Background: Obesity increases the risk for cancer development, suggesting that adipose tissue dysfunctions might play a crucial role therein. Macrophages infiltrate adipose tissue as well as tumors.
Results: Human adipose tissue macrophages (ATM) resemble human tumor-associated macrophages (TAM).
Conclusion: ATM may modulate cancer cell function.
Significance: ATM may be potential contributors to cancer development in obese subjects.
Keywords: Adipose Tissue, Cancer, Gene Expression, Macrophages, Obesity
Abstract
Obesity is associated with a significantly increased risk for cancer suggesting that adipose tissue dysfunctions might play a crucial role therein. Macrophages play important roles in adipose tissue as well as in cancers. Here, we studied whether human adipose tissue macrophages (ATM) modulate cancer cell function. Therefore, ATM were isolated and compared with monocyte-derived macrophages (MDM) from the same obese patients. ATM, but not MDM, were found to secrete factors inducing inflammation and lipid accumulation in human T47D and HT-29 cancer cells. Gene expression profile comparison of ATM and MDM revealed overexpression of functional clusters, such as cytokine-cytokine receptor interaction (especially CXC-chemokine) signaling as well as cancer-related pathways, in ATM. Comparison with gene expression profiles of human tumor-associated macrophages showed that ATM, but not MDM resemble tumor-associated macrophages. Indirect co-culture experiments demonstrated that factors secreted by preadipocytes, but not mature adipocytes, confer an ATM-like phenotype to MDM. Finally, the concentrations of ATM-secreted factors related to cancer are elevated in serum of obese subjects. In conclusion, ATM may thus modulate the cancer cell phenotype.
Introduction
Obesity is a low-grade chronic inflammatory disease due to the infiltration of immune-inflammatory cells including monocytes, which differentiate in macrophages and form aggregates in crown-like structures usually surrounding dead adipocytes. The number of adipose tissue macrophages (ATM)3 correlates with the body mass index (1). Within adipose tissue (AT), ATM are a major source of chemokines and inflammatory cytokines, such as IL-6 and TNFα, which may on their turn promote the recruitment of additional immune inflammatory cells. In animal models, AT macrophage infiltration and ensuing inflammation precedes the development of insulin resistance (2). Surface marker analysis revealed that human ATM present an anti-inflammatory phenotype (3) but are capable of producing pro-inflammatory mediators (4). ATM are surrounded by preadipocytes and adipocytes, and it is likely that paracrine loops exist via the production of adipokines, free fatty acids, and derived mediators, leading to inflammatory changes. In particular, preadipocytes produce higher levels of proinflammatory cytokines compared with adipocytes, suggesting that they play an important role in the induction and maintenance of inflammation (5, 6) and may thus operate as ATM activators.
Interestingly, there are now epidemiological evidences establishing obesity as a risk factor for the development of cancer such as colon, breast, esophagus, kidney, liver, and pancreas cancer (7). Whereas weight gain is accompanied by higher cancer incidence rates, recent longitudinal studies on metabolic surgery revealed that weight loss results in lower cancer rates (8). During obesity, the expanded AT could contribute to cancer development via a deregulated secretion of proinflammatory cytokines, chemokines, and adipokines (9). Based on these observations, we thus hypothesized that ATM could be involved in the development of cancer related to obesity. Interestingly, we found that conditioned medium from ATM, but not from autologous monocyte-derived macrophages (MDM) isolated from the same morbidly obese patients, induced phenotypic changes and activation of human breast cancer cells. To characterize the phenotypical differences between human ATM and MDM, a global gene expression analysis was performed on ATM and MDM isolated from the same morbidly obese patients. Several genes over-represented in ATM, compared with MDM, were identified to belong to cytokine-cytokine receptor interaction pathways (including chemokine, hematopoietin, PDGF, interleukin, TNF, interferon, and TGFβ pathways) identifying a specific ATM signature. Interestingly, ATM displayed a gene expression profile sharing similarities with human tumor-associated macrophages (TAM) (10, 11). Accordingly, ATM were found to produce a repertoire of growth factors, cytokines, chemokines, and proteolytic enzymes involved in the regulation of tumor growth, angiogenesis, invasion, and/or promotion of cancer metastasis, as observed in TAM (12). Finally, we show that the ATM phenotype is programmed by the cellular environment as demonstrated by indirect co-culture experiments applying the preadipocyte-conditioned media to MDM from lean subjects, resulting in an ATM-like phenotype.
These data indicate that the TAM-like human ATM phenotype is directed by surrounding cell types and suggest that factors released by ATM may contribute to cancer initiation and progression in obese patients. These observations suggest that ATM may be potential contributors to cancer development in obese humans.
EXPERIMENTAL PROCEDURES
Cell Culture
Visceral AT biopsies (n = 11) were obtained from non-diabetic morbidly obese patients undergoing bariatric surgery at the Internal Surgery Department, Hospital of Lille (Lille, France) (see supplemental Table 1 for patient characteristics). This study was approved by the ethics committee of the CHRU of Lille under the Atlas Biologique de l′Obésité Sévère (ABOS) and OMENTectomie au Cours de la Gastrectomie en Manchon Chez l′OBèse Sévère: Étude Pilote Contrôlée et Randomisee (OMENTOB) frameworks. None of the patients had any clinical symptoms of systemic inflammation or cancer. All patients gave informed consent. To avoid contamination of ATM by blood monocytes, the vessels were dissected carefully, and blood was removed by extensive PBS washes before AT digestion. AT samples were digested in Krebs buffer (pH 7.4) containing collagenase (1.5 mg/ml, Roche Diagnostics). After filtration through a 200-μm filter (Spectra Mesh; Biovalley) and centrifugation, floating adipocytes were washed with PBS, and released lipids were collected. The stromal vascular fraction was collected as described (13). ATM-CD14+ cells were isolated using CD14-labeled magnetic beads (Miltenyi Biotec). ATM were cultured for 24 h in endothelial cell basal medium (Promocell) supplemented with 0.1% BSA to dampen possible inflammation responses due to magnetic bead separation and isolation procedures (13). Medium was then changed, cells were cultured for another 24 h with fresh medium, and supernatants were collected thereafter.
The CD14− fraction (containing endothelial cells, fibroblasts, lymphocytes, stem cells, and preadipocytes) was cultured in preadipocyte basal medium (Promocell) for 24 h. Adherent preadipocytes were then cultured in selective preadipocyte growth medium (PGM, Promocell) during 3 days before collecting the preadipocyte-conditioned medium (PCM). Human monocytes were isolated from fresh blood of the same obese or from lean patients, differentiated for 9 days in the presence of 10% pooled human serum (14) and treated for 72h with indicated PGM or PCM.
Human TAM were isolated as described (10, 15) from solid tumors of untreated patients with histologically confirmed epithelial ovarian carcinoma (n = 7) admitted to the San Gerardo Hospital (Monza, Italy). Human T47D breast ductal carcinoma and HT-29 colon adenocarcinoma cells were obtained from ATCC (Rockville, MD).
Immunohistochemical Analysis and Laser Capture Microdissection of Human Atherosclerotic Plaques
Human atherosclerotic plaques (n = 5) were removed from patients eligible for surgical carotid endarterectomy recruited at the Cardiovascular Surgery Department (Hospital of Lille, Lille, France). Informed consent was obtained from all patients. Proinflammatory CD68-positive and mannose receptor-negative macrophages were isolated from lipid-rich areas within human atherosclerotic plaques by laser capture microdissection as described previously (16).
Oil Red O Staining
T47D cells were treated with ATM or MDM-conditioned media, fixed with 4% paraformaldehyde in PBS, stained using Oil Red O and nuclei counterstained with hematoxylin (40× magnification).
RNA Extraction and Real Time Quantitative Polymerase Chain Reaction (Q-PCR) Analysis
Cellular RNA was extracted using RNeasy kits (Qiagen). Total RNA was reverse transcribed using the high capacity cDNA reverse transcription kit (Applied Biosystems). cDNAs were quantified by Q-PCR on an MX4000 apparatus (Agilent Biotechnologies) using specific primers (supplemental Table 2). Statistical differences were analyzed by Student's t tests and considered significant when p < 0.05.
Sample Preparation for Gene Expression and Data Analysis of Affymetrix Oligonucleotide Arrays
Total RNA was checked for quantity and quality using the Agilent 2100 Bioanalyzer (Agilent Biotechnologies). ATM, MDM, and TAM RNA samples were processed using HG-U133 plus 2.0 (ATM and MDM) and HG-U133A (TAM) Affymetrix Genechips. The MatchProbes package (42) was used to match the probe sequences between the GeneChips. Quality assurance of the hybridization was assessed by constructing a virtual image of the chip by plotting each residual obtained with each probe at the corresponding position using the affyPLM package. Raw gene expression data were processed and normalized with the Affymetrix GCOS software (version 1.4) and by multiarray analysis (Robust Multichip Analysis) (17).
Statistical and Informatic Methods
The raw microarray data were preprocessed using Bioconductor tools to generate a single expression value for each probe set in each sample. To identify expressed genes, a statistical analysis based on individual probe information was applied using the R2.6 software. A statistical t test was used to assess how likely changes in differential expression occurred by chance (threshold = 0.05). Signal intensity ratios and p values were calculated for each probe set using the linear model library (Limma), allowing identification of genes differentially expressed between ATM and MDM. Genes showing statistically significant differences of p ≤ 10−5 were selected for further analysis. Based on previous literature, the minimum cut off in intensities per gene was set at 1.5-fold change (FC) (18). Differential gene expression was verified by Q-PCR on ATM and MDM obtained from seven additional donors.
Functional Classification, Visualization, and Pathway Analysis
Selected gene lists were uploaded in the DAVID (database for annotation, visualization, and integrated discovery) website (19). Bioinformatics resource and the functional annotation and classification gene tools were used to generate clusters of related genes (20). Integrative data analysis, including multiple graphical visualizations, was performed using GenespringGX10 (Agilent Biotechnologies) and pathway analysis by the Kyoto Encyclopedia of Genes and Genomes (KEGG). The profile of functional pathways was analyzed using ProfCom software (21). This software allows comparing the proportion of genes related to specific gene ontology categories (biological process, cellular component, and molecular function) among the ATM higher expressed genes to the proportion of genes related to the same category within the gene ontology reference set of all genes from Homo sapiens (21).
Prediction of ATM Protein Secretion
Secreted protein prediction was conducted by interrogating the list of the highly expressed ATM genes (FC ≥ 1.5; p ≤ 10−5) using the SignalP3.0 (22, 23) and SecretomeP2.0 (22) software to identify genes with the highest probability of encoding either signal peptides or protein sequences predictive of non-classic secretion, respectively, using a cut-off N score of 0.7 (22). The corresponding genes were marked as predicted or not predicted. Gene encoding sequences that met the cut-off value for either classic or non-classic secretion were marked as classical or non-classical secretory genes.
Protein Extraction and Western Blot Analysis
MDM were harvested in ice-cold protein lysis buffer (radioimmune precipitation assay buffer). Cell homogenates were collected by centrifugation, and proteins were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane (Amersham Biosciences). Membranes were then subjected to immunodetection using rabbit polyclonal antibodies against Stat3, Stat1, p44/p42 MAPK (ERK1/2), and their phosphorylated forms, Stat3 (Tyr-705, Ser-727), Stat1 (Tyr-701, Ser-727), p44/p42 MAPK (Thr-202/Tyr-204) (all from Cell Signaling) and β-actin (Santa Cruz Biotechnology). After incubation with appropriate secondary antibodies, immunoreactive bands were revealed using a chemiluminescence HRP substrate detection kit (ImmobilonTM Western, Millipore).
Measurement of Circulating CXCL Proteins
Sera were collected from obese and lean subjects (20 per group) and CXCL1, 2, 3 (Gro), CXCL6, CXCL7, CXCL9, CXCL10, CXCL11, sgp130, and TGFβ1 concentrations were measured by Luminex technology using commercially available kits (Millipore for all, except TGFβ1 (R&D Systems)). CXCL16 was measured by ELISA (R&D Systems). Statistical differences between groups were analyzed by Mann-Whitney rank-sum test and were considered significant at p < 0.05.
RESULTS
ATM-conditioned Medium Induces Lipid Accumulation and Expression of Inflammatory Markers in Human Breast Cancer Cells
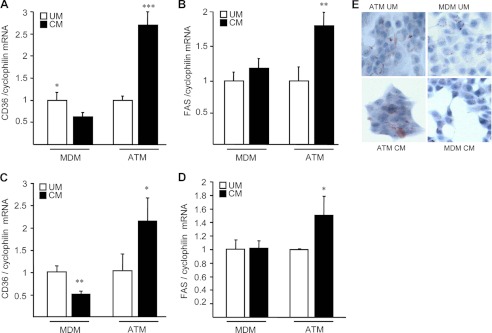
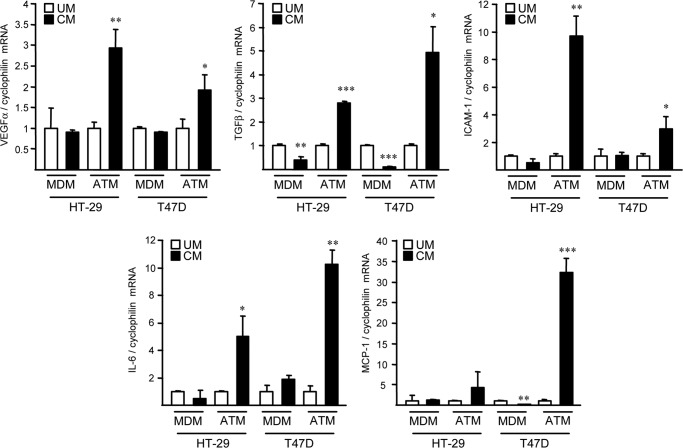
Because several lines of evidences suggest a potential link between obesity and cancer and as macrophages play important roles in both pathologies, we determined whether secreted factors from ATM can impact on tumor cells. Therefore, indirect co-culture experiments were performed on human T47D breast cancer and HT-29 colon adenocarcinoma cells in the presence of conditioned media (CM) from ATM or MDM isolated from the same obese subjects. Interestingly, ATM-CM, but not MDM-CM, increased the expression of CD36 and fatty acid synthase in T47D and HT-29 cells and enhanced lipid accumulation in T47D cells, as determined by Oil Red O staining (Fig. 1), a phenotype associated to a poor cancer prognosis (24). Moreover, ATM-CM and not MDM-CM also modulated the expression of several genes involved in angiogenesis and inflammation in HT-29 and T47D cells, such as VEGFα, TGFβ, ICAM-1, IL-6, and MCP-1 (Fig. 2). Taken together, these results indicate that ATM-CM, but not MDM-CM can modulate the phenotype of human cancer cells.
FIGURE 1.
ATM-conditioned medium induces lipid accumulation in human cancer cells. T47D (A, B, and E) and HT-29 (C and D) cells were treated for 72 h with ATM or MDM-unconditioned medium (UC) or CM, and mRNA levels of CD36 (A and C) and fatty acid synthase (FAS; B and D) were analyzed by Q-PCR and normalized to cyclophilin mRNA. Results are expressed relative to the levels of cells in the presence of ATM or MDM unconditioned medium set at 1. Each bar is the mean value ± S.D. of triplicate determinations. Statistically significant differences are indicated (*, p < 0.05; **, p < 0.01; ***, p < 0.001). E, Oil Red O staining performed on T47D cells. Results represent a single experiment repeated four times with similar results.
FIGURE 2.
Different gene expression regulation by ATM versus MDM-conditioned medium in human cancer cells. HT-29 and T47D cancer cells were treated for 72 h with unconditioned (UM) or ATM- or MDM-CM. mRNA levels of VEGFA, TGF-β1, ICAM-1, MCP-1, and IL-6 subsequently were analyzed by Q-PCR and normalized to cyclophilin mRNA. Results are expressed relative to the levels of cells in the presence of unconditioned medium set at 1. Each bar is the mean value ± S.D. of triplicate determinations. Statistically significant differences are indicated (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Identification of Genes Differentially Expressed between ATM and MDM
To characterize the differences in phenotype between ATM and MDM, a whole genome microarray analysis was performed on ATM and MDM from the same donors. To specifically evaluate the effect of obesity and not of other metabolic components such as diabetes, only obese non-diabetic subjects were included. Hierarchical clustering of the entire Affymetrix chip and after probe sets (PSN) filtration revealed a normal gene expression distribution and a strong similarity in the global gene expression profile between ATM and MDM (supplemental Fig. 1). Based on our criteria of selection stringency, >80% of PSN were similarly expressed and <20% of PSN showed differences. Normalized intensity values of gene expression in ATM were higher than the 75th percentile, whereas those of MDM were rather lower than the 25th percentile, indicating that gene expression is globally higher in ATM than in MDM (supplemental Fig. 2A). The numbers of transcript abundance differences (increased or decreased) between ATM and MDM are shown by a volcano plot (supplemental Fig. 2B). 1371 PSN were identified as higher expressed (Log2Ratio > 0.58 corresponding to a FC > 1.5; p < 0.01), and 1771 PSN were lower expressed in ATM compared with MDM (log2 ratio < 0.58 corresponding to a FC < −1.5; p < 0.01).
Functional pathway analysis revealed that the cytokine-cytokine receptor interaction (CCRI) pathway is the most significantly over-represented pathway in ATM (based on the p value) (supplemental Fig. 2C). Moreover, a large number of other pathways, including the NOD-like receptor, chemokine, MAPK, Toll-like receptor, RIG-I-like receptor, complement and coagulation cascades, Jak-STAT, focal adhesion, extracellular matrix-receptor interaction and tight junction pathways, were overexpressed in ATM. Surprisingly, several cancer signaling pathways (acute myeloid leukemia, small cell lung, and bladder cancer) also were found to be higher expressed in ATM (supplemental Fig. 2C). Gene ontology analysis identified a number of genes associated with certain gene ontology categories (biological process, cellular components, molecular function) to be higher expressed in ATM. ATM most prominently expressed genes related to transcription regulation, cell adhesion and signaling, inflammation, and immune response. Genes implicated in the activity of transcription factors, signal transduction, growth factors, and chemokines also were significantly enriched in ATM (supplemental Fig. 2D). By contrast, genes related to metabolism, ion binding, transferase, and oxidoreductase activities were higher expressed in MDM, whereas genes related to general macrophage functions such as phagocytosis and cell cytotoxicity were expressed at the same level in ATM and MDM (data not shown).
Genes Belonging to CCRI Pathways Are Highly Expressed in ATM
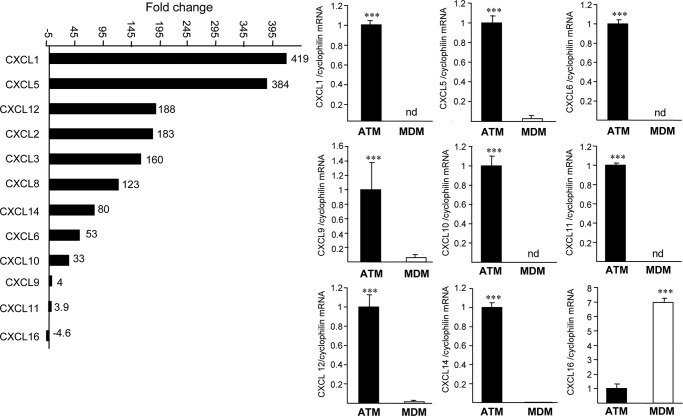
Because the CCRI pathways are overexpressed most significantly in ATM, we focused our attention on these pathways, which include the chemokine (from the CC and C-X-C subfamilies), hematopoietin (oncostatin M (OSM), gp130 (IL6ST)), PDGF, interleukin, TNF, interferon, and TGFβ families. Interestingly, several members of the CC chemokine ligand family, such as CCL2, -3, -4, -5, -7, -8, -11, -18, and -20 (Fig. 3 and supplemental Fig. 3) and the majority of CXCL ligands (CXCL1, -2, -3, -5, -6, -7, -9, -10, -11, -12, and -14), but not their receptors (except for CCR6 and CCR7), were higher expressed in ATM than in MDM (supplemental Fig. 3), whereas only CXCL16 scavenger receptor for phosphatidyl serine and oxidized low density lipoprotein (SR-PSOX) was higher expressed in MDM (Fig. 3). Q-PCR analysis performed on ATM and MDM preparations from seven additional obese individuals confirmed these results (Fig. 3).
FIGURE 3.
Genes belonging to the CC and CXC chemokine subfamily are expressed highly in ATM. Histogram of Affymetrix FC values of CXC chemokine ligand genes differently expressed between ATM and MDM and Q-PCR analysis of CXCL1, CXCL5, CXCL6, CXCL9, CXCL10, CXCL11, CXCL12, CXCL14, and CXCL16 in ATM and MDM from seven donors. mRNA levels were normalized to cyclophilin mRNA, and results were expressed as mean ± S.D. of triplicate determinations relative to the levels in ATM set at 1. Statistically significant differences are indicated (***, p < 0.001). nd, non-detectable.
Within the CCRI pathway, major differences were observed in the expression of genes of the hematopoietin family, such as IL-6, IL6-ST, IL-11, leukemia inhibitory factor, OSM, and their receptors (supplemental Fig. 3C). Q-PCR analysis confirmed the different expression pattern of OSM, OSM-R, and IL6-ST (supplemental Fig. 3D). Gene expression profiling and Q-PCR analysis also confirmed the differential expression of several members of the TGFβ signaling pathways, which play an important role in the regulation of macrophage immune functions (supplemental Fig. 3, E–G). To determine whether the expression profile of CXCL genes was a specific signature of ATM or a common characteristic of inflammatory macrophages, their expression levels were analyzed in proinflammatory CD68+ macrophages isolated from lipid-rich areas within human atherosclerotic plaques by laser capture microdissection and compared with levels in ATM.
Our results indicate that the expression level of CXCL1, CXCL10, and CXCL11 was much lower in CD68+ atherosclerotic plaque macrophages compared with ATM. Expression of CXCL9 and CXCL12 was highly variable between samples also because of the relatively low expression levels of these two chemokines (supplemental Fig. 4). Moreover, CXCL5 mRNA expression was not detectable in CD68+ atherosclerotic plaque macrophages. Overall, these data indicate that the expression profile of CXCL members in ATM is different from those obtained in atherosclerotic plaque inflammatory macrophages.
Large Number of Highly Expressed Genes in ATM Encode Predicted Secreted Proteins
SignalIP (version 3.0) and SecretomeP (version 2.0) software were used to predict secreted proteins from ATM (supplemental Table 3). 57 genes of the 1066 genes overexpressed in ATM were not identified by the software and are thus marked as unknown (supplemental Fig. 5). A total of 320 of the remainder 1009 genes (30%) were predicted to encode secreted proteins. Among these 30%, ∼17% were classically secreted proteins, and ∼13% corresponded to non-classical secreted proteins. Proteins with anchor signals, which represent ∼4% of the gene list, also were considered because they can be processed by cleavage to soluble factors with biological functions. Thus, 34% of ATM-enriched genes encode predicted secreted proteins, suggesting an over-representation of secreted proteins in ATM relative to their abundance in the human genome (15–20%). Grouping of genes in classes according to the FC (1 to 200; 201 to 400; 401 to 600; 601 to 800; 801 to 1066) (supplemental Fig. 5 and Table 3) showed that the highest percentage of predicted secreted proteins was found in the gene class showing the highest FC, again suggestive of an enrichment of predicted secreted proteins in ATM, compared with MDM.
Preadipocytes Determine ATM Phenotype
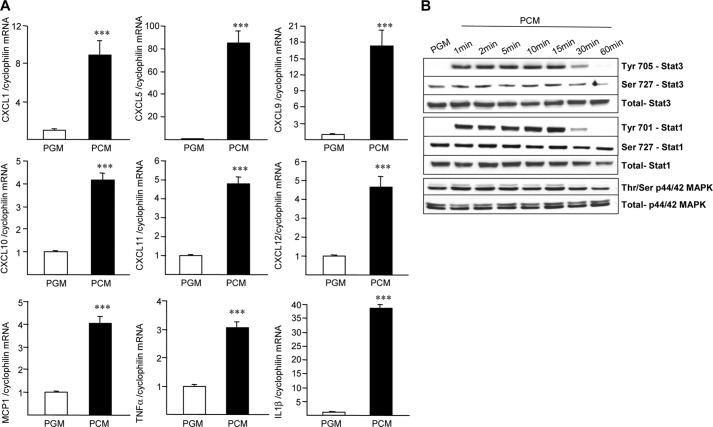
To address whether and how the AT environment (cells and lipids) can affect the ATM characteristics, indirect co-culture experiments were performed on MDM from lean subjects incubated with conditioned media from preadipocytes from obese subjects (PCM). A substantial increase of CXCL1, -5, -10, -11, and -12 as well as MCP-1, TNFα, and IL-1β gene expression, resembling the ATM profile, was observed in lean MDM treated with PCM compared with control medium (PGM) (Fig. 4A). By contrast, no modification or a decreased expression of most CXCL genes was observed after incubation with released lipids from mature adipocytes directly isolated from AT (supplemental Fig. 6). These results indicate that preadipocytes secretion products determine the ATM phenotype. Western blot analysis of total and phosphorylated forms of Stat3, Stat1, and p44/p42 MAPK revealed that incubation of MDM with PCM very rapidly induced tyrosine phosphorylation of Stat3 and Stat1 as well as threonine/serine phosphorylation of p44/p42 MAPK (Fig. 4B).
FIGURE 4.
PCM induces an ATM-like phenotype in MDM. MDM from lean subjects were treated with PCM or unconditioned PGM during 72 h (A). mRNA levels of CXCL1, CXCL5, CXCL9, CXCL10, CXCL11, CXCL12, MCP-1, TNFα, and IL-1b were analyzed by Q-PCR and normalized to cyclophilin mRNA. Results are representative of three experiments and are expressed relative to the levels of cells in the presence of PGM set at 1. Each bar is the mean value ± S.D. of triplicate determinations. Statistically significant differences are indicated (***, p < 0.001). MDM were treated with PCM for the indicated time periods (B). Western blot analysis was performed using antibodies against total or phosphorylated forms of Stat3, Stat1, and p44/p42 MAPK (ERK1/2).
ATM Phenotype Resembles Phenotype of TAM
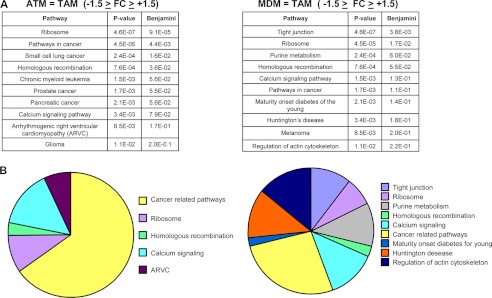
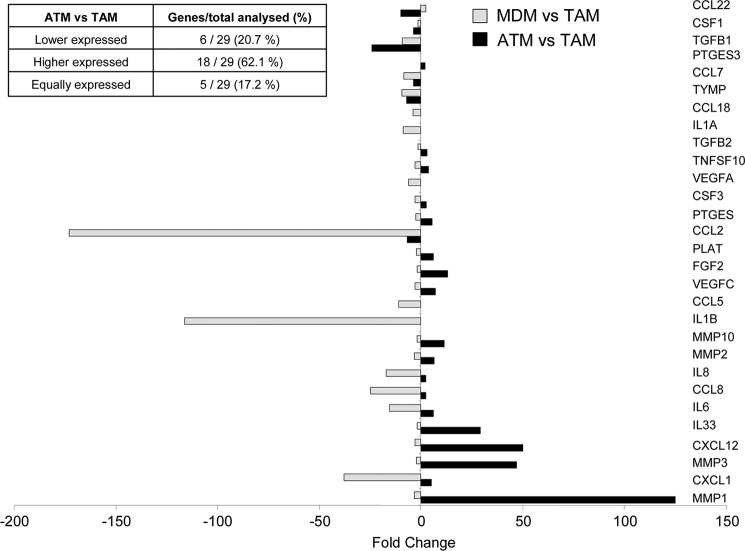
Because ATM but not MDM were able to modify the cancer cell phenotype, we decided to determine whether a possible resemblance could exist between ATM and TAM by comparing the expression profile of ATM and MDM to that of TAM isolated from human tumors (10). Based on our stringency criteria, ∼80% of PSN were similarly expressed between the three types of macrophages, and ∼20% of PSN showed differences (supplemental Fig. 7). Next, it was analyzed which genes are expressed similarly between ATM and TAM and between MDM and TAM (−1.5 > FC > +1.5). KEGG analysis revealed that among the 10 most significantly represented pathways shared between ATM and TAM (based on the p value), the majority of them are cancer-related pathways (small lung cancer, chronic myeloid leukemia, prostate cancer, pancreatic cancer, and glioma). By contrast, the cancer-related pathways in MDM represented only a small number of the pathways shared with TAM (Fig. 5), indicating an enrichment of cancer-related pathways in ATM compared with MDM. Moreover, based on the literature (11), 29 genes highly expressed by TAM, including angiogenic factors, chemokines, cytokines, proteases, and growth factors, were also higher in ATM compared with MDM (supplemental Fig. 7). Interestingly, the majority of these genes were higher or equally expressed in ATM compared with TAM (Fig. 6). By contrast, the expression of the same genes was generally lower in MDM compared with TAM (Fig. 6). These data demonstrate that human ATM display a TAM-like phenotype.
FIGURE 5.
ATM exhibit a gene expression profile similar to TAM. A, top 10 of the most significant KEGG molecular pathways, including genes similarly expressed between ATM/TAM and MDM/TAM, respectively. B, pie chart representations of the top 10 KEGG pathways; cancer-related pathways have been considered together.
FIGURE 6.
TAM-expressed genes are enriched in ATM compared with MDM. Shown is a histogram of Affymetrix FC values of selected TAM genes differently expressed between ATM/TAM and MDM/TAM, respectively.
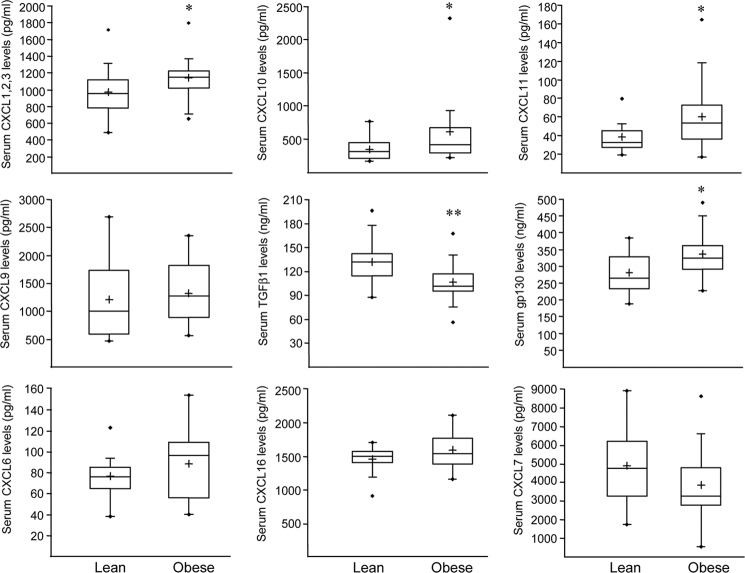
Serum Concentrations of CXCL1, -2, -3, -10, and 11 Are Elevated in Obese Subjects
To determine whether circulating concentrations of CXCL chemokines and some factors belonging to the CCRI pathways, such as sgp130 and TGFβ1, which are over-represented in ATM, are altered in vivo in obese individuals, serum from obese and lean subjects was analyzed. The serum levels of total CXCL1, -2, -3 (Gro), CXCL 10, -11, and sgp130 were elevated significantly in obese compared with lean individuals, whereas the level of the anti-proliferative factor TGFβ1 was decreased significantly in obese individuals (Fig. 7). Altogether, these results indicate that the expression of several factors highly expressed in ATM and related to cancer correlates with circulating levels in obese subjects without malignancies.
FIGURE 7.
Obesity is associated with altered serum concentrations of ATM-expressed factors. Serum protein concentrations of CXCL1, -2, -3 (Gro), CXCL6 (GCP-2), CXCL7 (NAP-2), CXCL9 (MIG), CXCL10 (IP-10), CXCL11 (I-TAC), CXCL16 (S-PROX), TGFβ1, and sgp130 were quantified in obese and lean subjects. Statistically significant differences are indicated (*, p < 0.05; **, p < 0.01).
DISCUSSION
Obesity is a low-grade chronic inflammatory state characterized by an increased number of infiltrated macrophages and altered adipocytokine production in AT (1). ATM are a major source of inflammatory cytokines and mediators involved in obesity induced-insulin resistance, such as IL-6 and TNFα. Interestingly, experimental and epidemiological studies have shown an association between excessive adiposity and increased cancer incidence and death, probably as a consequence of the obesity-associated metabolic and endocrine perturbations (7). Together, these observations raise the hypothesis that ATM can contribute to the link between obesity and cancer development.
Here, we show that CM from ATM, but not from MDM isolated from the same obese donors, increase the expression of fatty acid synthase and CD36, leading to lipid accumulation in cancer cells. Interestingly, tumors overexpressing fatty acid synthase display a more aggressive behavior compared with those with normal fatty acid synthase levels because fatty acid synthase plays a pivotal role in cancer cell survival and is directly involved in the maintenance or enhancement of the malignant phenotype (25). Fatty acid synthase is overexpressed in breast cancers with poor prognosis (24, 26–28). In addition, several inflammatory molecules known to be dysregulated in cancer were enhanced in T47D and HT-29 cells by ATM-CM. Altogether, these results indicate that factors released by ATM, but not by MDM, could affect tumor cell phenotype. Many tumors have increased levels of obesity-related factors, both adipokines and inflammatory components, in their microenvironment, rendering, in some cases, the tumors more aggressive (29). Thus, macrophages in peritumoral adipose tissue, which release obesity-related factors, could be involved locally in enhancing carcinogenesis.
Therefore, we compared global gene expression profiles of ATM to MDM isolated from the same obese patients. Functional cluster analysis revealed that genes involved in inflammatory pathways were found to be enriched in ATM compared with MDM, among which the CCRI pathway was the most significant. Surprisingly, several cancer-related signaling pathways were also higher expressed in ATM versus MDM. This phenotype was confirmed by comparing the MDM and ATM gene profiles to those of TAM isolated from solid human tumors (10).
Within the AT, ATM interact in a paracrine way with surrounding cells (T cells, preadipocytes, adipocytes, and endothelial and stromal cells) via the production of hormone-like and angiogenic factors, which can promote survival of damaged cells and thus can actively participate in the promotion of tumors by endocrine and/or paracrine mechanisms (30). Within tumors, TAM participate in the progression/evolution of cancer and their infiltration correlates with cancer metastasis and poor prognosis in a variety of human carcinomas (31, 32). TAM promote cancer metastasis in established advanced tumor stages through mechanisms such as stimulation of angiogenesis (31, 32) via a wide variety of factors which stimulate blood vessel growth and/or maturation. Our analysis revealed that the expression of angiogenic factors, chemokines, cytokines, proteases, and growth factors (CCLs (MCP-1 … ), CXCLs (Gro, CXCL7 … ), IL-1α/β, IL-6, FGF2), which are also actively secreted by TAM (11), are higher expressed in ATM versus MDM. The majority of CC and CXC chemokine ligands (CCL, CXCL) are highly expressed in ATM. Chemokines mediate recruitment of immune cells (neutrophils, dendritic cells, lymphocytes, and monocytes) at the site of inflammation, play a role in cell proliferation, inhibition of apoptosis as well as angiogenesis, both in AT and in the tumor microenvironment (33–35). CCL and CXCL are induced by the typical Th1 cytokine IFNγ alone or in combination with other inflammatory cytokines (36). Interestingly, different members of the IFNγ subfamily also are higher expressed in ATM (supplemental Fig. 4), suggesting a possible amplification of the Th1 immune response (12). It is worth noting that ATM overexpressed the ligands, but not the receptors of these chemokines, with an exception for CXCR6, CX3CR1, and CCR7, suggesting that the ligands produced by ATM may exert paracrine effects on surrounding cells possessing their cognate receptors, such as mature adipocytes (37). Surprisingly, the only CXCL lower expressed in ATM was CXCL16, also involved in leukocyte migration and adhesion (38). Importantly, high expression levels of CXCL16 in renal cancer tissue has been correlated with a better survival rate of patients (39, 40). Our study also reveals that circulating levels of Gro (CXCL1, -2, -3), CXCL9, -10, -11 (CXCR3 ligands), and sgp130, key players in the maintenance and amplification of autoimmunity-related inflammatory processes (35), are elevated in obese subjects, whereas anti-proliferative TGFβ1 concentrations are lower (41). ATM overexpressed both OSM family members (IL-6, LIF, OSM, IL-11) and their receptors. Indirect co-culture experiments on MDM indicated that the phenotype of ATM is related specifically to factors released by the preadipocytes, whereas lipids released from mature adipocytes do not induce these effects. These data are in line with previous observations, suggesting the high proinflammatory potential of preadipocytes (5, 6).
Collectively, our data indicate that ATM may contribute to the link between obesity and cancer and revealed for these cells a gene expression profile comparable to those of TAM.
Acknowledgments
We are grateful to the patients who participated in this study. Members of the F. Pattou surgery team, particularly M. F. Six, C. Eberle, and L. Arnalsteen are acknowledged. We are grateful to P. Gelé (Centre d'Investigation Clinique, CHRU, Lille) for blood and serum collection and to J. Berthout, A. Blondy, E. Vallez, C. Rommens, A. Lucas, C. Copin, A. Patrice, V. Gmyr, S. Deledicque, D. Hot, and L. Huot for technical assistance.
This work was supported by the “Nouvelle Société Française d'Athérosclérose/Sanofi-Aventis” (to T. H. M.), the “Société Française de Cardiologie/Sanofi-Aventis,” the European Concerted Research Action BM0602, and the European Community's 7th Framework Programme (FP7/2007-2013) under Grant 201608.

This article contains supplemental Tables 1–3, Figs. 1–7, and data.
- ATM
- adipose tissue macrophage(s)
- MDM
- monocyte-derived macrophage(s)
- TAM
- tumor-associated macrophage(s)
- AT
- adipose tissue
- PGM
- preadipocyte growth medium
- PCM
- preadipocyte-conditioned medium
- Q-PCR
- quantitative PCR
- CM
- conditioned media
- CCRI
- cytokine-cytokine receptor interaction
- OSM
- oncostatin M
- PSN
- probe set(s)
- FC
- fold change
- CHRU
- Centre Hospitalier Regionale Universitaire.
REFERENCES
- 1. Curat C. A., Wegner V., Sengenès C., Miranville A., Tonus C., Busse R., Bouloumié A. (2006) Macrophages in human visceral adipose tissue: Increased accumulation in obesity and a source of resistin and visfatin. Diabetologia 49, 744–747 [DOI] [PubMed] [Google Scholar]
- 2. Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aron-Wisnewsky J., Tordjman J., Poitou C., Darakhshan F., Hugol D., Basdevant A., Aissat A., Guerre-Millo M., Clément K. (2009) Human adipose tissue macrophages: m1 and m2 cell surface markers in subcutaneous and omental depots and after weight loss. J. Clin. Endocrinol. Metab. 94, 4619–4623 [DOI] [PubMed] [Google Scholar]
- 4. Zeyda M., Farmer D., Todoric J., Aszmann O., Speiser M., Gyori G., Zlabinger G., Stulnig T. (2007) Human adipose tissue macrophages are of an anti-inflammatory phenotype but capable of excessive pro-inflammatory mediator production. Int. J. Obes. 31, 1420–1428 [DOI] [PubMed] [Google Scholar]
- 5. Poulain-Godefroy O., Froguel P. (2007) Preadipocyte response and impairment of differentiation in an inflammatory environment. Biochem. Biophys. Res. Commun. 356, 662–667 [DOI] [PubMed] [Google Scholar]
- 6. Mack I., BelAiba R. S., Djordjevic T., Görlach A., Hauner H., Bader B. L. (2009) Functional analyses reveal the greater potency of preadipocytes compared with adipocytes as endothelial cell activator under normoxia, hypoxia, and TNFα exposure. Am. J. Physiol. Endocrinol. Metab. 297, E735–748 [DOI] [PubMed] [Google Scholar]
- 7. Calle E. E., Kaaks R. (2004) Overweight, obesity, and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 4, 579–591 [DOI] [PubMed] [Google Scholar]
- 8. Adams T. D., Hunt S. C. (2009) Cancer and obesity: Effect of bariatric surgery. World J. Surg. 33, 2028–2033 [DOI] [PubMed] [Google Scholar]
- 9. Prieto-Hontoria P. L., Pérez-Matute P., Fernández-Galilea M., Bustos M., Martínez J. A., Moreno-Aliaga M. J. (2011) Role of obesity-associated dysfunctional adipose tissue in cancer: A molecular nutrition approach. Biochim. Biophys. Acta 1807, 664–678 [DOI] [PubMed] [Google Scholar]
- 10. Solinas G., Schiarea S., Liguori M., Fabbri M., Pesce S., Zammataro L., Pasqualini F., Nebuloni M., Chiabrando C., Mantovani A., Allavena P. (2010) Tumor-conditioned macrophages secrete migration-stimulating factor: A new marker for M2 polarization, influencing tumor cell motility. J. Immunol. 185, 642–652 [DOI] [PubMed] [Google Scholar]
- 11. Ono M. (2008) Molecular links between tumor angiogenesis and inflammation: Inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 99, 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mantovani A., Sica A. (2010) Macrophages, innate immunity, and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 22, 231–237 [DOI] [PubMed] [Google Scholar]
- 13. Curat C. A., Miranville A., Sengenès C., Diehl M., Tonus C., Busse R., Bouloumié A. (2004) From blood monocytes to adipose tissue-resident macrophages: Induction of diapedesis by human mature adipocytes. Diabetes 53, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 14. Chinetti G., Griglio S., Antonucci M., Torra I. P., Delerive P., Majd Z., Fruchart J. C., Chapman J., Najib J., Staels B. (1998) Activation of proliferator-activated receptors α and γ induces apoptosis of human monocyte-derived macrophages. J. Biol. Chem. 273, 25573–25580 [DOI] [PubMed] [Google Scholar]
- 15. Saccani A., Schioppa T., Porta C., Biswas S. K., Nebuloni M., Vago L., Bottazzi B., Colombo M. P., Mantovani A., Sica A. (2006) p50 nuclear factor-κB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 66, 11432–11440 [DOI] [PubMed] [Google Scholar]
- 16. Chinetti-Gbaguidi G., Baron M., Bouhlel M. A., Vanhoutte J., Copin C., Sebti Y., Derudas B., Mayi T., Bories G., Tailleux A., Haulon S., Zawadzki C., Jude B., Staels B. (2011) Human atherosclerotic plaque alternative macrophages display low cholesterol handling but high phagocytosis because of distinct activities of the PPARγ and LXRα pathways. Circ. Res. 108, 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 18. Shah R., Lu Y., Hinkle C. C., McGillicuddy F. C., Kim R., Hannenhalli S., Cappola T. P., Heffron S., Wang X., Mehta N. N., Putt M., Reilly M. P. (2009) Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes 58, 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 20. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 21. Antonov A. V., Schmidt T., Wang Y., Mewes H. W. (2008) ProfCom: A web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 36, W347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 23. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8, 581–599 [DOI] [PubMed] [Google Scholar]
- 24. Alo' P. L., Visca P., Marci A., Mangoni A., Botti C., Di Tondo U. (1996) Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer 77, 474–482 [DOI] [PubMed] [Google Scholar]
- 25. Menendez J. A., Lupu R. (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 [DOI] [PubMed] [Google Scholar]
- 26. Esslimani-Sahla M., Thezenas S., Simony-Lafontaine J., Kramar A., Lavaill R., Chalbos D., Rochefort H. (2007) Increased expression of fatty acid synthase and progesterone receptor in early steps of human mammary carcinogenesis. Int. J. Cancer 120, 224–229 [DOI] [PubMed] [Google Scholar]
- 27. Di Lorenzo D., Giannì M., Savoldi G. F., Ferrari F., Albertini A., Garattini E. (1993) Progesterone induced expression of alkaline phosphatase is associated with a secretory phenotype in T47D breast cancer cells. Biochem. Biophys. Res. Commun. 192, 1066–1072 [DOI] [PubMed] [Google Scholar]
- 28. Ronen D., Altstock R. T., Firon M., Mittelman L., Sobe T., Resau J. H., Vande Woude G. F., Tsarfaty I. (1999) Met-HGF/SF mediates growth arrest and differentiation in T47D breast cancer cells. Cell Growth Differ. 10, 131–140 [PubMed] [Google Scholar]
- 29. Kulbe H., Thompson R., Wilson J. L., Robinson S., Hagemann T., Fatah R., Gould D., Ayhan A., Balkwill F. (2007) The inflammatory cytokine tumor necrosis factor-α generates an autocrine tumor-promoting network in epithelial ovarian cancer cells. Cancer Res. 67, 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao Y. (2010) Adipose tissue angiogenesis as a therapeutic target for obesity and metabolic diseases. Nat. Rev. Drug Discov. 9, 107–115 [DOI] [PubMed] [Google Scholar]
- 31. Steidl C., Lee T., Shah S. P., Farinha P., Han G., Nayar T., Delaney A., Jones S. J., Iqbal J., Weisenburger D. D., Bast M. A., Rosenwald A., Muller-Hermelink H. K., Rimsza L. M., Campo E., Delabie J., Braziel R. M., Cook J. R., Tubbs R. R., Jaffe E. S., Lenz G., Connors J. M., Staudt L. M., Chan W. C., Gascoyne R. D. (2010) Tumor-associated macrophages and survival in classic Hodgkin lymphoma. N. Engl. J. Med. 362, 875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 33. Mantovani A., Savino B., Locati M., Zammataro L., Allavena P., Bonecchi R. (2010) The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. 21, 27–39 [DOI] [PubMed] [Google Scholar]
- 34. Koch A. E., Polverini P. J., Kunkel S. L., Harlow L. A., DiPietro L. A., Elner V. M., Elner S. G., Strieter R. M. (1992) Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science 258, 1798–1801 [DOI] [PubMed] [Google Scholar]
- 35. Lacotte S., Brun S., Muller S., Dumortier H. (2009) CXCR3, inflammation, and autoimmune diseases. Ann. N.Y. Acad. Sci. 1173, 310–317 [DOI] [PubMed] [Google Scholar]
- 36. Martinez F. O., Gordon S., Locati M., Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 37. Gerhardt C. C., Romero I. A., Cancello R., Camoin L., Strosberg A. D. (2001) Chemokines control fat accumulation and leptin secretion by cultured human adipocytes. Mol. Cell. Endocrinol. 175, 81–92 [DOI] [PubMed] [Google Scholar]
- 38. Shimaoka T., Nakayama T., Fukumoto N., Kume N., Takahashi S., Yamaguchi J., Minami M., Hayashida K., Kita T., Ohsumi J., Yoshie O., Yonehara S. (2004) Cell surface-anchored SR-PSOX/CXC chemokine ligand 16 mediates firm adhesion of CXC chemokine receptor 6-expressing cells. J. Leukoc. Biol. 75, 267–274 [DOI] [PubMed] [Google Scholar]
- 39. Gutwein P., Schramme A., Sinke N., Abdel-Bakky M. S., Voss B., Obermüller N., Doberstein K., Koziolek M., Fritzsche F., Johannsen M., Jung K., Schaider H., Altevogt P., Ludwig A., Pfeilschifter J., Kristiansen G. (2009) Tumoral CXCL16 expression is a novel prognostic marker of longer survival times in renal cell cancer patients. Eur. J. Cancer 45, 478–489 [DOI] [PubMed] [Google Scholar]
- 40. Meijer J., Ogink J., Kreike B., Nuyten D., de Visser K. E., Roos E. (2008) The chemokine receptor CXCR6 and its ligand CXCL16 are expressed in carcinomas and inhibit proliferation. Cancer Res. 68, 4701–4708; Retraction; Ogink, J., Kreike, B., Nuyten, D., de Visser, K. E., and Roos, E. (2011) Cancer Res.3, 1196 [DOI] [PubMed] [Google Scholar]
- 41. James A. W., Xu Y., Lee J. K., Wang R., Longaker M. T. (2009) Differential effects of TGF-β1 and TGF-β3 on chondrogenesis in posterofrontal cranial suture-derived mesenchymal cells in vitro. Plast. Reconstr. Surg. 123, 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huber W., Gentleman R. (2004) Matchprobes: A Bioconductor package for the sequence-matching of microarray probe elements. Bioinformatics 20, 1651–1652 [DOI] [PubMed] [Google Scholar]