Background: Complete Lys-for-Arg substitutions in α-defensins Crp4 and RMAD4 affect microbicidal activities very differently.
Results: The peptide-lipid phase behavior of Crp4, RMAD4, and associated mutants correlated with differential biological activities.
Conclusion: A stringent agreement exists between α-defensin bactericidal effects and an induced negative Gaussian curvature model of membrane disruption.
Significance: These findings provide a basis for molecular engineering of novel peptide mimetic microbicides.
Keywords: Antimicrobial Peptides, Defensins, Innate Immunity, Membrane, X-ray Scattering, Membrane Curvature, Peptide-Lipid Interactions
Abstract
The conserved tridisulfide array of the α-defensin family imposes a common triple-stranded β-sheet topology on peptides that may have highly diverse primary structures, resulting in differential outcomes after targeted mutagenesis. In mouse cryptdin-4 (Crp4) and rhesus myeloid α-defensin-4 (RMAD4), complete substitutions of Arg with Lys affect bactericidal peptide activity very differently. Lys-for-Arg mutagenesis attenuates Crp4, but RMAD4 activity remains mostly unchanged. Here, we show that the differential biological effect of Lys-for-Arg replacements can be understood by the distinct phase behavior of the experimental peptide-lipid system. In Crp4, small-angle x-ray scattering analyses showed that Arg-to-Lys replacements shifted the induced nanoporous phases to a different range of lipid compositions compared with the Arg-rich native peptide, consistent with the attenuation of bactericidal activity by Lys-for-Arg mutations. In contrast, such phases generated by RMAD4 were largely unchanged. The concordance between small-angle x-ray scattering measurements and biological activity provides evidence that specific types of α-defensin-induced membrane curvature-generating tendencies correspond directly to bactericidal activity via membrane destabilization.
Introduction
Mammalian α-defensins are mediators of the host innate immune response. Their common characteristics include an ∼4.5-kDa size, a triple-stranded β-sheet secondary structure conferred by three invariant disulfide bonds, a cationic net charge, and an amphipathic character (1–3). In vitro, α-defensins display collective broad-spectrum microbicidal activities against a variety of pathogens, including Gram-positive and Gram-negative bacteria (4–6), fungi (7), protozoa (8), and enveloped viruses (9). Although certain α-defensins have been implicated in adaptive and innate immune responses (10–12), most are direct microbicides that lack mammalian cell cytotoxicity. The well established mechanism for α-defensin-mediated antibacterial effects is selective membrane disruption (13–15) that leads to target cell permeabilization, depolarization, dissipation of electrochemical gradients, leakage, and eventual cell death.
α-Defensin sequences destabilize bacterial cell membranes preferentially as a function of the phospholipid composition of microbial cell membranes (3, 16). Therefore, certain alterations of α-defensin primary structure are likely to alter determinants of maximal membrane disruption with consequent attenuation of bactericidal activity. For example, complete Lys-for-Arg substitutions in mouse Paneth cell α-defensin cryptdin-4 (Crp4)2 and rhesus myeloid α-defensin-4 (RMAD4) (supplemental Fig. S1) affected their in vitro microbicidal activities differentially. These peptides were selected for these analyses because they have canonical α-defensin attributes and the same overall cationic charge, but the distribution of electropositive charge differs in the two molecules. In (R/K)-Crp4 (where R/K indicates Lys-for-Arg substitution), Lys replacements attenuated activity and required longer exposure times to affect survival of Escherichia coli (see Fig. 1), but analogous substitutions in RMAD4 ((R/K)-RMAD4) had little or no effect on bactericidal peptide activity (5). These results posed a challenge to the current understanding of membrane disruption in which Lys-for-Arg mutations in pore-forming peptides lead to a monotonic decrease in activity by reducing peptide interactions with lipid headgroups. Moreover, because both native α-defensins permeabilize live E. coli and disrupt lipid membranes in vitro, these results provided a rationale for a stringent analysis of the physical mechanisms of the differential effects of Lys-for-Arg mutagenesis on the antimicrobial action of Crp4 and RMAD4.
FIGURE 1.
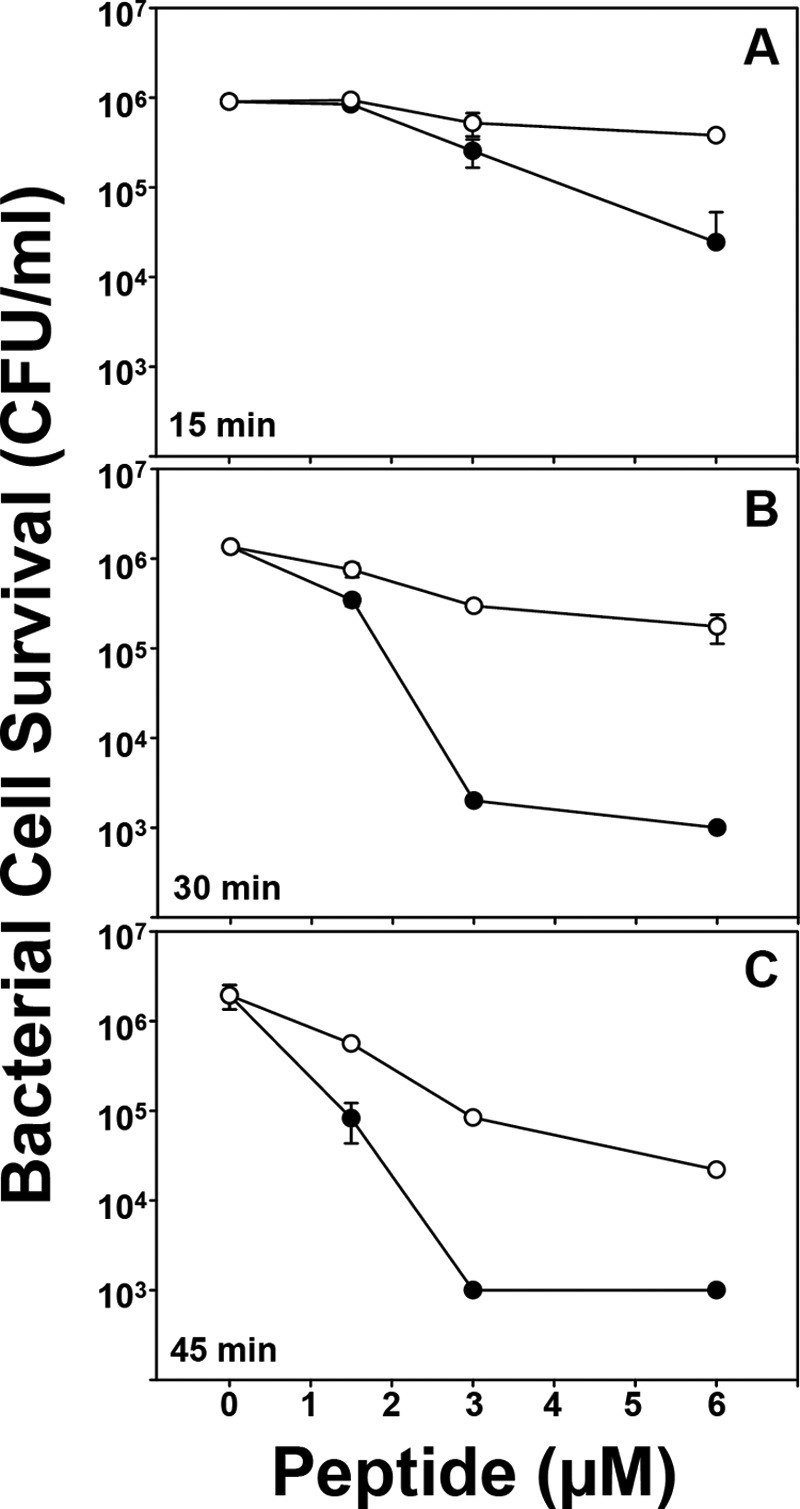
E. coli ML35 cell survival in response to Crp4 and (R/K)-Crp4 exposure time. E. coli ML35 cells were exposed to peptides at 37 °C in 50 μl of PIPES/Trypticase soy broth buffer. Following peptide exposure for 15 min (A), 30 min (B), or 45 min (C), the bacteria were plated on Trypticase soy broth-agar plates and incubated overnight at 37 °C. Surviving bacteria were counted as colony-forming units (CFU)/ml at each peptide concentration, and values at or below 1 × 103 colony-forming units/ml signify no detectable colonies. Each condition was performed in triplicate. Error bars denote S.D. ●, Crp4; ○,(R/K)-Crp4.
To approach this biological question, we used synchrotron small-angle x-ray scattering (SAXS) to characterize the structures and phase behavior of peptide-lipid complexes produced by wild-type and Lys-for-Arg variants of Crp4 and RMAD4 in model bacterial and eukaryotic membranes. By modifying the biophysical properties of the membranes by changing lipid composition, the curvature deformations produced by each of the four α-defensins could be probed as a function of membrane properties and as to whether cationicity is contributed by Arg or Lys side chains. In particular, we assayed for the existence of cubic phases, which require negative Gaussian (or “saddle-splay”) curvature, the curvature necessary for pore formation and other membrane destabilization mechanisms.
In agreement with their in vitro bactericidal activities (5), both native α-defensins exhibited a tendency to produce cubic phases rich in negative Gaussian membrane curvature. In Crp4, Arg-to-Lys substitutions shifted the incidence of negative Gaussian curvature formation significantly compared with the native peptide. However, Lys-for-Arg replacements had little effect on curvature generation in the context of the RMAD4 primary structure. The diminished strength of Lys side chain interactions with lipid headgroups has been considered to be the basis for attenuation of Lys-substituted peptides (17, 18). Although this may be so for certain cell-penetrating peptides, a parallel channel of action exists for Lys-substituted α-defensins, in which geometric differences in Lys and Arg lipid binding change the peptide-induced curvature deformations, resulting in differential microbicidal activities. Our results suggest an explanation for the differential effects of Lys-for-Arg substitutions in individual α-defensins on bactericidal peptide activity based on the topological requirements for membrane destabilization.
EXPERIMENTAL PROCEDURES
Preparation of α-Defensin and Lysine-substituted α-Defensin Variants
Peptides were prepared as described (5). Briefly, Crp4, (R/K)-Crp4, and RMAD4 peptides were expressed as recombinant His6 fusion proteins in E. coli BL21-CodonPlus (DE3)-RIL cells using the pET-28a vector and affinity-purified by nickel-nitrilotriacetic acid resin affinity chromatography (19). The N-terminal fusion partner was removed by cyanogen bromide cleavage reaction, and α-defensin peptides were purified by sequential C18 reverse-phase HPLC. Peptide homogeneity was confirmed by acid/urea-PAGE, a sensitive method for resolving α-defensins and misfolded peptide isomers. Molecular masses were confirmed by MALDI-TOF-MS and quantified by absorbance at 280 nm and by Bradford analysis (19). (R/K)-RMAD4 (supplemental Fig. S1) was synthesized in solid phase and purchased from CPC Scientific, Inc. (Sunnyvale, CA).
Lysine-substituted variants were refolded after reduction with dithiothreitol at a molar ratio 18:1 to peptide (i.e. 3 mol of reducing agent/mol of cysteine) in 6 m guanidine HCl, 0.2 m Trizma (Tris base), and 2 mm EDTA (pH 8.2) for 4 h at 50 °C. Reduction was terminated by adjustment of the reduction mixture to 5% acetic acid, and the reduced forms were purified by analytical C18 reverse-phase HPLC. Reduced peptides were refolded with stirring in 0.1 m NH4HCO3, 2.0 mm EDTA, 0.1 mg/ml cystine, and 0.1 mg/ml cysteine (pH 7.8) at concentrations of 100–300 μg/ml under N2 at 4 °C. Peptide refolding was monitored by analytical C18 reverse-phase HPLC at 24-h intervals, followed by a final purification performed by analytical C18 reverse-phase HPLC and acid/urea-PAGE analyses. Properly folded α-defensins have fewer exposed hydrophobic side chains and thus shorter retention times than reduced or misfolded peptides. The quality of peptides prepared by these methods also has been verified by NMR.
Liposome Preparation for X-ray Measurements
The procedure was similar to that described previously (20). Briefly, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (sodium salt; DOPS) lyophilized lipids (Avanti Polar Lipids) were used without further purification. Small unilamellar vesicles were prepared by sonication. DOPS was dissolved in 90:10 chloroform/methanol, and DOPC and DOPE were dissolved in chloroform for stock solutions at ∼20 mg/ml. Ternary mixtures of DOPS, DOPE, and DOPC were prepared at mass ratios, e.g. 20:60:20 DOPS/DOPE/DOPC is equivalent to a 1:3:1 mass ratio. Lipid mixtures were dried under N2, desiccated overnight under vacuum, and resuspended at a final concentration of 20 mg/ml in Millipore H2O. Solutions were incubated overnight at 37 °C and then sonicated until clear. Small unilamellar vesicles were obtained via extrusion (0.2-μm pore Nucleopore filter).
SAXS Experiments
Lipid solutions were thoroughly mixed with α-defensins at specific peptide/lipid ratios and sealed in quartz capillaries (Mark tubes, code no. 4017515, Hilgenberg GmbH). All samples were prepared in 100 mm NaCl. SAXS experiments were performed at the Stanford Synchrotron Radiation Lightsource (beamline 4-2) and the Advanced Light Source (beamline 7.3.3). Monochromatic x-rays with energies of 10–11 and 10 keV were used at the Stanford Synchrotron Radiation Lightsource and the Advanced Light Source, respectively. Scattered radiation was collected using a Rayonix MX225-HE detector (pixel size, 73.2 μm) at the Stanford Synchrotron Radiation Lightsource and a Pilatus 1M detector (pixel size, 172 μm) at the Advanced Light Source. The SAXS data from both setups were checked for mutual consistency. The two-dimensional SAXS powder patterns were azimuthally integrated, i.e. I(Q)  I(Qr) = ∫I(Qr,Qθ)QrdQθ/(2πQr), using the Nika 1.2 package (Argonne National Laboratory, usaxs.xor.aps.anl.gov/staff/ilavsky/nika.html) for IGOR Pro and FIT2D.
I(Qr) = ∫I(Qr,Qθ)QrdQθ/(2πQr), using the Nika 1.2 package (Argonne National Laboratory, usaxs.xor.aps.anl.gov/staff/ilavsky/nika.html) for IGOR Pro and FIT2D.
Calculation of Cubic and Inverted Hexagonal Phase Lattice Parameters
The procedure was identical to that described previously (20). Q positions of the diffraction peaks were determined by visual inspection of the integrated log(I(Q)) versus Q SAXS data using OriginLab graphing software. These measured peak Q positions were tabulated, and their ratios were compared against the ratios of the permitted reflections for different crystal phases to determine which phases were present in the sample. For example, a Pn3m cubic phase has permitted reflections: (110), (111), (200), (211). For powder averaged cubic phases, Q(hkl) = 2π√(h2 + k2 + l2)/a, where h, k, and l, are the Miller indices, so Q(111)/Q(110) = √3/√2, Q(200)/Q(111) = √4/√2, and Q(211)/Q(110) = √6/√2. Once the crystal phase was determined, its lattice parameter was calculated by a linear trend line fit of the measured assigned peak Q positions, Q(hkl)meas, versus their assigned reflections. For a cubic phase, Q(hkl) = 2π√(h2 + k2 + l2)/a, and for a hexagonal phase, Q(hk) = (4π/√3)√(h2 + k2 + hk)/a. Therefore, we performed linear fits of Q(hkl)meas versus √(h2 + k2 + l2) for bicontinuous cubic (QII) phases or Q(hk)meas versus √(h2 + k2 + hk) for inverted hexagonal (HII) phases. The slope of the linear fit (M) was used to determine cubic (M = 2π/a) and hexagonal (M = 4π/√3a) lattice parameters.
Generation of Solution Structures of Native α-Defensins and Lysine-substituted α-Defensin Variants
The solution structure of Crp4 is from Protein Data Bank code 2GW9 (24). (R/K)-Crp4 was threaded onto the Crp4 template. RMAD4 and (R/K)-RMAD4 were threaded onto the rhesus oral α-defensin-1 template (Protein Data Bank code 2K1I) (25). Threaded structures were generated using SWISS-MODEL (26–28) and analyzed using UCSF Chimera.
RESULTS
In a prior study (5), (R/K)-Crp4 exhibited attenuated bactericidal peptide activity against wild-type strains of Salmonella enterica serovar Typhimurium, and (R/K)-Crp4 required longer peptide exposure times than Crp4 to reduce E. coli cell survival. The bactericidal effects of Crp4, RMAD4, and their respective Lys-for-Arg variants had been compared against Staphylococcus aureus, Listeria monocytogenes, Vibrio cholerae, E. coli ML35, wild-type S. enterica serovar Typhimurium strain 14028S, and phoPc S. enterica serovar Typhimurium strain CS022 (5). Attenuation of (R/K)-Crp4 activity was most evident in assays against S. enterica serovar Typhimurium. In addition, little evident bacterial cell killing occurred when E. coli cells were exposed for 30 min to (R/K)-Crp4, but 99% of E. coli cells were killed when exposed to Crp4 under the same conditions. To confirm and refine this finding, E. coli ML35 cells were exposed in triplicate to 1.5, 3, and 6 μm Crp4 or (R/K)-Crp4 for 15, 30, and 45 min, and surviving colony-forming units were measured as a function of exposure time (Fig. 1). In addition to reproducing the previous 30-min exposure outcomes, the additional 15- and 45-min exposures to peptide illustrated the consistency of the result, confirming the conclusion that (R/K)-Crp4 requires longer peptide exposure times to affect bacterial cell survival. These findings prompted an analysis using SAXS to characterize the phase behavior of these four peptides to investigate whether biophysical correlates exist to these differential α-defensin bactericidal peptide activities.
Small unilamellar vesicles composed of negatively charged DOPS and zwitterionic DOPE and DOPC lipids were incubated with α-defensins. Because the intrinsic curvature of both DOPS and DOPC is approximately zero, whereas DOPE has an intrinsic curvature of <0, ternary mixtures of these lipid types allow the membrane anionic surface charge density and monolayer intrinsic curvature to be independently varied. α-Defensins are thought to target cell membranes with specific phospholipid distributions, such as elevated levels of both anionic and negative intrinsic curvature phospholipids, which model bacterial membranes compared with those of host cells (16, 21). Representative spectra of native Crp4 and (R/K)-Crp4 show that both peptides induced a variety of structure factor correlation peaks, demonstrating that both peptides restructured DOPS/DOPE/DOPC membranes substantially (Fig. 2; see supplemental Fig. S2 for analogous SAXS spectra from anionic 1,2-dioleoyl-sn-glycero-3-phosphoglycerol lipids, which displayed identical phase behavior). For example, in 20:60:20 DOPS/DOPE/DOPC membranes (Fig. 2A), Crp4 induced reflections with peak Q positions at a ratio of 1:2, indicating that Crp4 reorganized 60% of the DOPE liposomes into multilamellar (Lα) phases, stacks of locally flat membranes, with lattice spacing d = 5.80 nm. In membranes with higher levels of negative intrinsic curvature lipids (20:80 DOPS/DOPE), two additional sets of correlation peaks appeared with ratios of 1:√3:√4:√7 and √2:√3:√4:√6:√8, which were indexed (see “Experimental Procedures”) to coexisting inverted hexagonal (HII) and Pn3m cubic (QII) phases with lattice parameters ahex = 7.73 nm and aPn3m = 16.4 nm, respectively. The Pn3m is a QII phase, where the center of the lipid bilayer traces out a minimal surface that has negative Gaussian curvature at every point. Geometrically, the surface will curve upwards in one direction and downwards in a perpendicular direction, locally giving it the shape of a saddle. Negative Gaussian curvature generation is a potent way to disrupt membranes because it is necessary for many different membrane destabilization processes (20, 29). The HII phase is composed of lipid monolayers that have a negative mean curvature. Because negative curvature is necessary (but not sufficient) for negative Gaussian curvature, negative mean curvature deformations are also topologically active and assist in membrane disruption as shown in previous studies on the antibacterial activities of synthetic membrane active antimicrobials (30–32). Therefore, the observed liquid crystalline phases strongly depend on membrane phospholipid composition. This correlates well with specific activity against bacterial membranes rich in PE headgroup lipids and is consistent with earlier reports on both natural (20) and synthetic (30, 31) antimicrobials.
FIGURE 2.
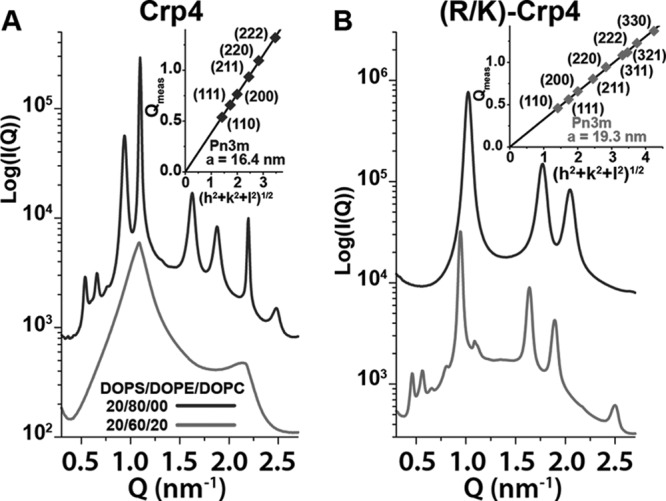
SAXS shows that substitution of lysine for arginine in α-defensin Crp4 substantially changes phase behavior. A, Crp4 induced non-lamellar Pn3m QII and HII phases in PE-rich 20:80 DOPS/DOPE membranes, but not in membranes with reduced amounts of DOPE. B, (R/K)-Crp4 generated a pure HII phase at 20:80 DOPS/DOPE and coexisting QII and HII phases in 20:60:20 DOPS/DOPE/DOPC membranes. For both Crp4 and (R/K)-Crp4, the upper curves correspond to 20:80 DOPS/DOPE membranes, and the lower curves correspond to 20:60:20 DOPS/DOPE/DOPC membranes. SAXS spectra in A and B are at a 1:45 peptide/lipid molar ratio, except for Crp4 at 20:80 DOPS/DOPE, which is at a ratio of 1:67. The insets show indexation of the Pn3m cubic phases and their calculated lattice parameters. The inset for Crp4 corresponds to cubic peaks observed for the 20:80 DOPS/DOPE spectra (upper curve), whereas the inset for (R/K)-Crp4 corresponds to cubic peaks in the 20:60:20 DOPS/DOPE/DOPC spectra (lower curve).
The analogous scattering spectra for (R/K)-Crp4 showed that, in 20:80 DOPS/DOPE membranes, (R/K)-Crp4 induced a pure HII phase with lattice parameter a = 7.09 nm, indicating that the lysine-rich variant peptide generated only negative mean curvature in membranes enriched in negative intrinsic curvature lipids (Fig. 2B). When the DOPE concentration was reduced to 20:60:20 DOPS/DOPE/DOPC, coexisting Pn3m QII (a = 19.3 nm) and HII (a = 7.68 nm) phases were observed. Therefore, (R/K)-Crp4 generated negative Gaussian curvature only in membranes with reduced content of negative intrinsic curvature lipids.
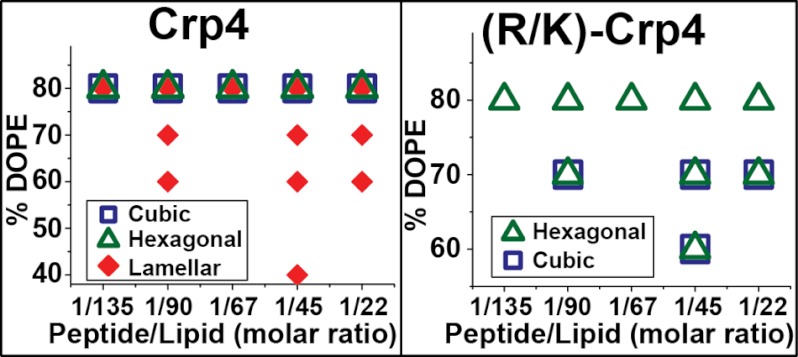
When the phase diagrams (Fig. 3) were compared, substantial effects of Lys substitution on the curvature-generating abilities of Crp4 were observed. For Crp4, non-lamellar QII and HII phases occurred only at the highest (80%) DOPE membrane composition, consistent with its previously reported phase behavior (20). Conversely, (R/K)-Crp4 induced only an HII phase at 80% DOPE and generated both negative Gaussian and negative mean curvatures in membranes with DOPE as low as 60%. In ternary 20:X:(80 − X) DOPS/DOPE/DOPC membranes, the phase progression produced by Crp4 with increasing DOPE content is Lα → QII → HII, a sequence observed in previous experimental and theoretical studies on lyotropic systems (33–36). The effect of complete Arg-to-Lys substitutions in Crp4 is to reduce the quantity of negative intrinsic curvature lipids required for topological transitions to non-lamellar phases.
FIGURE 3.
Crp4 and (R/K)-Crp4 phase diagrams show significant differences in phase behavior. The diagrams show which phases were observed as a function of lipid membrane DOPE content (vertical axis) and peptide/lipid molar ratio (horizontal axis). Ternary lipid compositions containing 20/X/(80 − X) DOPS/DOPE/DOPC (where X = % DOPE) were used in all experiments. Replacement of arginine with lysine in Crp4 decreased the threshold DOPE necessary to induce non-lamellar QII and HII phases with negative Gaussian and negative mean curvatures, respectively. The difference between phase diagrams indicated that Crp4 and (R/K)-Crp4 had different membrane curvature-generating abilities.
In contrast to the behavior of Crp4 and (R/K)-Crp4, the spectra for RMAD4 closely resembled those of (R/K)-RMAD4 (Fig. 4). Native RMAD4 generated coexisting Pn3m (a = 13.3 nm) and Im3m (a = 17.0 nm) QII phases in addition to an HII phase (a = 7.43 nm) in 20:80 DOPS/DOPE membranes (Fig. 4A). Like Pn3m, Im3m is a QII phase rich in negative Gaussian curvature. The ratio of the Im3m and Pn3m lattice parameters (aIm3m/aPn3m = 1.278) is close to the theoretically predicted Bonnet ratio of 1.279 (37), indicating that both QII phases have identical negative Gaussian curvature distributions on their surfaces. In membranes with the same lipid composition, (R/K)-RMAD4 generated coexisting Pn3m (a = 18.2 nm) and HII (a = 7.42 nm) phases (Fig. 4B). The Pn3m cubic phase lattice parameter for RMAD4 is smaller than that for (R/K)-RMAD4, with respective values of 13.3 and 18.2 nm. Because the average negative Gaussian curvature is inversely proportional to the square of the lattice parameter (38), this indicated that the Lys-for-Arg substitutions decreased the amount of negative Gaussian curvature generated by RMAD4. However, the phase behavior of the two peptides was very similar; they both induced negative Gaussian and negative mean curvatures in PE-rich membranes. Furthermore, when the concentration of negative intrinsic curvature lipids was reduced to 20:60:20 DOPS/DOPE/DOPC, Lα phases were observed for both RMAD4 (d = 7.01 nm) and (R/K)-RMAD4 (d = 7.04 nm). These results show that Lys-for-Arg substitutions affect the membrane curvature deformations induced by RMAD4 much less than those induced by Crp4, despite their similar topologies and electropositive charges.
FIGURE 4.
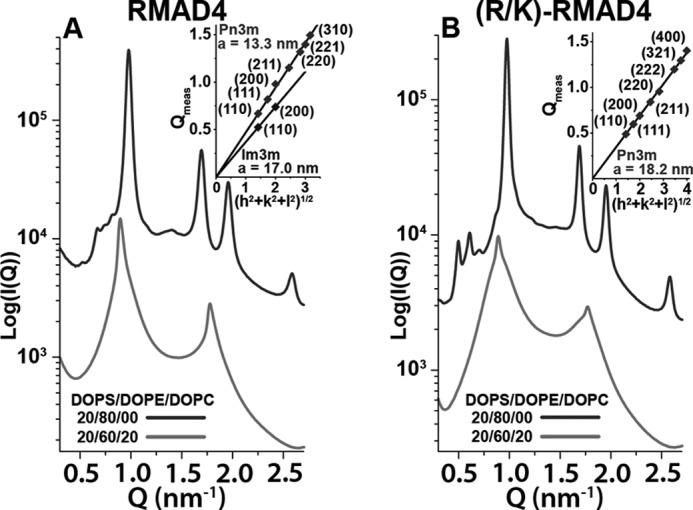
Lys-for-Arg substitutions in α-defensin RMAD4 do not significantly affect phase behavior. Both RMAD4 (A) and (R/K)-RMAD4 (B) generated coexisting non-lamellar QII and HII phases in 20:80 DOPS/DOPE membranes (upper curves), but not in 20:60:20 DOPS/DOPE/DOPC membranes (lower curves). SAXS spectra in A and B are at a peptide/lipid molar ratio of 1:80, and the insets show indexation of QII phases along with their calculated lattice parameters. For both RMAD4 and (R/K)-RMAD4, the insets correspond to the cubic peaks observed in the 20:80 DOPS/DOPE samples (upper curves).
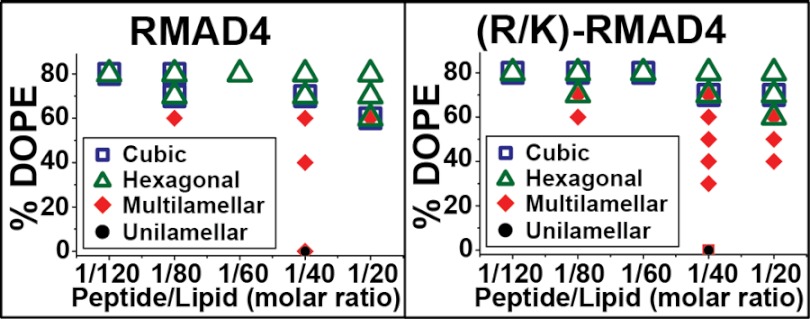
Fig. 5 shows the phase diagrams for RMAD4 and (R/K)-RMAD4 (see supplemental Fig. S3 for similar data for (R1K/R2K/R5K/R33K)-RMAD4). As observed for the Crp4 peptides, the general phase progression of Lα → QII → HII with increasing DOPE membrane content was also seen for RMAD4. Moreover, the peptide/lipid ratio also affected phase behavior, as increasing the peptide/lipid ratio tended to promote non-lamellar QII and HII phase formation. In striking contrast to (R/K)-Crp4, Lys-for-Arg substitutions in RMAD4 did not affect the phase behavior of the molecule. For the three RMAD4 peptides, the presence of non-zero curvature phases began at ∼60% DOPE content. QII and HII phases dominated at 70 and 80% DOPE, respectively. Taken together, the x-ray data show that, although Arg-to-Lys substitutions drastically affect the membrane curvature deformations induced by Crp4, similar cationic residue substitutions in RMAD4 have little effect on its membrane curvature-generating abilities.
FIGURE 5.
Phase diagrams of RMAD4 and (R/K)-RMAD4 are very similar. The diagrams show the observed phases as a function of membrane % DOPE content and peptide/lipid molar ratio. Ternary lipid compositions containing 20/X/(80 − X) DOPS/DOPE/DOPC (where X = % DOPE) were used. The similarity between phase diagrams indicated that replacement of arginine with lysine had little effect on the curvature-generating abilities of RMAD4.
DISCUSSION
All α-defensins examined in this study induced non-lamellar QII and HII phases in membranes enriched in PE lipids, compositions that resemble bacterial cell membranes. By specifically generating membrane-destabilizing negative Gaussian and negative mean curvatures in model bacterial membranes, Crp4, RMAD4, and their Arg-to-Lys variants exploit innate compositional differences between bacterial and mammalian cell membranes that render bacteria selectively more vulnerable to membrane disruption.
The shift of non-lamellar phase generation by (R/K)-Crp4 to membranes with reduced negative intrinsic curvature lipid content is due to the differing membrane curvature-generating tendencies of Arg and Lys. Electrostatic interactions favor a state where the anionic membrane wraps around the peptide to maximize contact and thereby maximize counterion release (39, 40). The entropy gain from such counterion release drives the self-assembly of opposite charged objects in water (41–43). In this way, both Arg and Lys generate negative mean curvature, which takes the form of cylindrical curvature. However, because the guanidinium group of arginine can robustly maintain H-bonds with multiple lipid headgroups, a local deployment of multiple arginines can bind to a high density of lipids along the peptide chain and generate positive curvature from steric interactions. This leads to negative Gaussian curvature (or saddle-shaped curvature). Compared with guanidine, the amine group of lysine has reduced H-bonding ability and thus cannot generate the positive curvature component necessary for negative Gaussian curvature. Therefore, arginine supports generation of negative Gaussian curvature, but lysine substitutions limit peptide capability because lysine favors generation of negative mean curvature only (29, 44). The electrostatically driven generation of HII phases by (R/K)-Crp4 demonstrates that the lysine-rich variant strongly interacts with and binds to membranes, just like arginine-rich native Crp4. The primary effect of Arg-to-Lys substitution in Crp4 is to reduce the amount of total positive curvature generated via removal of negative Gaussian curvature-inducing amino acids, so (R/K)-Crp4 produces HII phases instead of QII phases in PE-rich bacterial membranes. Negative Gaussian curvature generation by different antimicrobial peptides is optimized for specific lipid compositions in the target membrane. In (R/K)-Crp4, this optimal range has shifted away from typical bacterial compositions to lower concentrations of membrane PE, implying that the curvature-producing abilities of (R/K)-Crp4 are not matched with bacterial membranes, so they are no longer able to optimally destabilize them. The result is a peptide that retains some activity but is attenuated compared with the wild type.
We propose that differences between the reliance of Crp4 on arginine to generate negative Gaussian curvature and the insensitivity of RMAD4 to Arg-to-Lys substitutions can be attributed to differences in the distribution of peptide surface cationic residues when interacting with membranes (Fig. 6). Arginine generates negative Gaussian curvature in anionic membranes by inducing negative curvature along one direction via electrostatic interactions (20, 44) and positive curvature along a perpendicular direction via directed lipid headgroup coordination and resultant steric “crowding” effects (22). If the dense presentation of the guanidine group of arginine with lipid headgroups is sufficiently diluted, its bidentate H-bonding (23) ability may diminish to the point that it behaves like lysine. Consistent with this, we have recently shown that alleviation of steric strain in polyarginine by incorporation of flexible PEG chains within polyarginine can make polyarginine behave like polylysine and thereby suppress negative Gaussian curvature generation and Pn3m phases and result in HII phases with negative mean curvature (22). Conversely, we hypothesize that a clustering of lysine residues with reduced H-bonding ability may be able to mimic the multidentate H-bonding of arginine to generate the required negative Gaussian curvature as long as the lysines do not interfere with one another. This is the case for (R/K)-RMAD4, where a relatively dense spatial presentation of Arg is replaced with a similarly dense presentation of Lys.
FIGURE 6.
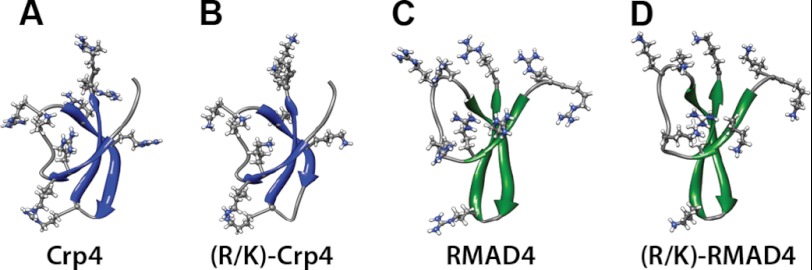
Distribution of electropositive residues in solution structures and homology-modeled threaded structures of native α-defensins and lysine-substituted α-defensin variants. A, Crp4. B, (R/K)-Crp4. C, RMAD4. D, (R/K)-RMAD4. β-Sheet regions in Crp4 peptides are shown in green, and those in RMAD4 peptides are shown in blue. Electropositive residues are represented in a ball-and-stick configuration.
In summary, the changes in phase behavior of the peptide-lipid system resulting from Lys-for-Arg substitutions in Crp4 and RMAD4 correspond directly and with remarkable consistency to their in vitro microbicidal peptide activities. That such a detailed agreement between biophysical results and activity data is possible suggests that specific types of membrane curvature-generating tendencies of antimicrobial peptides provide a useful index of antibacterial activity via membrane destabilization. Therefore, recently proposed design rules for antimicrobial peptides based on the topological requirements of membrane curvature generation (20) constitute an important step toward deterministic molecular engineering of novel peptide mimetic microbicides.
Acknowledgments
We thank the Aspen Center for Physics for promoting this multidisciplinary collaboration. X-ray research was carried out at the Stanford Synchrotron Radiation Lightsource; at the SLAC National Accelerator Laboratory, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Stanford University; at the Advance Light Source, supported by the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract DE-AC02-05CH11231; and at the UCLA California NanoSystems Institute.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK044632 and R01 AI059346 and USC Norris Cancer Center Support Grant P30 CA014089 (to A. J. O.) and Grant 1UO1 AI082192-01. This work was also supported by National Science Foundation Grants DMR0846582, DMR1106106, and DMR0409769.

This article contains supplemental Figs. S1–S3.
- Crp4
- cryptdin-4
- RMAD4
- rhesus myeloid α-defensin-4
- SAXS
- small-angle x-ray scattering
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- QII
- bicontinuous cubic
- HII
- inverted hexagonal
- Lα
- lamellar.
REFERENCES
- 1. Ganz T. (2003) Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 2. Lehrer R. I. (2004) Primate defensins. Nat. Rev. Microbiol. 2, 727–738 [DOI] [PubMed] [Google Scholar]
- 3. Selsted M. E., Ouellette A. J. (2005) Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 4. Ericksen B., Wu Z., Lu W., Lehrer R. I. (2005) Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 49, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Llenado R. A., Weeks C. S., Cocco M. J., Ouellette A. J. (2009) Electropositive charge in α-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect. Immun. 77, 5035–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lehrer R. I., Ganz T. (2002) Defensins of vertebrate animals. Curr. Opin. Immunol. 14, 96–102 [DOI] [PubMed] [Google Scholar]
- 7. Cunliffe R. N. (2003) α-Defensins in the gastrointestinal tract. Mol. Immunol. 40, 463–467 [DOI] [PubMed] [Google Scholar]
- 8. Aley S. B., Zimmerman M., Hetsko M., Selsted M. E., Gillin F. D. (1994) Killing of Giardia lamblia by cryptdins and cationic neutrophil peptides. Infect. Immun. 62, 5397–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanabe H., Qu X., Weeks C. S., Cummings J. E., Kolusheva S., Walsh K. B., Jelinek R., Vanderlick T. K., Selsted M. E., Ouellette A. J. (2004) Structure-activity determinants in Paneth cell α-defensins: loss-of-function in mouse cryptdin-4 by charge reversal at arginine residue positions. J. Biol. Chem. 279, 11976–11983 [DOI] [PubMed] [Google Scholar]
- 10. Territo M. C., Ganz T., Selsted M. E., Lehrer R. (1989) Monocyte chemotactic activity of defensins from human neutrophils. J. Clin. Invest. 84, 2017–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chertov O., Michiel D. F., Xu L., Wang J. M., Tani K., Murphy W. J., Longo D. L., Taub D. D., Oppenheim J. J. (1996) Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J. Biol. Chem. 271, 2935–2940 [DOI] [PubMed] [Google Scholar]
- 12. Befus A. D., Mowat C., Gilchrist M., Hu J., Solomon S., Bateman A. (1999) Neutrophil defensins induce histamine secretion from mast cells: mechanisms of action. J. Immunol. 163, 947–953 [PubMed] [Google Scholar]
- 13. Hristova K., Selsted M. E., White S. H. (1996) Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry 35, 11888–11894 [DOI] [PubMed] [Google Scholar]
- 14. Satchell D. P., Sheynis T., Kolusheva S., Cummings J., Vanderlick T. K., Jelinek R., Selsted M. E., Ouellette A. J. (2003) Quantitative interactions between cryptdin-4 amino-terminal variants and membranes. Peptides 24, 1795–1805 [DOI] [PubMed] [Google Scholar]
- 15. Satchell D. P., Sheynis T., Shirafuji Y., Kolusheva S., Ouellette A. J., Jelinek R. (2003) Interactions of mouse Paneth cell α-defensins and α-defensin precursors with membranes. Prosegment inhibition of peptide association with biomimetic membranes. J. Biol. Chem. 278, 13838–13846 [DOI] [PubMed] [Google Scholar]
- 16. Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 17. Su Y., Doherty T., Waring A. J., Ruchala P., Hong M. (2009) Roles of arginine and lysine residues in the translocation of a cell-penetrating peptide from 13C, 31P, and 19F solid-state NMR. Biochemistry 48, 4587–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rothbard J. B., Jessop T. C., Wender P. A. (2005) Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells. Adv. Drug Delivery Rev. 57, 495–504 [DOI] [PubMed] [Google Scholar]
- 19. Figueredo S., Mastroianni J. R., Tai K. P., Ouellette A. J. (2010) in Antimicrobial Peptides (Giuliani A., Rinaldi A. C., eds) pp. 47–60, Humana Press, Totowa, NJ [Google Scholar]
- 20. Schmidt N. W., Mishra A., Lai G. H., Davis M., Sanders L. K., Tran D., Garcia A., Tai K. P., McCray P. B., Ouellette A. J., Selsted M. E., Wong G. C. (2011) Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133, 6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Epand R. M., Epand R. F. (2009) Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta 1788, 289–294 [DOI] [PubMed] [Google Scholar]
- 22. Mishra A., Lai G. H., Schmidt N. W., Sun V. Z., Rodriguez A. R., Tong R., Tang L., Cheng J., Deming T. J., Kamei D. T., Wong G. C. (2011) Translocation of HIV TAT peptide and analogues induced by multiplexed membrane and cytoskeletal interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schug K. A., Lindner W. (2005) Noncovalent binding between guanidinium and anionic groups: focus on biological and synthetic based arginine/guanidinium interactions with phosph(on)ate and sulf(on)ate residues. Chem. Rev. 105, 67–114 [DOI] [PubMed] [Google Scholar]
- 24. Rosengren K. J., Daly N. L., Fornander L. M., Jönsson L. M, Shirafuji Y., Qu X., Vogel H. J., Ouellette A. J., Craik D. J. (2006) Structural and functional characterization of the conserved salt bridge in mammalian Paneth cell α-defensins. Solution structures of mouse cryptdin-4 and (E15D)-cryptdin-4. J. Biol. Chem. 281, 28068–28078 [DOI] [PubMed] [Google Scholar]
- 25. Vasudevan S., Yuan J., Ösapay G., Tran P., Tai K., Liang W., Kumar V., Selsted M. E., Cocco M. J. (2008) Synthesis, structure, and activities of an oral mucosal α-defensin from rhesus macaque. J. Biol. Chem. 283, 35869–35877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peitsch M. C. (1995) Protein modeling by e-mail. Nat. Biotechnol. 13, 658–660 [Google Scholar]
- 27. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 28. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt N., Mishra A., Lai G. H., Wong G. C. (2010) Arginine-rich cell-penetrating peptides. FEBS Lett. 584, 1806–1813 [DOI] [PubMed] [Google Scholar]
- 30. Yang L., Gordon V. D., Mishra A., Som A., Purdy K. R., Davis M. A., Tew G. N., Wong G. C. (2007) Synthetic antimicrobial oligomers induce a composition-dependent topological transition in membranes. J. Am. Chem. Soc. 129, 12141–12147 [DOI] [PubMed] [Google Scholar]
- 31. Yang L., Gordon V. D., Trinkle D. R., Schmidt N. W., Davis M. A., DeVries C., Som A., Cronan J. E., Jr., Tew G. N., Wong G. C. (2008) Mechanism of a prototypical synthetic membrane-active antimicrobial: efficient hole punching via interaction with negative intrinsic curvature lipids. Proc. Natl. Acad. Sci. U.S.A. 105, 20595–20600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Som A., Yang L., Wong G. C., Tew G. N. (2009) Divalent metal ion-triggered activity of a synthetic antimicrobial in cardiolipin membranes. J. Am. Chem. Soc. 131, 15102–15103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shyamsunder E., Gruner S. M., Tate M. W., Turner D. C., So P. T., Tilcock C. P. (1988) Observation of inverted cubic phase in hydrated dioleoylphosphatidylethanolamine membranes. Biochemistry 27, 2332–2336 [DOI] [PubMed] [Google Scholar]
- 34. Anderson D. M., Gruner S. M., Leibler S. (1988) Geometrical aspects of the frustration in the cubic phases of lyotropic liquid crystals. Proc. Natl. Acad. Sci. U.S.A. 85, 5364–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. So P. T., Gruner S. M., Erramilli S. (1993) Pressure-induced topological phase transitions in membranes. Phys. Rev. Lett. 70, 3455–3458 [DOI] [PubMed] [Google Scholar]
- 36. Siegel D. P., Kozlov M. M. (2004) The Gaussian curvature elastic modulus of N-monomethylated dioleoylphosphatidylethanolamine: relevance to membrane fusion and lipid phase behavior. Biophys. J. 87, 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shearman G. C., Ces O., Templer R. H., Seddon J. M. (2006) Inverse lyotropic phases of lipids and membrane curvature. J. Phys. Condens. Matter 18, S1105–S1124 [DOI] [PubMed] [Google Scholar]
- 38. Harper P. E., Gruner S. M. (2000) Electron density modeling and reconstruction of infinite periodic minimal surfaces (IPMS)-based phases in lipid-water systems. I. Modeling IPMS-based phases. Eur. Phys. J. E 2, 217–228 [Google Scholar]
- 39. May S., Ben-Shaul A. (1997) DNA-lipid complexes: stability of honeycomb-like and spaghetti-like structures. Biophys. J. 73, 2427–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koltover I., Salditt T., Rädler J. O., Safinya C. R. (1998) An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 281, 78–81 [DOI] [PubMed] [Google Scholar]
- 41. Wong G. C, Tang J. X., Lin A., Li Y., Janmey P. A., Safinya C. R. (2000) Hierarchical self-assembly of F-actin and cationic lipid complexes: stacked three-layer tubule networks. Science 288, 2035–2039 [DOI] [PubMed] [Google Scholar]
- 42. Yang L., Liang H., Angelini T. E., Butler J., Coridan R., Tang J. X., Wong G. C. (2004) Self-assembled virus-membrane complexes. Nat. Mater. 3, 615–619 [DOI] [PubMed] [Google Scholar]
- 43. Sanders L. K., Xian W., Guáqueta C., Strohman M. J., Vrasich C. R., Luijten E., Wong G. C. (2007) Control of electrostatic interactions between F-actin and genetically modified lysozyme in aqueous media. Proc. Natl. Acad. Sci. U.S.A. 104, 15994–15999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mishra A., Gordon V. D., Yang L., Coridan R., Wong G. C. (2008) HIV TAT forms pores in membranes by inducing saddle-splay curvature: potential role of bidentate hydrogen bonding. Angew. Chem. Int. Ed. Engl. 47, 2986–2989 [DOI] [PubMed] [Google Scholar]