Background: Heparan sulfate (HS) has important roles in cellular signaling that depend on the arrangement of sulfated domains.
Results: Drosophila HS contains an essentially single sulfated domain.
Conclusion: The domain organization of Drosophila HS is novel and differs from previously described HS structures.
Significance: The findings may provide novel insight into structure-function relationships for HS.
Keywords: Biosynthesis, Carbohydrate Structure, Drosophila, Glycosaminoglycan, Heparan Sulfate, Disaccharide Composition, Domain Organization
Abstract
Heparan sulfate (HS) proteoglycans play critical roles in a wide variety of biological processes such as growth factor signaling, cell adhesion, wound healing, and tumor metastasis. Functionally important interactions between HS and a variety of proteins depend on specific structural features within the HS chains. The fruit fly (Drosophila melanogaster) is frequently applied as a model organism to study HS function in development. Previous structural studies of Drosophila HS have been restricted to disaccharide composition, without regard to the arrangement of saccharide domains typically found in vertebrate HS. Here, we biochemically characterized Drosophila HS by selective depolymerization with nitrous acid. Analysis of the generated saccharide products revealed a novel HS design, involving a peripheral, extended, presumably single, N-sulfated domain linked to an N-acetylated sequence contiguous with the linkage to core protein. The N-sulfated domain may be envisaged as a heparin structure of unusually low O-sulfate content.
Introduction
Heparan sulfate (HS)4 glycosaminoglycans constitute a family of polysaccharides structurally related to (but less sulfated than) heparin and characterized by extensive structural heterogeneity and variability. In contrast to heparin, which occurs exclusively in connective tissue-type mast cells, HS is produced by most cells in the body and is expressed, largely in proteoglycan form, at cell surfaces and in the extracellular matrix. HS has been ascribed diverse and fundamental roles in development and homeostasis due to interactions, more or less electrostatic in nature, with a multitude of proteins. The polysaccharide occurs throughout the evolutionary system, from Cnidaria onward (1, 2). Several reviews dealing with the structural, metabolic, and functional aspects of HS proteoglycans have been published (3–8).
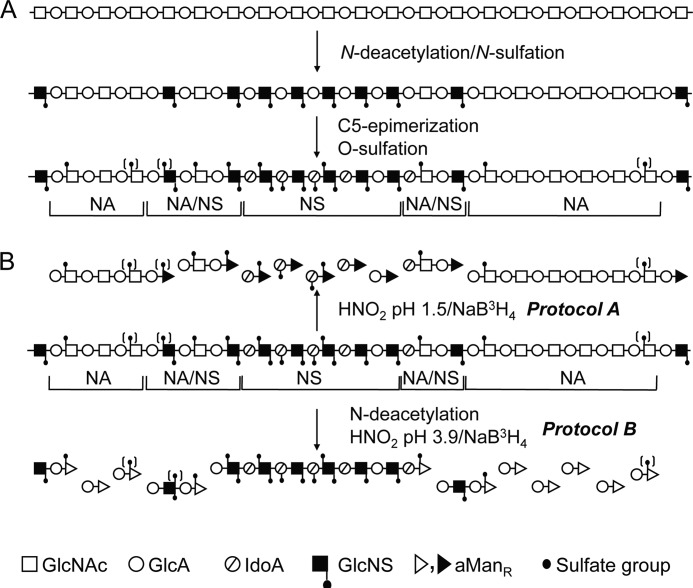
Biosynthesis of HS involves polymerization of alternating d-glucuronic acid (GlcUA) and 2-deoxy-2-acetamido-d-glucose (N-acetyl-d-glucosamine; GlcNAc) residues, yielding a (GlcUAβ1,4GlcNAcα1,4)n precursor polymer covalently attached to a proteoglycan core protein through a distinct “linkage region” (8–10). The product is modified through a complex series of reactions involving N-deacetylation/N-sulfation of GlcNAc (yielding GlcNS, where NS is an N-sulfate group) residues, C-5 epimerization of GlcUA to iduronic acid (IdoUA) units, and O-sulfation of hexuronic acids (HexUA) at C-2 and glucosamine residues at C-3 and C-6. Because these reactions generally engage only a fraction of potentially available target residues, the final products have heterogeneous structures that vary with tissue source (11). The bifunctional N-deacetylation/N-sulfation enzymes (N-deacetylase/N-sulfotransferase (NDST); four mammalian isoforms described in mammals) have a key role in the overall polymer modification process because subsequent GlcUA C-5 epimerization and O-sulfation reactions generally depend on the presence of GlcNS residues. Due to selective NDST action, vertebrate HS chains typically display domains of consecutive N-sulfated disaccharide units (NS-domains), alternating N-acetylated and N-sulfated units (NA/NS-domains), and essentially unmodified N-acetylated sequences (NA-domains) (see Fig. 1A). IdoUA and O-sulfate residues are confined to the NS- and NA/NS-domains (12). Heparin may be considered an unusually extended and highly O-sulfated NS-domain.
FIGURE 1.
Structure of HS chain and approaches for analysis. A, the structural domains (NS-, NA/NS-, and NA-domains) are defined with regard to the distribution of GlcNS and GlcNAc units. B, schematic representation of methods used to elucidate HS domain structure. For further information, see “Results” and “Experimental Procedures.” The symbols used are defined below the scheme. GlcA, GlcUA; IdoA, IdoUA.
Available information regarding HS structure derives predominantly from analysis of vertebrate samples. Characterization of HS/heparin from non-vertebrate sources revealed some unusual disaccharide units (1, 13) but, so far, no information regarding domain structure. Manipulation of genes encoding enzymes involved in biosynthesis of Drosophila HS defined roles of HS in various developmental events, again, however, only in terms of overall disaccharide composition (14–19). We therefore decided to approach Drosophila HS from a different angle, concentrating on domain organization as revealed by selective deaminative cleavage. The results point to a glycosaminoglycan domain of novel design, best envisaged as a low-sulfated heparin.
EXPERIMENTAL PROCEDURES
Purification of Drosophila HS
Batches of 15 or 30 g of dechorionated wild-type (Oregon R) Drosophila embryos from different embryonic stages (0–24 h) were digested with Pronase, followed by treatment with NaOH/NaBH4 essentially as described (1). Briefly, embryos were digested with 100 mg of Pronase E (Merck) in 20 ml of 0.1 m Tris-HCl (pH 8.0) containing 0.002 m CaCl2 at 55 °C for 24 h. Thereafter, an additional aliquot (50 mg) of enzyme in 2.5 ml of digestion buffer was added, and the digestion was continued for another 24 h. To release O-linked sugar chains (including HS) from the attached peptide generated by the protease digestion and to reduce the liberated O-linked saccharides, 4 m NaOH (final concentration, 0.5 m) and 20 mg of NaBH4 were added to the digest, and the mixture was kept overnight at 4 °C. After neutralization with 4 m HCl, the mixture was centrifuged at 100,000 × g for 1 h. Supernatants were adjusted to 0.15 m NaCl and pH 8.0 by the addition of Tris-HCl and applied to a 10-ml column of DEAE-Sephacel (GE Healthcare) equilibrated with 0.15 m NaCl and 50 mm Tris-HCl (pH 8.0). The column was first washed with equilibrium buffer and then with 50 mm acetate buffer (pH 4.0) containing 0.15 m NaCl. The pH was increased by washing with 0.15 m NaCl and 50 mm Tris-HCl (pH 8.0), and finally, DEAE-bound material was eluted with 2 m NaCl and 50 mm Tris-HCl (pH 8.0). Two-ml fractions were collected and analyzed for uronic acid content by the carbazole reaction (20). Carbazole-positive fractions were pooled and dialyzed against water. The dialyzed material was digested with 125 units of Benzonase (Merck) and then applied to an ∼1-ml column of DEAE-Sephacel, which either was subsequently eluted as described above, followed by collection of 1-ml fractions, or was subjected to stepwise elution with 1-ml portions of 0.2, 0.4, 0.5, 0.75, 1.0, and 2.0 m NaCl in 50 mm Tris-HCl (pH 8.0). Uronic acid-positive fractions were pooled and desalted on PD-10 gel filtration columns (Sephadex G-25, GE Healthcare) or dialyzed against water.
Structural Analysis of Drosophila HS
Characterization of Drosophila HS domain structure was approached by two complementary protocols (see Fig. 1B). Deamination of GlcNS residues by reaction with nitrous acid at pH 1.5 followed by reduction of the products with NaB3H4 (0.5 mCi for 5–10 μg of polysaccharide) yielded disaccharides from contiguous NS-domains or N-acetylated oligosaccharides with reducing terminal 3H-labeled 2,5-anhydro-d-mannitol (aManR) residues (Protocol A) (12). Conversely, HS samples digested with chondroitinases ABC and AC (Seikagaku), hence devoid of chondroitin sulfate, were chemically N-deacetylated by hydrazinolysis (treatment with 70% (w/v) aqueous hydrazine (Fluka) containing 1% (w/v) hydrazine sulfate at 96 °C for 4 h) (21), and the products were treated with nitrous acid at pH 3.9 followed by reduction with NaB3H4 (Protocol B). Cleavage at N-unsubstituted GlcN residues converted the original NA-domains to labeled disaccharides and NS-domains to N-sulfated oligosaccharides (see Fig. 1B). Labeled saccharides were separated from unincorporated radioactivity by gel chromatography on a Sephadex G-15 column (1 × 170 cm) in 0.2 m NH4HCO3. Fractions corresponding to di- and oligosaccharides were pooled and desalted by lyophilization. Labeled saccharides were analyzed for molecular size by gel chromatography on a Bio-Gel P-10 column (1.5 × 175 cm, fine grade, Bio-Rad) in 0.5 m NaCl.
The compositions of labeled disaccharides obtained by either Protocol A or B were determined by strong anion-exchange HPLC using a Whatman Partisil 10 SAX column (22) in combination, when required, with high-voltage paper electrophoresis in 0.83 m pyridine and 0.5 m acetic acid (pH 5.3) and paper chromatography as described (23). A schematic outline of the overall experimental protocol is shown in supplemental Fig. S1.
RESULTS
Frozen mixed-stage Drosophila embryos were digested with Pronase and treated with alkali to isolate free saccharide chains as described under “Experimental Procedures.” Carbazole-positive material bound to DEAE and eluted with ≥0.4 m NaCl was considered to represent free glycosaminoglycan chains. Most of the carbazole-positive material was eluted with 0.4 and 0.75 m NaCl. Approximately 2–3 μg of uronic acid, corresponding to 5–8 μg of polysaccharide, was obtained per gram of starting material.
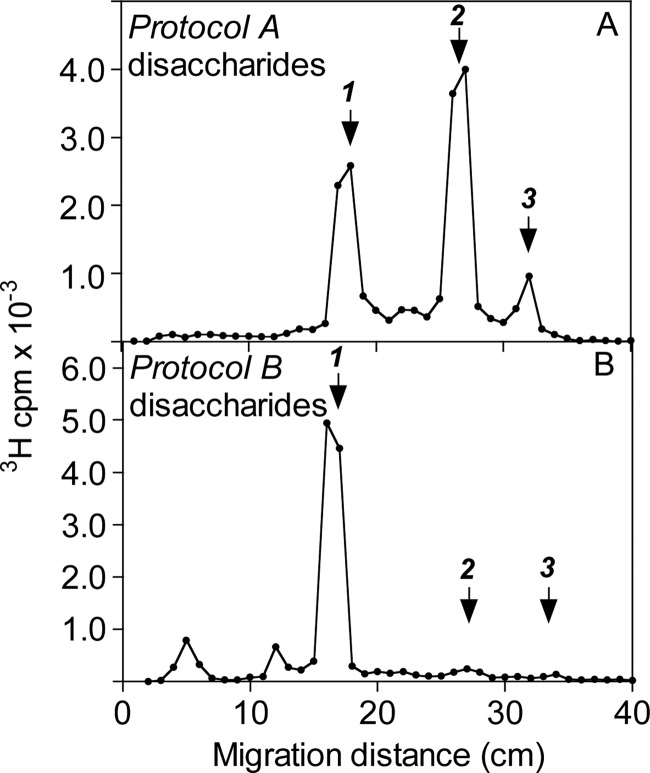
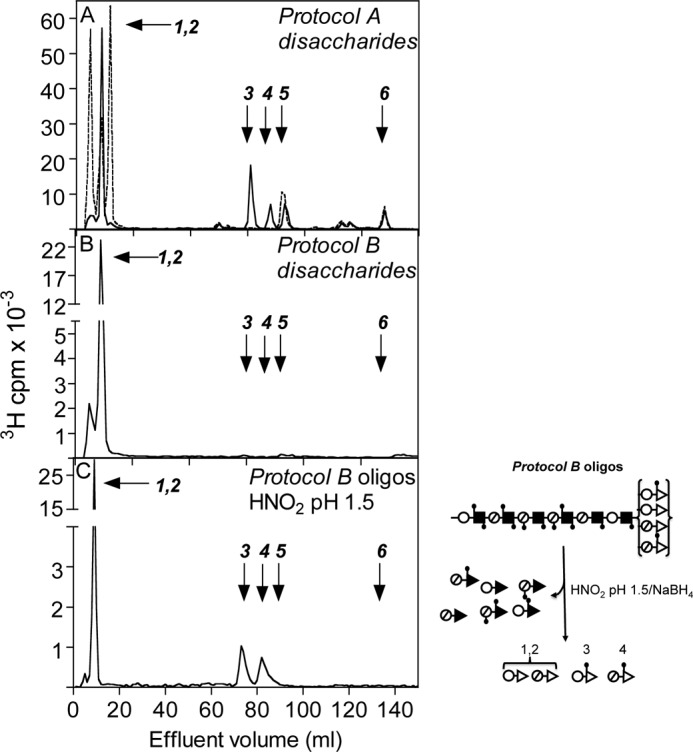
The domain organization of Drosophila HS was investigated by selective deaminative cleavage, as outlined in Fig. 1B. Deamination/reduction according to Protocol A will cleave glucosaminidic linkages emanating from N-sulfated GlcN residues and thus convert NS-domains into hexuronyl-[3H]aManR disaccharides. The mixed NA/NS-domains will yield labeled tetrasaccharides, whereas NA-domains will remain intact. A reference sample of HS isolated from pig intestinal mucosa was subjected to Protocol A treatment, and the reduced products were analyzed by gel chromatography on Bio-Gel P-10 (Fig. 2A). The expected pattern of fragments was observed, with predominant di- and tetrasaccharide peaks along with smaller peaks of larger N-acetylated oligosaccharides. (Note that these oligosaccharides are end-labeled; hence, peak areas need to be multiplied by the number of disaccharide units to reflect relative abundance.) By contrast, Drosophila HS yielded disaccharides as the only product of significance, with only trace amounts of material emerging in the position of tetrasaccharides (Fig. 2B). The generation of disaccharides and the lack of larger oligomers pointed to a parent polysaccharide containing significant extended N-sulfated sequence, uninterrupted by N-acetylated units (Fig. 1). In fact, the degradation pattern of Drosophila HS suggested an N-sulfate content similar to that of heparin.
FIGURE 2.
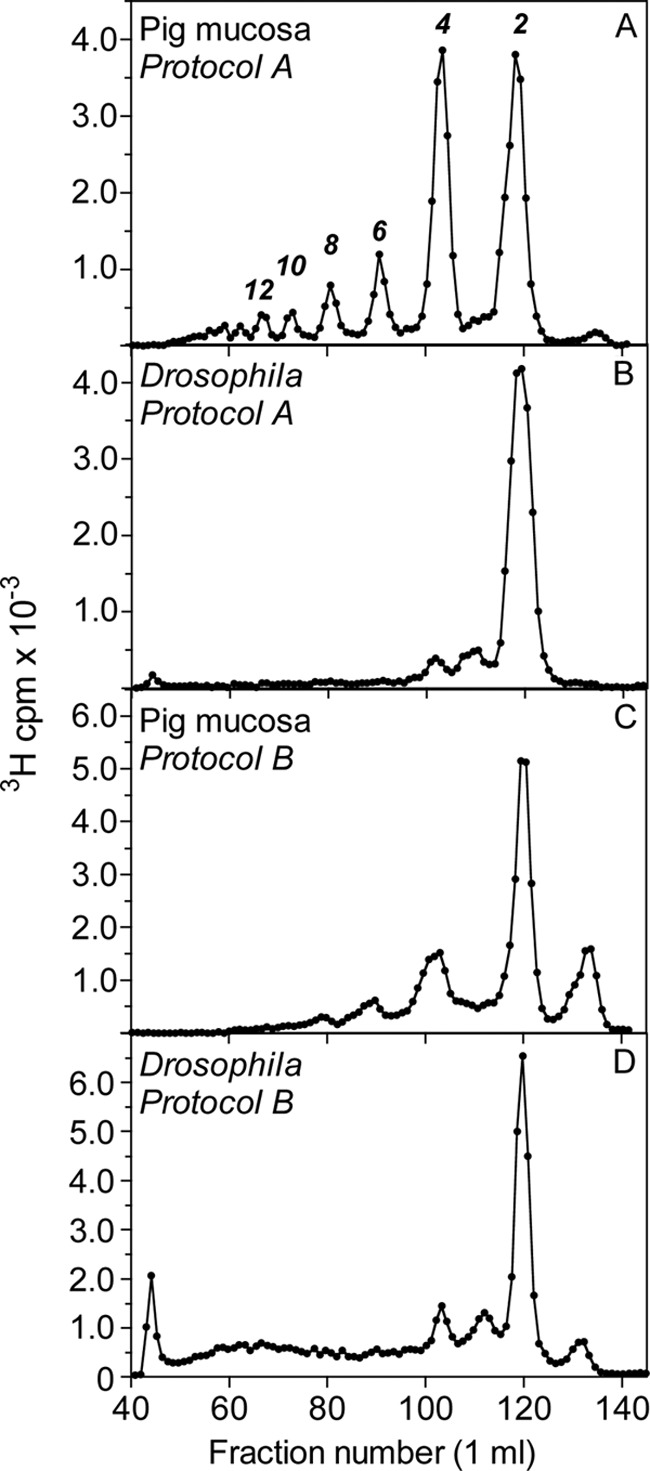
Gel chromatography of 3H-labeled saccharides obtained after deaminative cleavage of HS. HS samples from Drosophila or pig intestinal mucosa were deaminated and radiolabeled according to Protocol A (A and B) or Protocol B (C and D) (see “Experimental Procedures,” Fig. 1, and supplemental Fig. S1), and the products were analyzed by Bio-Gel P-10 gel chromatography. The numbers above each peak in A indicate oligosaccharide size.
To confirm these findings, we applied inverse-mode deaminative cleavage of N-deacetylated material according to Protocol B. This approach is expected to generate variously extended oligomers from NS-domains, tetrasaccharides from NA/NS-domains, and disaccharides from NA-domains (Fig. 1B). In our experience, some depolymerization of polysaccharide during the extended hydrazinolysis is unavoidable,5 and the fairly modest yield of higher oligosaccharides released from the intestinal HS standard (Fig. 2C) probably underestimates the actual contribution of N-sulfated structure to the intact polysaccharide. By contrast, the corresponding elution profile relating to Drosophila HS covered the entire separation range of the P-10 column (Fig. 2D), indicating oligosaccharides composed of consecutive N-sulfated disaccharide units. In addition, Protocol B yielded significant amounts of labeled disaccharide (Fig. 2D), presumably derived from NA-domain(s) that escaped detection by Protocol A (Fig. 2B) (see “Discussion”). The size of the NA-domain(s) in the parent HS chain was not defined through this experiment, nor was the precise extension of the N-sulfated sequence(s). Small oligosaccharides (in particular, tetrasaccharides) could be due to incomplete N-deacetylation of N-acetylated structure, but could also reflect partial depolymerization of NS-domains during hydrazinolysis. Nevertheless, the results demonstrate the occurrence of NS-domains in Drosophila HS that exceed the size of the corresponding domains in vertebrate HS. The N-sulfated nature of this material was confirmed by release of reducing terminal labeled disaccharides through deamination at pH 1.5 (see Fig. 4C and supplemental Fig. S2C). (The peak of excluded material in Fig. 2D presumably represents 3H-labeled macromolecules not related to HS judging by its resistance to deamination at pH 1.5 and enzymatic digestion with heparin lyases (data not shown).)
FIGURE 4.
Anion-exchange HPLC of 3H-labeled disaccharides. Disaccharides generated by deamination/radiolabeling according to Protocol A (A) or Protocol B (B) were analyzed by anion-exchange HPLC on a Partisil 10 SAX column. C, end-labeled, N-sulfated oligosaccharides obtained through Protocol B were deaminated at pH 1.5, and the released labeled disaccharide was isolated and subjected to anion-exchange HPLC. (Note that the [3H]aManR residues here represent a GlcNAc unit in the intact HS chains; the corresponding reaction is illustrated to the right.) The dashed line in A refers to a sample digested with liver β-glucuronidase/α-iduronidase prior to chromatography. Arrows indicate the elution positions of disaccharide standards (with the corresponding disaccharide units in the intact HS chain shown in parentheses): 1,2, GlcUA-aManR, IdoUA-aManR (-GlcUA/IdoUA-GlcNAc/NS-), aManR, and aManR6S (generated by β-glucuronidase/α-iduronidase digestion of disaccharides) (peaks not individually identified); 3, GlcUA-aManR6S (-GlcUA-GlcNS6S-); 4, IdoUA-aManR6S (-IdoUA-GlcNS6S-); 5, IdoUA2S-aManR (-IdoUA2S-GlcNS-); 6, IdoUA2S-aManR6S (-IdoUA2S-GlcNS6S-). Note that only the aManR residues indicated by open triangles were radiolabeled (derived from GlcNAc residues in the intact polysaccharide).
The disaccharides obtained according to the different cleavage procedures were identified by high-voltage paper electrophoresis, paper chromatography, and anion-exchange HPLC (which separated the various O-sulfated but not the non-sulfated species). High-voltage paper electrophoresis of the labeled disaccharides released by deamination according to Protocol A (Fig. 2B) showed major non-sulfated and mono-O-sulfated fractions, along with smaller amounts of di-O-sulfated disaccharide (Fig. 3A), compatible with a relatively low-sulfated parent NS-domain. Similar analysis of disaccharides obtained through Protocol B (Fig. 2D) revealed a single non-sulfated component (Fig. 3B), as predicted for a (GlcUA-GlcNAc)n NA-domain; the lack of O-sulfated species was verified also by anion-exchange HPLC (Fig. 4B). Paper chromatography of the non-O-sulfated NS-domain-derived disaccharide fraction indicated GlcUA-aManR and IdoUA-aManR, representing -GlcUA-GlcNS- and -IdoUA-GlcNS-, respectively, in intact HS, at an ∼8:2 ratio (supplemental Fig. S2A), whereas the NA-domain-derived disaccharide lacked IdoUA, as expected (supplemental Fig. S2B). Further separation of the O-sulfated disaccharides by anion-exchange HPLC (Fig. 4A) enabled the identification of all NS-domain-derived disaccharide units (Table 1). Results were confirmed by repeated HPLC following digestion with a mixture of β-glucuronidase and α-iduronidase, which selectively eliminated all disaccharides containing non-sulfated HexUA units. For comparison, the results are related to previous compositional analysis of Drosophila HS (based on complete lyase digestion) by Toyoda et al. (18). The composition of the resultant pool of N-sulfated disaccharides showed reasonable agreement with our present results (Table 1). Notably, contrary to our current protocol, their approach enabled assessment also of N-acetylated units that amounted to ∼30% of total disaccharides residues. The NS-domain(s) of Drosophila HS, although reminiscent of heparin, contained much less O-sulfate as shown by comparative disaccharide analysis (Table 1). Particularly notable, the trisulfated -IdoUA2S-GlcNS6S- unit (where 2S and 6S are 2-O-sulfate and 6-O-sulfate groups, respectively), most abundant in heparin (Table 1), is a minor component of the fly NS-domain. The labeled disaccharides released by low-pH deamination of oligosaccharides generated through Protocol B represent the transition zone between NS- and NA-domains (Fig. 1 and scheme to the right of Fig. 4C). Due to the influence of GlcN N-substituents on O-sulfotransferase reactions, 2-O-sulfate groups are essentially lacking outside NS-domains in HS, and the trisulfated -IdoUA2S-GlcNS6S- unit is entirely restricted to NS-domains (12, 24). Accordingly, ∼85% of the labeled disaccharides released by deamination at pH 1.5 of end-labeled Protocol B oligosaccharides were non-O-sulfated, the remainder being mono-O-sulfated exclusively at the 6-O-position (Fig. 4C). Further separation of the non-O-sulfated disaccharides by paper chromatography (supplemental Fig. S2C) showed a similar ratio of GlcUA-aManR to IdoUA-aManR as seen after direct low-pH deamination (supplemental Fig. S2A).
FIGURE 3.
High-voltage paper electrophoresis of 3H-labeled disaccharides. 3H-Labeled disaccharides isolated by gel chromatography (Sephadex G-15) after deamination and radiolabeling of Drosophila HS according to Protocol A (A) or Protocol B (B) (see “Experimental Procedures,” Fig. 1, and supplemental Fig. S1) were fractionated by high-voltage electrophoresis on Whatman No. 3MM paper at pH 5.3. After drying, papers were cut into 1-cm segments and analyzed for radioactivity. Arrows indicate the migration positions of non-sulfated (1), mono-O-sulfated (2), and di-O-sulfated (3) HexUA-aManR disaccharide standards.
TABLE 1.
Disaccharide composition of NS-domains in Drosophila heparan sulfate
Data are based on combined results of paper electrophoresis, paper chromatography, and anion-exchange HPLC. Each analysis was performed at least three times using material from two separate radiolabelings. The values are given as means ± S.D. Values in parenthesis refer to corresponding data for mucosal heparin, assembled from (47).
DISCUSSION
Drosophila has been increasingly applied as a model organism in studies of developmental processes dependent on HS proteoglycan involvement (25, 26). The diverse functions ascribed to HS include scaffold generation in basement membranes (4), stabilization of morphogen gradients (27, 28), and co-receptor functions in various signaling systems (14, 29, 30). Structural analysis of Drosophila HS has been invariably based on exhaustive lyase digestion followed by identification of the generated unsaturated disaccharides. Compositional data seemed in agreement with the current understanding of HS structure, pointing to a polysaccharide with approximately one sulfate residue per disaccharide unit (16, 18). Distinctive features were the relatively low content of N-acetylated glucosamine residues, corresponding to ∼30% of the total disaccharide units, and low, close to negligible amounts of 6-O-sulfated GlcNAc residues (16, 18). Notably, analysis of lyase-generated disaccharides containing 4,5-unsaturated HexUA residues did not allow a distinction between GlcUA and IdoUA units. The data provided no information regarding the domain organization of Drosophila HS.
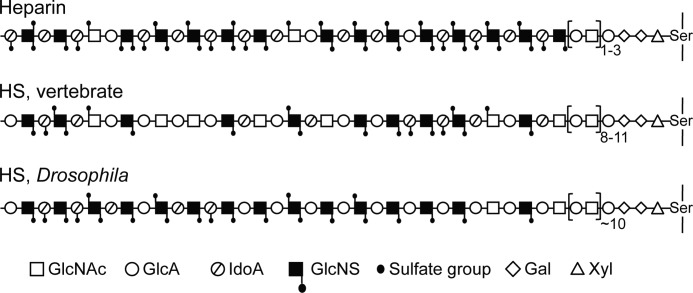
The results of this investigation are compatible with the previous compositional data (16, 18), yet indicate a novel HS design. The Drosophila HS chains were found to contain extended NS-domains with a relatively low degree of O-sulfation (Fig. 5). The number of such domains per chain is unclear but is presumably low; chains featuring a solitary NS-domain appear entirely plausible. The minor tetrasaccharide peak in Fig. 2B suggests that NS-domains might occasionally be separated by single GlcNAc residues, but could also reflect “anomalous ring contraction” during the deamination reaction (31). More abundant N-acetylated disaccharides, presumably corresponding to those previously observed after lyase digestion, were revealed through hydrazinolysis followed by deamination at pH 3.9 (Fig. 2D), but were not detected as labeled oligosaccharides after deamination at pH 1.5 and NaB3H4 reduction (Protocol A) (Fig. 2B) and therefore would seem not to be part of peripheral NA-domains. We tentatively propose that the disaccharides released through Protocol B were derived from the (GlcUA-GlcNAc)n sequence that occurs attached to the -GlcUA-Gal-Gal-Xyl- linkage region in HS (32, 33) and heparin (34) proteoglycans (Fig. 5). The released fly HS was reduced with unlabeled NaBH4 during isolation (see “Experimental Procedures”), and an N-acetylated section attached to the tetrasaccharide linkage region would therefore not be expected to incorporate any 3H label in the subsequent reduction steps of Protocol A. Notably, the predicted tetrasaccharide linkage region has been identified in Drosophila HS (19).
FIGURE 5.
Schematic presentation of HS/heparin chains. GlcA, GlcUA; IdoA, IdoUA.
Conversion of our structural data into a model of Drosophila HS depends on information regarding chain size. Assuming a 14-kDa chain (based on HS isolated from Drosophila S2 and Kc cells (35)) containing ∼30% -GlcUA-GlcNAc- disaccharide units (see legend to Table 1) (16, 18), all contiguous with the linkage region, would point toward an NA-domain of ∼10 disaccharide residues interspersed between the linkage region and a (possibly single, nonreducing terminal) NS-domain (Fig. 5). An unmodified sequence of similar size was found in fibroblast HS (36), whereas heparin can be N-sulfated one to three disaccharides from the linkage sequence (34, 37). Drosophila HS of lower molecular size than assumed here would display similar domain arrangement as in Fig. 5 but with proportionally shorter NA- and NS-domains. The low content of 6-O-sulfated GlcNAc residues in Drosophila (represented by the labeled disaccharides in Fig. 4C) (18) compared with vertebrate HS (Fig. 2 in Ref. 12) is of particular significance because these units mark the transition zones between NA- and NS-domains; our proposed model for Drosophila HS features essentially one such zone per chain (Fig. 5). The extended NS-domain and the absence of internal N-acetylated sequence would qualify the Drosophila polysaccharide as heparin rather than HS had it not been for the low O-sulfate content (and the length of the NA-domain attached to the linkage region).
The functional importance of HS domain structure is generally poorly understood. Adjacent NS-domains may interact with distinct separate clusters of basic amino acid residues in single protein ligands or with distinct proteins in oligomeric complexes of different kinds (8). The domain arrangement may restrict complex formation to certain locations along a HS chain. It has been proposed that NS-domains occupying the nonreducing terminal portion of HS chains are more highly sulfated than their internal counterparts and hence are particularly prone to interaction with various proteins (38–40). The significance of the proposed single-domain arrangement of Drosophila HS with regard to topology of HS-protein complexes, formation of morphogen gradients, and various HS-dependent signaling activities is unknown.
Finally, our findings highlight intriguing aspects of regulation in HS biosynthesis. Several studies, including comparative structural analysis of HS species isolated from different mouse organs, indicate that enzymatic polymer modification, including domain formation, is strictly regulated (8, 11). The N-sulfate groups added by the NDST enzymes not only define domain organization but also set the limits for subsequent modification reactions. Mammalian cells that synthesize HS generally contain more than one NDST species. In particular, NDST1 and NDST2 appear to operate in concerted mode, although the precise role of each individual enzyme remains to be defined (11). Notably, incubation of the appropriate (GlcUA-GlcNAc)n polysaccharide with either NDST recombinant isoform in vitro in the presence of the sulfate donor adenosine 3′-phosphate 5′-phosphosulfate resulted in formation of extended N-sulfated sequence rather than the interrupted domain structure typical of HS (41). These products thus show clear similarity to the Drosophila HS described here. Drosophila expresses only one NDST gene, sulfateless. Similarly, mammalian mast cells, which express high levels of NDST2 but no or little NDST1 (42), produce heparin, i.e. an extended NS-domain (43, 44). Conversely, in cells that express more than one NDST isoform, HS domain structure appears to depend on the relative amounts of the enzymes (45, 46). On the other hand, genetic ablation of NDST2 from mouse liver cells, which normally express both NDST1 (dominant) and NDST2, had a marginal effect on HS domain structure (46). Caenorhabditis elegans, again with only a single NDST enzyme (hst-1), synthesizes HS devoid of 6-O-sulfated GlcNAc residues (18) and is thus similar to Drosophila HS, which also lacks NA/NS-domains. Further comparative studies of HS fine structure and NDST expression in species at diverse evolutionary stages are required to better understand the organization and regulation of the biosynthetic machinery.
This work was supported in part by grants from the University of Bergen (to M. K.-G.) and Polysackaridforskning AB, Uppsala, Sweden (to U. L.).

This article contains supplemental Figs. S1 and S2.
U. Lindahl, unpublished data.
- HS
- heparan sulfate
- NDST
- N-deacetylase/N-sulfotransferase
- aManR
- 2,5-anhydro-d-mannitol (formed by reduction of terminal 2,5-anhydromannose with NaBH4).
REFERENCES
- 1. Feta A., Do A. T., Rentzsch F., Technau U., Kusche-Gullberg M. (2009) Molecular analysis of heparan sulfate biosynthetic enzyme machinery and characterization of heparan sulfate structure in Nematostella vectensis. Biochem. J. 419, 585–593 [DOI] [PubMed] [Google Scholar]
- 2. Medeiros G. F., Mendes A., Castro R. A., Baú E. C., Nader H. B., Dietrich C. P. (2000) Distribution of sulfated glycosaminoglycans in the animal kingdom: widespread occurrence of heparin-like compounds in invertebrates. Biochim. Biophys. Acta 1475, 287–294 [DOI] [PubMed] [Google Scholar]
- 3. Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 4. Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulfate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 5. Bülow H. E., Hobert O. (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 [DOI] [PubMed] [Google Scholar]
- 6. Casu B., Lindahl U. (2001) Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem. 57, 159–206 [DOI] [PubMed] [Google Scholar]
- 7. Lindahl U., Kusche-Gullberg M., Kjellén L. (1998) Regulated diversity of heparan sulfate. J. Biol. Chem. 273, 24979–24982 [DOI] [PubMed] [Google Scholar]
- 8. Lindahl U., Li J. P. (2009) Interactions between heparan sulfate and proteins–design and functional implications. Int. Rev. Cell Mol. Biol. 276, 105–159 [DOI] [PubMed] [Google Scholar]
- 9. Esko J. D., Selleck S. B. (2002) Order out of chaos: assembly of ligand-binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 10. Sugahara K., Kitagawa H. (2002) Heparin and heparan sulfate biosynthesis. IUBMB Life 54, 163–175 [DOI] [PubMed] [Google Scholar]
- 11. Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) Heparan sulfate structure in mice with genetically modified heparan sulfate production. J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 12. Maccarana M., Sakura Y., Tawada A., Yoshida K., Lindahl U. (1996) Domain structure of heparan sulfates from bovine organs. J. Biol. Chem. 271, 17804–17810 [DOI] [PubMed] [Google Scholar]
- 13. Pejler G., Danielsson A., Björk I., Lindahl U., Nader H. B., Dietrich C. P. (1987) Structure and antithrombin-binding properties of heparin isolated from the clams Anomalocardia brasiliana and Tivela mactroides. J. Biol. Chem. 262, 11413–11421 [PubMed] [Google Scholar]
- 14. Bornemann D. J., Duncan J. E., Staatz W., Selleck S., Warrior R. (2004) Abrogation of heparan sulfate synthesis in Drosophila disrupts the Wingless, Hedgehog, and Decapentaplegic signaling pathways. Development 131, 1927–1938 [DOI] [PubMed] [Google Scholar]
- 15. Kamimura K., Koyama T., Habuchi H., Ueda R., Masu M., Kimata K., Nakato H. (2006) Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J. Cell Biol. 174, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamimura K., Maeda N., Nakato H. (2011) In vivo manipulation of heparan sulfate structure and its effect on Drosophila development. Glycobiology 21, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toyoda H., Kinoshita-Toyoda A., Fox B., Selleck S. B. (2000) Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J. Biol. Chem. 275, 21856–21861 [DOI] [PubMed] [Google Scholar]
- 18. Toyoda H., Kinoshita-Toyoda A., Selleck S. B. (2000) Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J. Biol. Chem. 275, 2269–2275 [DOI] [PubMed] [Google Scholar]
- 19. Yamada S., Okada Y., Ueno M., Iwata S., Deepa S. S., Nishimura S., Fujita M., Van Die I., Hirabayashi Y., Sugahara K. (2002) Determination of the glycosaminoglycan-protein linkage region oligosaccharide structures of proteoglycans from Drosophila melanogaster and Caenorhabditis elegans. J. Biol. Chem. 277, 31877–31886 [DOI] [PubMed] [Google Scholar]
- 20. Bitter T., Muir H. M. (1962) A modified uronic acid carbazole reaction. Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 21. Shaklee P. N., Conrad H. E. (1984) Hydrazinolysis of heparin and other glycosaminoglycans. Biochem. J. 217, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smeds E., Habuchi H., Do A. T., Hjertson E., Grundberg H., Kimata K., Lindahl U., Kusche-Gullberg M. (2003) Substrate specificities of mouse heparan sulfate glucosaminyl 6-O-sulfotransferases. Biochem. J. 372, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kusche M., Lindahl U., Enerbäck L., Rodén L. (1988) Identification of oversulfated galactosaminoglycans in intestinal-mucosal mast cells of rats infected with the nematode worm Nippostrongylus brasiliensis. Biochem. J. 253, 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turnbull J. E., Gallagher J. T. (1991) Distribution of iduronate 2-sulfate residues in heparan sulfate. Evidence for an ordered polymeric structure. Biochem. J. 273, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin X., Perrimon N. (2002) Developmental roles of heparan sulfate proteoglycans in Drosophila. Glycoconj. J. 19, 363–368 [DOI] [PubMed] [Google Scholar]
- 26. Nybakken K., Perrimon N. (2002) Heparan sulfate proteoglycan modulation of developmental signaling in Drosophila. Biochim. Biophys. Acta 1573, 280–291 [DOI] [PubMed] [Google Scholar]
- 27. Kramer K. L., Yost H. J. (2003) Heparan sulfate core proteins in cell-cell signaling. Annu. Rev. Genet. 37, 461–484 [DOI] [PubMed] [Google Scholar]
- 28. Yan D., Lin X. (2009) Shaping morphogen gradients by proteoglycans. Cold Spring Harbor Perspect. Biol. 1, a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han C., Belenkaya T. Y., Khodoun M., Tauchi M., Lin X., Lin X. (2004) Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signaling and gradient formation. Development 131, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 30. Xian X., Gopal S., Couchman J. R. (2010) Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 339, 31–46 [DOI] [PubMed] [Google Scholar]
- 31. Guo Y. C., Conrad H. E. (1989) The disaccharide composition of heparins and heparan sulfates. Anal. Biochem. 176, 96–104 [DOI] [PubMed] [Google Scholar]
- 32. Gallagher J. T., Walker A. (1985) Molecular distinctions between heparan sulfate and heparin. Analysis of sulfation patterns indicates that heparan sulfate and heparin are separate families of N-sulfated polysaccharides. Biochem. J. 230, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyon M., Deakin J. A., Gallagher J. T. (1994) Liver heparan sulfate structure. A novel molecular design. J. Biol. Chem. 269, 11208–11215 [PubMed] [Google Scholar]
- 34. Lindahl U. (1966) Further characterization of the heparin-protein linkage region. Biochim. Biophys. Acta 130, 368–382 [DOI] [PubMed] [Google Scholar]
- 35. Kasevayuth K., Yanagishita M. (2004) Catabolism of heparan sulfate proteoglycans in Drosophila cell lines. Biochem. Biophys. Res. Commun. 324, 205–211 [DOI] [PubMed] [Google Scholar]
- 36. Lyon M., Steward W. P., Hampson I. N., Gallagher J. T. (1987) Identification of an extended N-acetylated sequence adjacent to the protein linkage region of fibroblast heparan sulfate. Biochem. J. 242, 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugahara K., Tsuda H., Yoshida K., Yamada S., de Beer T., Vliegenthart J. F. (1995) Structure determination of the octa- and decasaccharide sequences isolated from the carbohydrate-protein linkage region of porcine intestinal heparin. J. Biol. Chem. 270, 22914–22923 [DOI] [PubMed] [Google Scholar]
- 38. Naimy H., Buczek-Thomas J. A., Nugent M. A., Leymarie N., Zaia J. (2011) Highly sulfated nonreducing end-derived heparan sulfate domains bind fibroblast growth factor-2 with high affinity and are enriched in biologically active fractions. J. Biol. Chem. 286, 19311–19319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Staples G. O., Shi X., Zaia J. (2010) Extended N-sulfated domains reside at the nonreducing end of heparan sulfate chains. J. Biol. Chem. 285, 18336–18343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Z. L., Zhang L., Yabe T., Kuberan B., Beeler D. L., Love A., Rosenberg R. D. (2003) The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J. Biol. Chem. 278, 17121–17129 [DOI] [PubMed] [Google Scholar]
- 41. Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellén L. (2008) Heparin/heparan sulfate biosynthesis: processive formation of N-sulfated domains. J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 42. Kusche-Gullberg M., Eriksson I., Pikas D. S., Kjellén L. (1998) Identification and expression in mouse of two heparan sulfate glucosaminyl N-deacetylase/N-sulfotransferase genes. J. Biol. Chem. 273, 11902–11907 [DOI] [PubMed] [Google Scholar]
- 43. Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., Kjellén L. (1999) Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 400, 773–776 [DOI] [PubMed] [Google Scholar]
- 44. Humphries D. E., Wong G. W., Friend D. S., Gurish M. F., Qiu W. T., Huang C., Sharpe A. H., Stevens R. L. (1999) Heparin is essential for the storage of specific granule proteases in mast cells. Nature 400, 769–772 [DOI] [PubMed] [Google Scholar]
- 45. Dagälv A., Holmborn K., Kjellén L., Abrink M. (2011) Lowered expression of heparan sulfate/heparin biosynthesis enzyme N-deacetylase/N-sulfotransferase 1 results in increased sulfation of mast cell heparin. J. Biol. Chem. 286, 44433–44440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ledin J., Ringvall M., Thuveson M., Eriksson I., Wilén M., Kusche-Gullberg M., Forsberg E., Kjellén L. (2006) Enzymatically active N-deacetylase/N-sulfotransferase 2 is present in liver but does not contribute to heparan sulfate N-sulfation. J. Biol. Chem. 281, 35727–35734 [DOI] [PubMed] [Google Scholar]
- 47. Kusche M., Torri G., Casu B., Lindahl U. (1990) Biosynthesis of heparin. Availability of glucosaminyl 3-O-sulfation sites. J. Biol. Chem. 265, 7292–7300 [PubMed] [Google Scholar]