---|
| WT | G90D | |
| N1 | 18 ± 3 | 18 ± 4 |
| N2 | 27 ± 3 | 27 ± 3 |
| H1-C1-H2 | 38 ± 4 | 37 ± 5 |
| E1 | 97 ± 4 | 96 ± 4 |
| H3-C2-H4-E2 | 110 ± 4 | 108 ± 4 |
| H5-C3-H6.1 | 125 ± 8 | 124 ± 8 |
| H6.2-E3-H7 | 176 ± 11 | 172 ± 14 |
| H8 | 218 ± 6 | 215 ± 7 |
| CT | 240 ± 9 | 239 ± 9 |
FIGURE 4.
Stable structural segments of wild-type and G90D mutant rhodopsin detected by SMFS. The most probable contour length of each major force peak was used to localize the corresponding structural segment in wild-type rhodopsin (Table 1). Within the accuracy of the measurements, the structural segments observed for wild-type and G90D mutant rhodopsin were identical (Table 1). Each structural segment is colored and named on the secondary structure. The boundary amino acid residues for each segment are numbered, and the contour length of the corresponding force peak is given in parentheses. Transmembrane α-helices (TMH) 1–7 are numbered.
Wild-type and G90D Mutant Rhodopsin Exhibit Different Energetic and Mechanical Properties
The unfolding energy barrier of each major structural segment stabilizing rhodopsin describes the energetic and mechanical properties of that structural segment (Fig. 5). To reveal the parameters that quantify these properties, we determined the most probable unfolding force and loading rate of every major structural segment at each pulling velocity (supplemental Figs. S2 and S3). DFS plots were generated by plotting the most probable unfolding force versus the logarithm of the loading rate (Fig. 6A). DFS plots for every structural segment exhibited a linear relationship. This linearity indicates a two-state unfolding process where a folded structural segment overcomes a single energy barrier to unfold (Fig. 5).
DFS plots (Fig. 6A) were analyzed by the Bell-Evans model (31) to determine parameters characterizing the unfolding energy barrier of each structural segment (Fig. 5). Fitting DFS data to the Bell-Evans model approximates the unfolding rate (ku) in the absence of applied force (F) and the distance separating the folded state and transition state (xu) (Fig. 5). The reciprocal of the unfolding rate (ku) describes the lifetime of a structural segment, whereas xu approximates the width of the energy barrier that separates the folded from the unfolded state. The number of conformational substates (i.e., conformational variability) that can be hosted by an energy valley depends on its width, xu. Hence, a structural segment characterized by a small xu will have lower conformational variability compared with the one having a larger xu. These parameters were determined for every stable structural segment of dark state wild-type and mutant G90D rhodopsin (Table 2).
TABLE 2.
Parameters describing the energy barriers (ku, xu, and ΔG) and mechanical rigidity (κ) of stable structural segments detected for WT and G90D mutant mouse rhodopsin
p values derived from F-tests are given in supplemental Table S2.
| Structural segment |
ku ± S.D. |
xu ± S.D. |
ΔG ± S.D. |
κ ± S.D. |
||||
|---|---|---|---|---|---|---|---|---|
| WT | G90D | WT | G90D | WT | G90D | WT | G90D | |
| s−1 | nm | kBT | N/m | |||||
| N1 | 0.00 ± 0.01 | 5.56 ± 10.0 | 0.77 ± 0.24 | 0.35 ± 0.09 | 24.36 ± 4.62 | 16.71 ± 1.80 | 0.33 ± 0.14 | 1.14 ± 0.48 |
| N2a | 0.01 ± 0.02 | 8.68 ± 9.99 | 0.67 ± 0.13 | 0.25 ± 0.05 | 23.58 ± 2.74 | 16.26 ± 1.15 | 0.44 ± 0.12 | 2.10 ± 0.64 |
| H1-C1-H2a | 0.10 ± 0.08 | 4.36 ± 4.71 | 0.55 ± 0.04 | 0.30 ± 0.05 | 20.69 ± 0.80 | 16.95 ± 1.08 | 0.57 ± 0.07 | 1.57 ± 0.43 |
| E1 | 0.15 ± 0.33 | 0.53 ± 1.11 | 0.43 ± 0.10 | 0.34 ± 0.09 | 20.29 ± 2.15 | 19.05 ± 2.08 | 0.89 ± 0.32 | 1.36 ± 0.57 |
| H3-C2-H4-E2 | 0.07 ± 0.21 | 1.16 ± 1.85 | 0.39 ± 0.12 | 0.25 ± 0.06 | 21.07 ± 2.93 | 18.27 ± 1.59 | 1.12 ± 0.50 | 2.43 ± 0.90 |
| H5-C3-H6.1 | 0.12 ± 0.13 | 0.41 ± 0.75 | 0.42 ± 0.05 | 0.33 ± 0.08 | 20.52 ± 1.04 | 19.31 ± 1.82 | 0.96 ± 0.17 | 1.43 ± 0.53 |
| H6.2-E3-H7 | 1.92 ± 1.91 | 0.78 ± 0.67 | 0.46 ± 0.09 | 0.52 ± 0.07 | 17.77 ± 1.00 | 18.67 ± 0.86 | 0.71 ± 0.23 | 0.58 ± 0.13 |
| H8 | 0.10 ± 0.20 | 0.22 ± 0.27 | 0.48 ± 0.11 | 0.42 ± 0.07 | 20.70 ± 1.99 | 19.94 ± 1.23 | 0.74 ± 0.27 | 0.93 ± 0.23 |
| CT | 0.08 ± 0.15 | 0.65 ± 0.06 | 0.50 ± 0.10 | 0.36 ± 0.01 | 20.90 ± 1.78 | 18.85 ± 0.09 | 0.69 ± 0.23 | 1.22 ± 0.03 |
a Significant differences (p < 0.01) between WT and G90D.
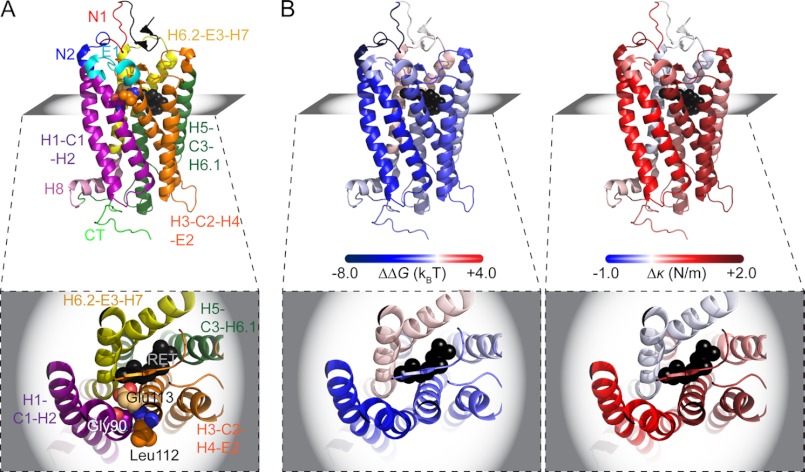
Mutant G90D rhodopsin generally exhibited higher ku and lower xu values than wild-type rhodopsin (Table 2). Using ku and xu, the height of the unfolding energy barrier (ΔG) and the mechanical spring constant (κ) were calculated for every structural segment (Table 2 and Figs. 5 and 6B). ΔG estimates the energetic stability of a structural segment, whereas κ describes its mechanical rigidity. With increasing κ, a structural segment decreases elasticity and increases rigidity. In wild-type rhodopsin, structural segments exhibited unfolding energy barrier heights ranging from 24.36 kBT ([N1]) to 17.77 kBT ([H6.2-E3-H7]) and mechanical spring constants ranging from a maximum rigidity of 1.12 n/m ([H3-C2-H4-E2]) to a maximum flexibility of 0.33 n/m ([N1]). In G90D rhodopsin, the structural segments had unfolding energy barrier heights ranging from 19.94 kBT ([H8]) to 16.26 kBT ([N2]) and mechanical spring constants ranging from a maximum rigidity of 2.43 N/m ([H3-C2-H4-E2]) to a maximum flexibility of 0.58 N/m ([H6.2-E3-H7]).
The DFS analyses revealed that the structural segments established in dark state wild-type and G90D rhodopsin exhibit different energetic and mechanical properties (Table 2). All of the structural segments in G90D rhodopsin showed a decreased energetic stability (ΔG) and increased mechanical rigidity (κ) compared with wild-type rhodopsin, except for segment [H6.2-E3-H7] (Figs. 6B and 7B and Table 2). The largest differences occurred in structural segments [N1], [N2], [H1-C1-H2], and [H3-C2-H4-E2]. The structural segments with the lowest energetic stability are found in the N-terminal region of G90D rhodopsin (16.71 kBT of [N1] and 16.26 kBT of [N2]), whereas the same region in wild-type rhodopsin exhibited the highest energetic stability (24.36 kBT of [N1] and 23.58 kBT of [N2]). Like wild-type rhodopsin (1.12 n/m), G90D rhodopsin (2.43 n/m) exhibited the highest mechanical rigidity in structural segment [H3-C2-H4-E2]. However, unlike wild-type rhodopsin, the next two most mechanically rigid structural segments in G90D rhodopsin were [N2] and [H1-C1-H2] rather than [E1] and [H5-C3-H6.1].
FIGURE 7.
Highlighting the structural, energetic stability and mechanical rigidity changes in G90D mutant rhodopsin. A, side view of rhodopsin (Protein Data Bank file 1U19) color-coded for stable structural segments: [N1] (red), [N2] (blue), [H1-C1-H2] (purple), [E1] (cyan), [H3-C2-H4-E2] (orange), [H5-C3-H6.1] (dark green), [H6.2-E3-H7] (yellow), [H8] (pink), anda [CT] (light green). Rhodopsin is shown in a side view with the extracellular surface at the top and the cytoplasmic surface at the bottom. The inset shows a top view in the plane indicated, which highlights the tight packing among Gly-90, Leu-112, and Glu-113. 11-cis-Retinal is shown as black spheres. B, the same rhodopsin structure is shown in a side and top view but colored to indicate changes of energetic stability (ΔΔG) and mechanical rigidity (Δκ) in G90D rhodopsin relative to wild-type rhodopsin. ΔΔG and Δκ were calculated using values reported in Table 2. The figures were generated using a PyMOL molecular graphics system (Version 1.2r3pre; Schrödinger).
An extra sum of squares F test was conducted (8) to determine the statistical significance of the difference between the two DFS data sets (supplemental Table S1). The most significant differences (p < 0.01) were observed between the data sets of wild-type and G90D rhodopsin for structural segments [N2] and [H1-C1-H2] (Table 2 and supplemental Table S1).
DISCUSSION
Structural Perturbations Promoted by G90D Mutation
G90D rhodopsin from a transgenic mouse model of congenital stationary night blindness was studied to determine the nature of perturbations occurring as a result of the point mutation. Gly-90 resides in transmembrane α-helix H2 of rhodopsin (Figs. 4 and 7A), facilitates packing with neighboring transmembrane α-helices, and affects the geometry of the chromophore-binding pocket (32). In wild-type rhodopsin, the absence of a side chain in Gly-90 allows α-helix H2 to pack tightly with Leu-112 and Glu-113 from α-helix H3 and form stable helix-helix interactions (Fig. 7A). Replacement of glycine with aspartic acid at this position would be expected to affect the tight packing between α-helices H2 and H3. This local perturbation is indicated by UV-visible spectroscopy of purified G90D rhodopsin revealing a blue-shifted maxima (Fig. 1A). The blue-shifted absorbance maximum of G90D rhodopsin is presumed to result, at least in part, by the Asp-90 mutation replacing Glu-113 as the counterion for the protonated Schiff base formed between 11-cis-retinal and Lys-296 (12, 33).
Structural changes of dark state G90D rhodopsin are indicated by our hydroxylamine accessibility assay. Unlike purified dark state wild-type rhodopsin, purified dark state G90D rhodopsin allowed hydroxylamine to access the chromophore-binding pocket (Fig. 1). Wild-type rhodopsin provides hydroxylamine access only in the light-activated state (28), termed metarhodopsin II, which is gained from the cytoplasmic side (34). This observation suggests that the G90D mutation introduces structural alterations in dark state rhodopsin that also occur in light-activated wild-type rhodopsin.
Previous in vitro studies of G90D rhodopsin expressed in heterologous expression systems using site-directed spin labeling and Fourier-transform infrared spectroscopy showed that the dark state mutant displays hallmarks of light-activated wild-type rhodopsin (27, 35, 36). These include neutralization of Glu-113 by protonation of its carboxylic acid group and rigid body movement of the cytoplasmic half of transmembrane α-helix H6 (27, 35, 36). Thus, the G90D mutation can promote structural alterations in the dark state receptor that has some resemblance to those occurring in wild-type rhodopsin upon light activation.
SMFS of rhodopsin embedded in native ROS disc membranes revealed that both wild-type and G90D rhodopsin stabilize similar structural segments (Fig. 3 and Table 1). Thus, the structural alterations in dark state G90D rhodopsin suggested by our hydroxylamine accessibility results and previous in vitro studies are likely relatively small. Moreover, the mutation does not change the receptor structure to an extent that would cause misfolding because G90D rhodopsin is properly folded, is integrated into ROS disc membranes, and binds 11-cis-retinal (Fig. 1A) (12, 15, 26, 27, 37, 38). Although structural changes in dark state G90D rhodopsin are predicted to be relatively small, these small changes may still be sufficient to switch the dark state mutant into an active state because large structural changes are not required for receptor activation (39).
G90D Mutation Changes Energetic Stability and Mechanical Rigidity of Rhodopsin
More detailed insights were gained about the perturbations promoted by the point mutation in dark state G90D rhodopsin from our DFS studies. A clear trend is observed in the differences in energetic stability and mechanical rigidity between dark state wild-type and G90D rhodopsin (Figs. 6B and 7B). In all structural segments except for segment [H6.2-E3-H7], G90D rhodopsin exhibits decreased energetic stability and increased mechanical rigidity (Table 2). Although only segments [N2] and [H1-C1-H2] revealed differences that were statistically significant in F tests (supplemental Table S1), the overall trend points to changes affecting the global structure of the receptor in addition to local changes.
The overall decrease in energetic stability observed in G90D rhodopsin is revealed by the decrease in the height of the energy barrier for structural segments. The active states of G protein-coupled receptors, including rhodopsin, are thermally less stable and more prone to irreversible denaturation compared with the inactive state (40, 41). Constitutively active G protein-coupled receptors also exhibit less thermal stability compared with inactive wild-type receptors (42, 43). Thus, the decrease in energetic stability observed in dark state G90D rhodopsin is consistent with properties expected for an active state. Moreover, the higher energetic stability of dark state wild-type rhodopsin may play a role in locking the receptor in an inactive state, which is required for maintaining low noise and single-photon detection capabilities in rod photoreceptor cells.
Because energetic stability (ΔG) is proportional to mechanical rigidity (κ) (see Equation 3 in “Experimental Procedures”), the lower energetic stability observed for structural segments of dark state G90D rhodopsin might lead to the assumption that its structure is more mechanically rigid compared with dark state wild-type rhodopsin. However, mechanical rigidity (κ) is only linearly proportional to the energetic stability (ΔG) but inversely proportional to the square of xu, which indicates conformational variability. The conformational variability of a structural segment can therefore have a greater impact on mechanical rigidity compared with energetic stability. Indeed, when taking a closer look at the xu and ΔG values (Table 2), it becomes evident that in dark state G90D rhodopsin, the mechanical rigidity of most structural segments increases, whereas their energetic stability decreases. Thus, the increase in mechanical rigidity of structural segments is caused by a decrease in conformational variability (Table 2).
A structural segment can increase mechanical rigidity by the introduction of additional chemical bonds. An extensive hydrogen bond network has been observed in crystal structures solved for the active metarhodopsin II state and for a mutant-promoted active state of rhodopsin (44, 45). This extensive hydrogen bond network is not observed in crystal structures of dark state wild-type rhodopsin (44, 46). The hydrogen bond network formed in the metarhodopsin II state of rhodopsin extends from the extracellular to the cytoplasmic side of the receptor and forms independently from conformational changes in the transmembrane region of the receptor (45). The formation of an additional hydrogen bond network in light-activated wild-type rhodopsin would be predicted to increase the mechanical rigidity of several structural segments. The increased mechanical rigidity observed for dark state G90D rhodopsin may indicate the formation of such a hydrogen bond network.
Conclusion
Our studies conducted on dark state G90D rhodopsin in affinity-purified and native ROS disc membrane preparations from a transgenic mouse model of congenital stationary night blindness have revealed the nature of perturbations promoted by the mutation. First, UV-visible absorbance spectroscopy detected that the mutant receptor can bind 11-cis-retinal but displays a blue-shifted absorbance maximum, thereby indicating an altered chromophore-binding pocket. Second, hydroxylamine accessibility to the chromophore-binding pocket in dark state G90D rhodopsin suggested that structural perturbations are caused by the mutation. These changes share similarities with those induced upon light activation of wild-type rhodopsin. Third, SMFS detected that wild-type and G90D rhodopsin stabilize the same structural segments, which suggests that the structural changes promoted by the mutation are relatively small. Fourth, DFS revealed considerable differences in the properties of structural segments in dark state G90D rhodopsin, which generally exhibited decreased energetic stability and increased mechanical rigidity.
Taken together, our studies revealed changes in the structural, energetic, and mechanical properties of dark state G90D rhodopsin. These changes observed in the dark state mutant are consistent with properties that have been described to change in light-activated wild-type rhodopsin. Thus, the G90D point mutation introduces perturbations that change dark state rhodopsin, so that it exhibits some of the properties expected for light-activated wild-type rhodopsin.
Acknowledgments
We thank Dr. Paul A. Sieving (National Eye Institute, Bethesda, MD) for generously providing G90D transgenic mice, H. Butler and K. Zongolowicz for maintaining our mouse colonies, and A. Yakubenko for genotyping mice.
This work was supported, in whole or in part, by National Institutes of Health Grants R00EY018085, R01EY021731, and P30EY011373. This work was also supported by the European Community's Seventh Framework Programme FP7/2007–2013 Grant 211800, Deutsche Forschungsgemeinschaft, Research to Prevent Blindness (an unrestricted grant and career development award), and the Ohio Lions Eye Research Foundation.

This article contains supplemental Table S1 and Figs. S1–S3.
- AFM
- atomic force microscopy
- DFS
- dynamic SMFS
- F-D curves
- force-distance curves
- ROS
- rod outer segment(s)
- SMFS
- single-molecule force spectroscopy.
REFERENCES
- 1. Mendes H. F., van der Spuy J., Chapple J. P., Cheetham M. E. (2005) Mechanisms of cell death in rhodopsin retinitis pigmentosa. Implications for therapy. Trends. Mol. Med. 11, 177–185 [DOI] [PubMed] [Google Scholar]
- 2. Kedrov A., Janovjak H., Sapra K. T., Müller D. J. (2007) Deciphering molecular interactions of native membrane proteins by single-molecule force spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 36, 233–260 [DOI] [PubMed] [Google Scholar]
- 3. Janovjak H., Sapra K. T., Kedrov A., Müller D. J. (2008) From valleys to ridges. Exploring the dynamic energy landscape of single membrane proteins. Chemphyschem. 9, 954–966 [DOI] [PubMed] [Google Scholar]
- 4. Tanuj Sapra K., Park P. S., Filipek S., Engel A., Müller D. J., Palczewski K. (2006) Detecting molecular interactions that stabilize native bovine rhodopsin. J. Mol. Biol. 358, 255–269 [DOI] [PubMed] [Google Scholar]
- 5. Sapra K. T., Park P. S., Palczewski K., Muller D. J. (2008) Mechanical properties of bovine rhodopsin and bacteriorhodopsin. Possible roles in folding and function. Langmuir 24, 1330–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park P. S., Sapra K. T., Jastrzebska B., Maeda T., Maeda A., Pulawski W., Kono M., Lem J., Crouch R. K., Filipek S., Müller D. J., Palczewski K. (2009) Modulation of molecular interactions and function by rhodopsin palmitylation. Biochemistry 48, 4294–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park P. S., Sapra K. T., Koliński M., Filipek S., Palczewski K., Muller D. J. (2007) Stabilizing effect of Zn2+ in native bovine rhodopsin. J. Biol. Chem. 282, 11377–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawamura S., Colozo A. T., Müller D. J., Park P. S. (2010) Conservation of molecular interactions stabilizing bovine and mouse rhodopsin. Biochemistry 49, 10412–10420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rader A. J., Anderson G., Isin B., Khorana H. G., Bahar I., Klein-Seetharaman J. (2004) Identification of core amino acids stabilizing rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 101, 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fanelli F., Seeber M. (2010) Structural insights into retinitis pigmentosa from unfolding simulations of rhodopsin mutants. FASEB J. 24, 3196–3209 [DOI] [PubMed] [Google Scholar]
- 11. Sieving P. A., Richards J. E., Naarendorp F., Bingham E. L., Scott K., Alpern M. (1995) Dark-light. Model for nightblindness from the human rhodopsin Gly-90 → Asp mutation. Proc. Natl. Acad. Sci. U.S.A. 92, 880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rao V. R., Cohen G. B., Oprian D. D. (1994) Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature 367, 639–642 [DOI] [PubMed] [Google Scholar]
- 13. Jin S., Cornwall M. C., Oprian D. D. (2003) Opsin activation as a cause of congenital night blindness. Nat. Neurosci. 6, 731–735 [DOI] [PubMed] [Google Scholar]
- 14. Dizhoor A. M., Woodruff M. L., Olshevskaya E. V., Cilluffo M. C., Cornwall M. C., Sieving P. A., Fain G. L. (2008) Night blindness and the mechanism of constitutive signaling of mutant G90D rhodopsin. J. Neurosci. 28, 11662–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sieving P. A., Fowler M. L., Bush R. A., Machida S., Calvert P. D., Green D. G., Makino C. L., McHenry C. L. (2001) Constitutive “light” adaptation in rods from G90D rhodopsin. A mechanism for human congenital nightblindness without rod cell loss. J. Neurosci. 21, 5449–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Florin E. L., Rief M., Lehmann H., Ludwig M., Dornmair C., Moy V. T., Gaub H. E. (1995) Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 17. Butt H. J., Jaschke M. (1995) Calculation of thermal noise in atomic force microscopy. Nanotechnology 6, 1–7 [Google Scholar]
- 18. Evans E. (1998) Energy landscapes of biomolecular adhesion and receptor anchoring at interfaces explored with dynamic force spectroscopy. Faraday Discuss. 111, 1–16 [DOI] [PubMed] [Google Scholar]
- 19. Evans E., Ritchie K. (1997) Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dietz H., Rief M. (2004) Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U.S.A. 101, 16192–16197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bieri O., Wirz J., Hellrung B., Schutkowski M., Drewello M., Kiefhaber T. (1999) The speed limit for protein folding measured by triplet-triplet energy transfer. Proc. Natl. Acad. Sci. U.S.A. 96, 9597–9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gräter F., Grubmüller H. (2007) Fluctuations of primary ubiquitin folding intermediates in a force clamp. J. Struct. Biol. 157, 557–569 [DOI] [PubMed] [Google Scholar]
- 23. Howard J. (2001) Mechanics of Motor Proteins and the Cytoskeleton, Sinauer Associates, Sunderland, MA [Google Scholar]
- 24. Zhukovsky E. A., Oprian D. D. (1989) Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science 246, 928–930 [DOI] [PubMed] [Google Scholar]
- 25. Sakmar T. P., Franke R. R., Khorana H. G. (1989) Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 86, 8309–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaushal S., Khorana H. G. (1994) Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 33, 6121–6128 [DOI] [PubMed] [Google Scholar]
- 27. Zvyaga T. A., Fahmy K., Siebert F., Sakmar T. P. (1996) Characterization of the mutant visual pigment responsible for congenital night blindness. A biochemical and Fourier-transform infrared spectroscopy study. Biochemistry 35, 7536–7545 [DOI] [PubMed] [Google Scholar]
- 28. Wald G., Brown P. K. (1953) The molar extinction of rhodopsin. J. Gen. Physiol. 37, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003) Atomic-force microscopy. Rhodopsin dimers in native disc membranes. Nature 421, 127–128 [DOI] [PubMed] [Google Scholar]
- 30. Azevedo A. W., Rieke F. (2011) Experimental protocols alter phototransduction. The implications for retinal processing at visual threshold. J. Neurosci. 31, 3670–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evans E. (1999) Looking inside molecular bonds at biological interfaces with dynamic force spectroscopy. Biophys. Chem. 82, 83–97 [DOI] [PubMed] [Google Scholar]
- 32. Javadpour M. M., Eilers M., Groesbeek M., Smith S. O. (1999) Helix packing in polytopic membrane proteins. Role of glycine in transmembrane helix association. Biophys. J. 77, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jäger S., Lewis J. W., Zvyaga T. A., Szundi I., Sakmar T. P., Kliger D. S. (1997) Time-resolved spectroscopy of the early photolysis intermediates of rhodopsin Schiff base counterion mutants. Biochemistry 36, 1999–2009 [DOI] [PubMed] [Google Scholar]
- 34. Jastrzebska B., Palczewski K., Golczak M. (2011) Role of bulk water in hydrolysis of the rhodopsin chromophore. J. Biol. Chem. 286, 18930–18937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim J. M., Altenbach C., Kono M., Oprian D. D., Hubbell W. L., Khorana H. G. (2004) Structural origins of constitutive activation in rhodopsin. Role of the K296/E113 salt bridge. Proc. Natl. Acad. Sci. U.S.A. 101, 12508–12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fahmy K., Zvyaga T. A., Sakmar T. P., Siebert F. (1996) Spectroscopic evidence for altered chromophore. Protein interactions in low-temperature photoproducts of the visual pigment responsible for congenital night blindness. Biochemistry 35, 15065–15073 [DOI] [PubMed] [Google Scholar]
- 37. Naash M. I., Wu T. H., Chakraborty D., Fliesler S. J., Ding X. Q., Nour M., Peachey N. S., Lem J., Qtaishat N., Al-Ubaidi M. R., Ripps H. (2004) Retinal abnormalities associated with the G90D mutation in opsin. J. Comp. Neurol. 478, 149–163 [DOI] [PubMed] [Google Scholar]
- 38. Toledo D., Ramon E., Aguilà M., Cordomí A., Pérez J. J., Mendes H. F., Cheetham M. E., Garriga P. (2011) Molecular mechanisms of disease for mutations at Gly-90 in rhodopsin. J. Biol. Chem. 286, 39993–40001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park P. S. (2012) Ensemble of G protein-coupled receptor active states. Curr. Med. Chem. 19, 1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogel R., Siebert F. (2002) Conformation and stability of α-helical membrane proteins. 2. Influence of pH and salts on stability and unfolding of rhodopsin. Biochemistry 41, 3536–3545 [DOI] [PubMed] [Google Scholar]
- 41. Sum C. S., Park P. S., Wells J. W. (2002) Effects of N-ethylmaleimide on conformational equilibria in purified cardiac muscarinic receptors. J. Biol. Chem. 277, 36188–36203 [DOI] [PubMed] [Google Scholar]
- 42. Gether U., Ballesteros J. A., Seifert R., Sanders-Bush E., Weinstein H., Kobilka B. K. (1997) Structural instability of a constitutively active G protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J. Biol. Chem. 272, 2587–2590 [DOI] [PubMed] [Google Scholar]
- 43. Wilson M. H., Limbird L. E. (2000) Mechanisms regulating the cell surface residence time of the *2A-adrenergic receptor. Biochemistry 39, 693–700 [DOI] [PubMed] [Google Scholar]
- 44. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 45. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 47. Bell G. I. (1978) Models for the specific adhesion of cells to cells. Science 200, 618–627 [DOI] [PubMed] [Google Scholar]