Background: Pathogenesis-related (PR-1-like) proteins are widely conserved in eukaryotes, but their biological function is unknown.
Results: Knockout or site-directed mutagenesis of fpr1 encoding a secreted PR-1-like protein in the fungal pathogen Fusarium oxysporum impairs virulence on mice.
Conclusion: Secreted PR-1-like proteins are important for fungal infection of mammals.
Significance: We show the first genetic evidence for a biological function of the predicted active site of PR-1-like proteins.
Keywords: Fungi, Infectious Diseases, Pathogen-associated Molecular Pattern (PAMP), Pathogenesis, Signaling, Virulence Factors, Fusarium, PR-1 Protein, Plant Pathogen
Abstract
The pathogenesis-related PR-1-like protein family comprises secreted proteins from the animal, plant, and fungal kingdoms whose biological function remains poorly understood. Here we have characterized a PR-1-like protein, Fpr1, from Fusarium oxysporum, an ubiquitous fungal pathogen that causes vascular wilt disease on a wide range of plant species and can produce life-threatening infections in immunocompromised humans. Fpr1 is secreted and proteolytically processed by the fungus. The fpr1 gene is required for virulence in a disseminated immunodepressed mouse model, and its function depends on the integrity of the proposed active site of PR-1-like proteins. Fpr1 belongs to a gene family that has expanded in plant pathogenic Sordariomycetes. These results suggest that secreted PR-1-like proteins play important roles in fungal pathogenicity.
Introduction
Fungi are an extremely versatile group of organisms. Most are saprophytes that thrive on dead organic material, but a number of species can infect and cause disease on other organisms, including plants and mammals (1). Fungal pathogens have evolved an array of mechanisms allowing them to recognize and penetrate a host, overcome its innate defenses, and exploit its nutrient resources. The combined action of these virulence factors determines both the infectious potential of a pathogen and the severity of disease it causes on a given host.
Hosts respond to fungal infection by mounting a robust immune response. Interestingly, plants and mammals share common principles of innate immunity, such as the ability to recognize pathogen-associated molecular patterns (PAMPs) (2) or the presence of pathogenesis-related 1 (PR-1) proteins, which have been implicated both in plant and animal immune systems. PR-1 proteins were originally identified in tobacco as part of the defense response to viral infection (3). PR-1 is the most highly expressed class of PR proteins and contributes up to 10% of total protein in infected leaves (4). PR-1-like proteins were subsequently found in a variety of eukaryotes, including fungi, insects, and mammals, and the term CAP protein superfamily was coined to encompass mammalian cysteine-rich secretory proteins (CRISPs)3, Ag5-antigens from insects and plant PR-1 proteins (5). Comparative structural analysis identified a putative active site of two histidine and two glutamate residues that is highly conserved among the members of the protein family (6). Despite their ubiquitous distribution, the biochemical function and biological roles of PR-1-like proteins have remained largely elusive (5).
Fusarium oxysporum is an important soilborne fungal pathogen that causes vascular wilt disease on more than a hundred different plant species (7). F. oxysporum can also provoke infections in humans, ranging from superficial and locally invasive to disseminated fusariosis with mostly lethal outcomes (8). The genus Fusarium now represents the second most frequent mold causing invasive fungal infections, and F. oxysporum, together with Fusarium solani and Fusarium verticillioides, is responsible for practically all cases of invasive fusariosis (8, 9). Human pathogenic isolates of F. oxysporum have polyphyletic origin and respond poorly to available antifungal agents (10, 11). We showed previously that a single isolate of F. oxysporum f. sp. lycopersici can cause disease both on tomato plants and immunodepressed mice (12) as well as on the invertebrate model host Galleria mellonella (13). In this work, we functionally characterized Fpr1, a secreted PR-1 like protein, from F. oxysporum. Using a genetic approach, we established that Fpr1 function is required for full virulence on a mammalian host but dispensable for virulence on plants. We provide evidence that Fpr1 is part of a gene family that has expanded in F. oxysporum and other plant pathogenic Sordariomycetes. Our results shed new light on the role of secreted PR-1-like proteins and suggest that they are key players in fungal virulence.
EXPERIMENTAL PROCEDURES
Fungal Isolates and Culture Conditions
F. oxysporum f.sp. lycopersici wild-type strain 4287 (race 2) was grown and maintained as reported (14). The generation of the following mutant strains was described previously: MAPK mutant Δfmk1 (15), G protein subunit β mutant Δfgb1, and Δfmk1Δfgb1 double mutant (16). Growth conditions for microconidia production, nucleic acid extraction, Western blot analysis, microscopic examination, and analysis of colony phenotypes are detailed in the supplemental Methods.
Nucleic Acid Manipulations, Construction of Plasmid Vectors, and Fungal Transformation
Total RNA and genomic DNA extraction from F. oxysporum mycelium, Southern and Northern blot analyses, and PCR amplification were performed as described (17, 18). For details on fpr1 gene cloning, construction of the gene knockout vector, gene knockout, and subsequent complementation with the wild type or the fpr1H170A,E177A allele see supplemental Methods. Transformation of fungal protoplasts to hygromycin or phleomycin resistance was performed as described (14). Gene knockout and complementation events were confirmed by Southern and Northern blot analysis (supplemental Fig. 2). The presence and correct expression of the different fpr1 alleles in the complemented strains was confirmed by PCR on genomic DNA and sequencing, as well as by Northern blot analysis.
Production and Characterization of Recombinant Fpr1 Protein
Cloning of a fpr1 cDNA clone lacking the predicted signal peptide into the pET-28c bacterial or the pPIC9 yeast expression vector, as well as purification of recombinant Fpr1 protein from Escherichia coli or Pichia pastoris, respectively, is reported in the supplemental Methods. Determination of putative proteolytic activity of purified Fpr1-His6 protein from E. coli or P. pastoris culture supernatants containing Fpr1 was done against azocaseine (19) using 25 μg of protein in phosphate buffer (pH 6.0, 7.0, and 8.0) at 37 °C for periods from 30 min to 2 h. For details on enzymatic assays and zymography for gelatinolytic activity see the supplemental Methods. To study proteolytic processing of Fpr1, 2 μg of recombinant Fpr1 from P. pastoris was incubated for the indicated time periods with 10 μl of dialyzed and concentrated supernatant of strains Δfpr1 or Δfgb1 in 50 mm phosphate buffer (pH 7.4) in a total volume of 25 μl and supplemented with 1 mm of different protease inhibitors when indicated. For Western blot analyses, protein samples were separated by electrophoresis in 14% (w/v) acrylamide-SDS gels and analyzed using a polyclonal α-Fpr1 antibody, obtained from rabbit as detailed in the supplemental Methods).
CD spectra were obtained on a Jasco 715 spectropolarimeter equipped with a thermostated cell holder and a NesLab-111 circulating water bath at 0.2 nm/s scanning speed. The instrument was calibrated with (+)-10-camphorsulfonic acid. CD spectra were recorded in cylindrical cells of 0.1 cm optical path. Mean residue weight ellipticities were expressed in units of degree × cm2 × dmol−1.
Protein Identification by MALDI-TOF-MS
Protein bands of interest were excised from the gel, subjected to tryptic digest, and analyzed on a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems) using α-cyano-4-hydroxycinnamic acid as a matrix. MALDI-MS spectra were internally calibrated using the singly protonated trypsin autodigestion peaks at m/z 2273.159 and 2163.056 and searched against the F. oxysporum database downloaded from the Broad Institute using Mascot software version 2.1 (Matrix Science) (20) (for details, see supplemental Methods).
RT-PCR Analysis
Fungal strains were germinated in potato dextrose broth for 24 h, washed in minimal medium (MM) (14), and transferred for 8 h either to MM, MM supplemented with 10% (v/v) bovine fetal serum (Sigma), or MM with submerged tomato roots. Details on reverse transcription and PCR reactions are provided in the supplemental Methods. As a control, the actin gene transcript was amplified. For quantitative real-time RT-PCR, the wild-type strain was germinated 16 h at 28 °C in MM, transferred for 2 h to MM at 37 °C, and then transferred for different time periods either to MM or to heparinized human whole blood (Dunn Labortechnik GmbH, Asbach, Germany) at 37 °C. PCR products were obtained using iQ SYBR Green Supermix (Bio-Rad) and an iCycler iQ real-time PCR system (Bio-Rad). Transcript levels were calculated by comparative ΔCt and normalized to act1.
Plant and Mouse Infection Assays
Tomato root inoculation assays with microconidia from different F. oxysporum strains were performed as described (14). Ten plants were used for each treatment. Assays for invasive growth on tomato fruits (cultivar Daniela) were carried out as described (15). Plant infection experiments were performed at least three times with similar results.
Mice were cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series, no. 123). Experimental conditions were approved by the Animal Welfare Committee at the Faculty of Medicine, Universitat Rovira i Virgili. Infection assays with immunodepressed OF-1 male mice (Charles River Laboratories, Criffa S.A., Barcelona, Spain) were performed as described (12). Briefly, groups of 10 immunosuppressed mice were infected by injecting 0.2 ml of an inoculum of 108 F. oxysporum microconidia/ml of sterile saline into a lateral vein of the tail. Survival was recorded each day for 13 days. Infection experiments with each individual strain were performed at least three times. Survival was estimated by the Kaplan-Meier method and compared among groups using the log-rank test. For analysis of fpr1 gene expression or determination of fungal tissue burden in organs, randomly chosen surviving mice were sacrificed 3 or 7 days after inoculation, respectively. Liver, spleen, kidneys and lungs were aseptically removed and immediately frozen in liquid nitrogen for RNA extraction or weighed, homogenized in sterile saline and 10-fold serial dilutions were spread onto Potato Dextrose Agar to calculate the number of Colony Forming Units per gram of organ. Fungal colony counts were converted to log10 and compared using analysis of variance. Calculations were performed using SPSS for Windows version 10.0.
Sequence Alignments and Phylogenetic Analysis
Members of the PR-1 protein family in different fungal genomes were identified by BLASTp searches on the Web server of the Broad Institute using the Fpr1 protein as bait (see supplemental Methods for details). Full-length sequences were aligned with Clustal W (21) and inspected manually. Selected plant PR-1 protein sequences obtained from GenBanktm were included in multiple alignments. A maximum likelihood tree was built from the alignment by PhyML version 4.0 using both parsimony and distance analysis (neighbor joining, NJ) with 1000 bootstrap replicates (22).
For identification of protein domains, full-length sequences were analyzed using InterProScan. Presence of a signal peptide was determined with SignalP version 3.0 (23) using a standardized threshold value of 0.5. Putative GPI consensus sequences were identified using the Fungal BigPi software (24).
RESULTS
Cloning, Mutation, and Expression Analysis of the F. oxysporum fpr1 Gene
The fpr1 gene was identified during analysis of a F. oxysporum Expressed Sequence Tag library. One of the sequenced clones showed homology with plant PR-1 proteins in the databases. The complete fpr1 genomic region was cloned from a λEMBL3 genomic library of F. oxysporum, and the sequence was deposited in GenBanktm under accession number GQ411527. fpr1 consists of an open reading frame encoding a putative 259-amino acid protein with a predicted molecular mass of 27.7 kDa and a pI of 4.9. Sequence alignment with the fpr1 cDNA revealed the presence of a single 52-bp intron. A BLAST search of the complete genome database of F. oxysporum produced a single high identity match (FOXG_09795), consistent with the presence of a single hybridizing band in Southern blot analysis.
The predicted F. oxysporum Fpr1 protein contains three distinct regions: an N-terminal signal peptide of 19 amino acids, a proline-rich region of unknown function, and a C-terminal domain with homology to the PR-1-like, SCP, or CAP protein family PF00188 (supplemental Fig. 1A). This family includes plant pathogenesis-related proteins group 1 (PR-1) (25), human glioma-associated protein GliPR (26), mammalian CRISP proteins (27), allergens of insect venoms (28), and snake or lizard venoms (29). Fpr1 contains two conserved histidine (H170, H217) and two glutamate (Glu-177, Glu-198) residues that constitute the proposed active site (30) (supplemental Fig. 1, A and B). On the basis of these data, we conclude that F. oxysporum fpr1 encodes a fungal homologue of PR-1-like pathogenesis-related proteins.
F. oxysporum mutants lacking a functional copy of fpr1 were generated by targeted gene disruption. For complementation experiments, a 2.7-kb DNA fragment encompassing either the wild-type fpr1 allele or a fpr1H170A,E177A allele in which two conserved residues at the predicted active site had been replaced by site-directed mutagenesis with alanines was introduced into the Δfpr1–1 mutant (supplemental Fig. 2).
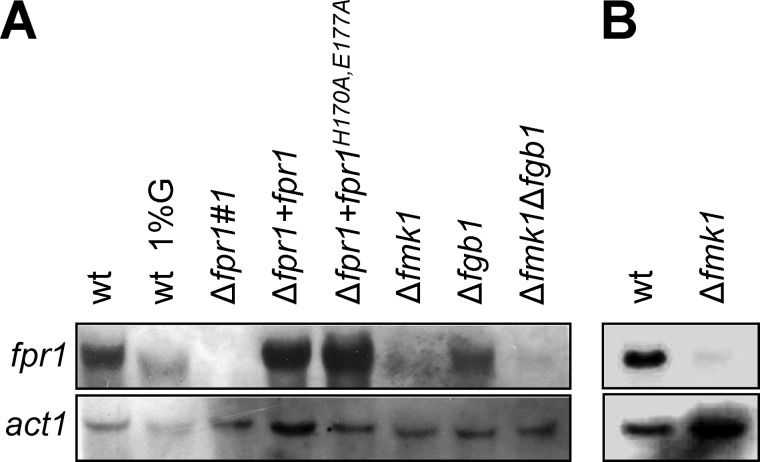
Northern blot analysis detected a single fpr1 transcript in F. oxysporum mycelium grown either in liquid or solid MM (Fig. 1). High concentrations of glucose (1% w/v) resulted in reduced transcript abundance (Fig. 1A). The transcript was not detected in the Δfpr1 mutant but was restored in the complemented Δfpr1+fpr1 and Δfpr1+fpr1H170A,E177A strains. A mutant lacking the MAPK Fmk1 (15) had drastically reduced fpr1 transcript levels both in liquid and on solid medium, whereas the Δfmk1Δfgb1 mutant lacking both Fmk1 and the heterotrimeric Gβ subunit Fgb1 (16) had further reduced transcript levels (Fig. 1). Thus, expression of fpr1 is promoted by the Fmk1 MAPK cascade and repressed by glucose.
FIGURE 1.
Expression of fpr1 is repressed by glucose and stimulated by the pathogenicity MAP kinase Fmk1. Shown is the Northern analysis for fpr1 transcript accumulation in fungal cultures grown either on liquid minimal medium supplemented with 0.1% (w/v) glucose (G) or with 1% w/v glucose where indicated (A) or on solid minimal medium (B). Total RNA was extracted, fractionated in an agarose gel, blotted onto a nylon membrane, and hybridized to the fpr1 probe or the actin probe (act1) as a control for RNA loading.
Fpr1 Forms a Dimer in Solution
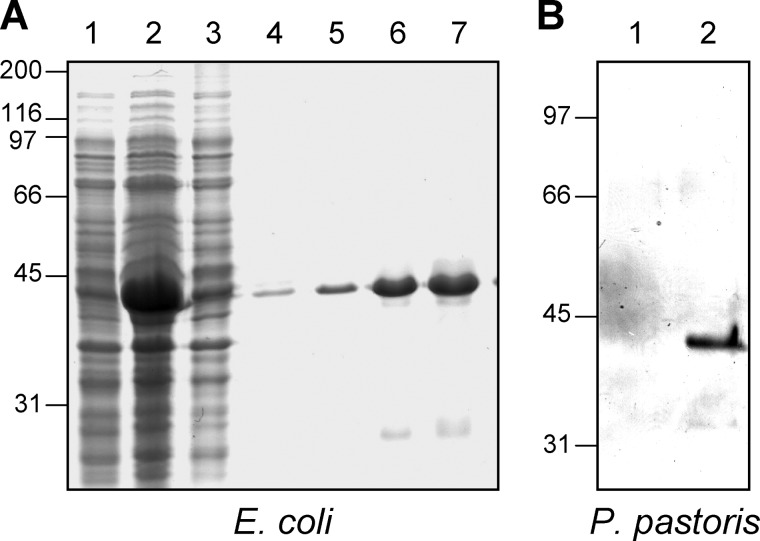
Heterologous expression of fpr1 or fpr1H170A,E177A cDNA in E. coli resulted in the presence of a major protein band in isopropyl 1-thio-β-d-galactopyranoside-induced cells that was absent from uninduced cells (Fig. 2A) or from cells carrying the empty expression vector (data not shown). The apparent mass of affinity-purified Fpr1 and Fpr1H170A,E177A protein deduced from SDS-PAGE was 40 kDa, which is significantly higher than predicted. By contrast, MALDI-MS analysis of the purified recombinant protein detected a major peak with a mass of 30 kDa, in line with the predicted mass of His-tagged Fpr1.
FIGURE 2.
Heterologous expression of Fpr1 in E. coli and P. pastoris. A, purification of recombinant Fpr1–6xHis protein from E. coli. Lanes 1 and 2, crude cell lysate of bacterial strain BL21 transformed with plasmid pet28c containing the fpr1 cDNA, either before (lane 1) or after induction with 1 mm IPTG (lane 2). Lanes 3–7, different fractions eluted from a Ni2+ NTA column. Shown are flow-through (lane 3) and eluates with 20 mm (lane 4), 40 mm (lane 5), and 250 mm imidazol (lanes 6 and 7). B, expression of Fpr1 in P. pastoris. Culture supernatants of yeast strain GS115 transformed either with the empty plasmid pPIC9 (lane 1) or with pPIC9 containing the fpr1 cDNA (lane 2). Proteins were separated by SDS-PAGE and visualized with Coomassie Blue (A) or silver staining (B). Relative positions of molecular weight markers (in kDa) are indicated at the left.
Recombinant Fpr1 protein was also obtained from the methylotrophic yeast P. pastoris. After induction with 0.5% methanol, culture supernatants of fpr1-expressing P. pastoris transformants contained a major protein band that was absent in the control strain transformed with the empty pPIC9 vector (Fig. 2B). Similar to E. coli-produced Fpr1, the recombinant protein band from P. pastoris had an estimated mass of 40 kDa.
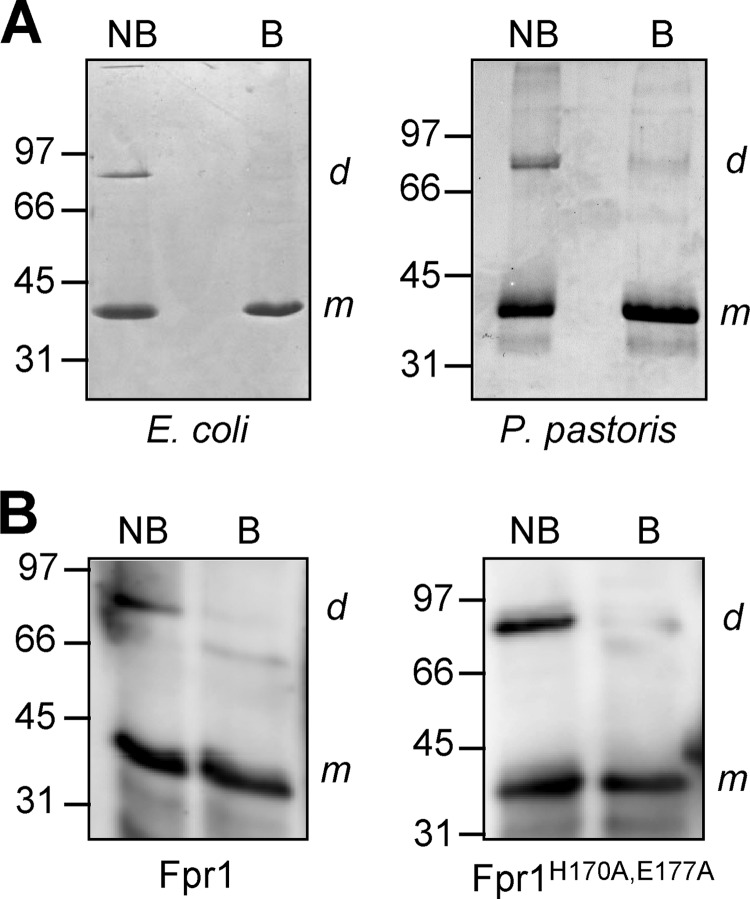
SDS-PAGE fractionation of recombinant Fpr1 from E. coli or P. pastoris without prior boiling of the sample revealed a second protein band with an apparent mass of ∼80 kDa, which is consistent with the expected mobility of a putative Fpr1 homodimer (Fig. 3A). Analysis of MALDI MS spectra of the high and low molecular weight bands confirmed that both correspond to Fpr1 (supplemental Fig. 3). Western blot analysis of unboiled Fpr1 and Fpr1H170A,E177A protein with a polyclonal α-Fpr1 antibody detected the presence of the dimer in both protein species, suggesting that the H170 and E177 are not essential for dimerization (Fig. 3B). Comparison of circular dichroism spectra of wild-type and mutant Fpr1 showed that the percentage of secondary structural elements was unchanged by the mutation, indicating that the Fpr1H170A,E177A protein is folded correctly (supplemental Fig. 4).
FIGURE 3.
Fpr1 forms a dimer in solution. A, 10-μg recombinant Fpr1 protein obtained from E. coli or P. pastoris was subjected to SDS-PAGE either without (NB) or with 5 min of boiling (B) prior to loading. Proteins were stained with Coomassie Blue. B, Western blot analysis of recombinant Fpr1 or FprH170A,E177A protein obtained from E. coli, subjected to SDS-PAGE either without (NB) or with 5 min of boiling (B) prior to loading and hybridized with α-Fpr1 antiserum. m and d indicate positions of hybridizing bands corresponding to the monomeric and dimeric form of Fpr1, respectively. Relative positions of molecular weight markers (in kDa) are indicated at the left.
A PR-1-like protein of the CRISP subfamily from the cone snail Conus textile, Tex31, was suggested previously to exhibit substrate-specific serine protease activity (31). Incubation of purified recombinant Fpr1 protein from E. coli with the general protease substrate azocasein buffered at different pHs in the absence or presence of metal ions (CaCl2, MgCl2, ZnCl2) or with a synthetic substrate containing the reported cleavage site of Tex31 (H-LVKA-pNA) failed to detect protease activity under any of the conditions tested (results not shown). Gelatinolytic activity zymograms of P. pastoris culture supernatants from the fpr1-expressing transformant and the control strain revealed similar clearing bands, suggesting that they originate from P. pastoris proteases (data not shown).
Fpr1 Is Secreted and Proteolytically Processed by F. oxysporum
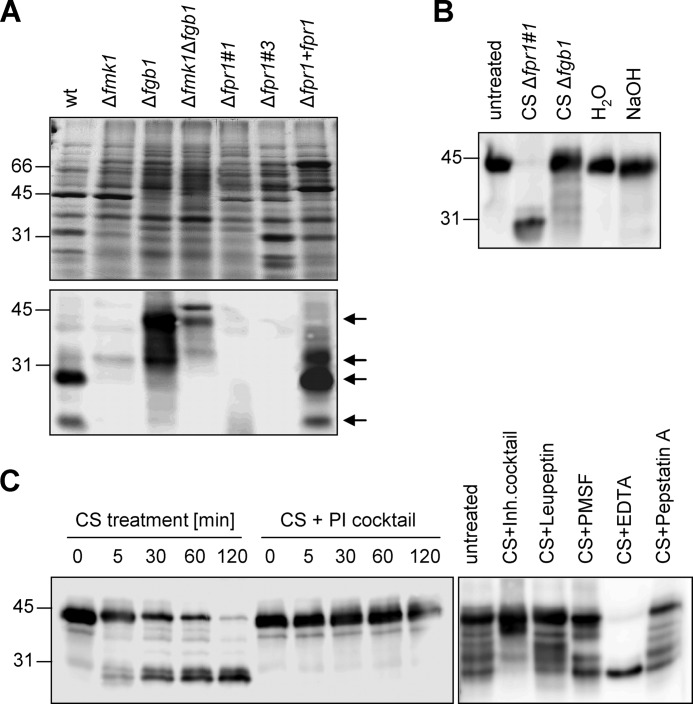
Sequence analysis of Fpr1 predicted the presence of an N-terminal signal peptide (supplemental Fig. 1A). To test whether Fpr1 is secreted by F. oxysporum, fungal cell lysate and culture supernatant were subjected to Western blot analysis with α-Fpr1 antibody. No signal was detected in the cell lysates, whereas culture supernatants contained a major hybridizing band of ∼30 kDa (Fig. 4A). The signal intensity was higher in culture supernatants from MM (containing 0.1% w/v glucose) than in those from potato dextrose broth (2% glucose) (Fig. 4B). In contrast to the wild-type and complemented strain, no hybridizing signal was detected in supernatants of the two knockout mutants Δfpr1#1 and Δfpr1#3 (Fig. 5A). Lack of background hybridization in the mutants indicates that the polyclonal α-Fpr1 antibody exhibits a high specificity toward the Fpr1 protein. Supernatants of the Δfmk1 and Δfmk1Δfgb1 mutants contained significantly lower amounts of Fpr1 than those of the wild-type strain, confirming the results from Northern blot analyses (see Fig. 1). Interestingly, supernatants of the Δfgb1 mutant contained very low amounts of the 30-kDa band but, instead, had a strong hybridizing band migrating at an apparent mass of 40 kDa, similar to that of recombinant Fpr1. Inspection of the Western blot analyses revealed at least four hybridizing bands with approximate molecular masses of 26, 30, 33, and 40 kDa (marked by arrows in Fig. 5A). All bands were absent in the Δfpr1 mutants, suggesting that they represent different isoforms of Fpr1.
FIGURE 4.
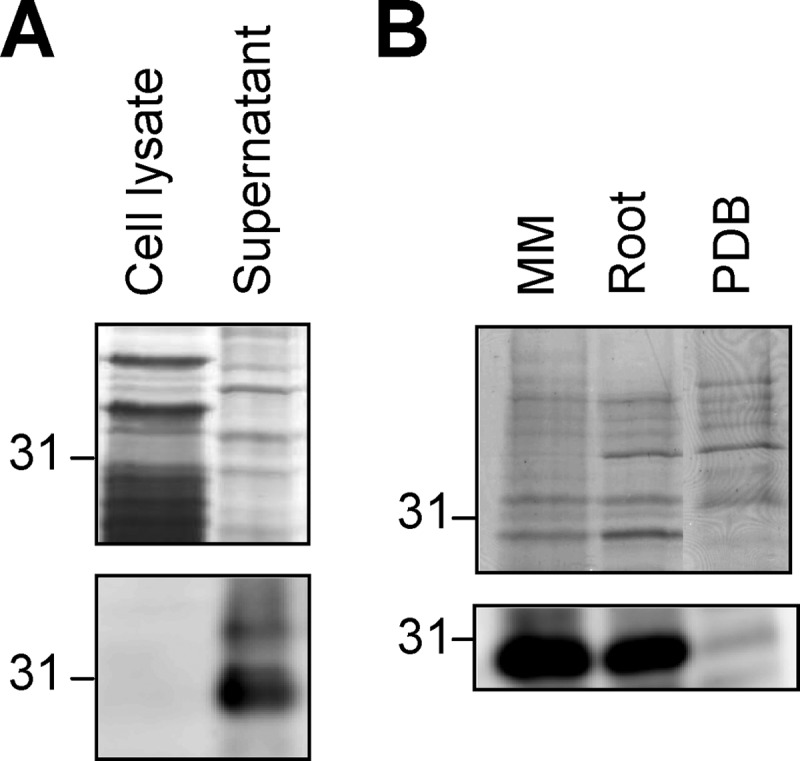
Fpr1 is secreted. A, Western blot analysis of cell lysate and culture supernatant of the F. oxysporum wild-type strain. Upper panel, samples separated by SDS-PAGE and stained with Coomassie Blue. Lower panel, immunoblot analysis with α-Fpr1 antiserum. B, Western analysis of culture supernatants of the wild-type strain grown on liquid minimal medium in the absence (MM) or presence of tomato roots (Root) or on potato dextrose broth (PDB). Relative positions of molecular weight markers are indicated at the left.
FIGURE 5.
Fpr1 is processed proteolytically. A, Western blot analysis of culture supernatants of different F. oxysporum strains with α-Fpr1 antiserum. The arrows point to hybridizing bands of different molecular weight. B, immunoblot of Fpr1 protein incubated for 16 h in the presence of culture supernatant (CS) of the indicated fungal strains, water, or 0.2 N NaOH. C, immunoblot of recombinant Fpr1 protein incubated with culture supernatant of strain Δfpr1–1 for the indicated time periods (left panel) or for 16 h (right panel) in the absence or presence of a protease inhibitor (PI) cocktail or of the indicated protease inhibitors. Relative positions of molecular weight markers are indicated at the left.
We next tested whether the 40-kDa band corresponds to an Fpr1 precursor which is processed into the major 30-kDa form by the wild-type strain but not by the Δfgb1 mutant. To this aim, recombinant Fpr1 protein from E. coli was incubated with culture supernatant from the Δfpr1#1 or the Δfgb1 strains. Supernatant of Δfpr1#1 was used instead of wild-type supernatant to circumvent hybridization interference from the native Fpr1 protein. As shown in Fig. 5B, the 40-kDa Fpr1 band was converted into the 30-kDa form upon incubation with culture supernatant of the Δfpr1#1 strain but not that of the Δfgb1 strain. Similar results were obtained with recombinant Fpr1 from P. pastoris (data not shown). Treatment with NaOH had no effect, suggesting that the mass shift is not caused by changes in O-glycosylation (Fig. 5B). However, addition of a protease inhibitor mixture completely abolished the mass shift of recombinant Fpr1 incubated with Δfpr1#1 culture supernatant (Fig. 5C, left panel). To gain insight into the nature of the proteolytic enzyme(s) responsible for Fpr1 processing, different protease inhibitors were added, including leupeptin (inhibits serine and cysteine proteases), PMSF (serine proteases), EDTA (metalloproteases), and pepstatin A (aspartyl proteases). All inhibitors except EDTA prevented the size shift of Fpr1 to different extents (Fig. 5C, right panel). This suggests that multiple proteolytic enzymes other than metalloproteases contribute to processing of secreted Fpr1 by F. oxysporum.
Fpr1 Is Dispensable for Vegetative Growth, Development and Virulence on Tomato Plants
Hyphal growth and conidiation of the Δfpr1 mutants was indistinguishable from the wild-type strain either on minimal or rich medium, in liquid or solid culture, as well as under conditions of osmotic (0.8 m NaCl), oxidative (10 μg ml−1 menadione), high temperature (37 °C), or cell wall stress (20 μg ml−1 Congo Red or 20 μg ml−1 Calcofluor White) (data not shown). The Δfpr1#1 mutant performed as efficiently as the wild type in colonization and maceration of tomato fruit tissue (supplemental Fig. 5A, only mutant Δfpr1#1 is shown). Root inoculation of tomato plants with microconidia of the wild type, Δfpr1#1, or Δfpr1#1+fpr1 strains caused similar extent of vascular wilt symptoms and plant mortality (supplemental Fig. 5B). In different virulence-related phenotypic assays, the Δfpr1 mutant was indistinguishable from of the wild-type strain, including secretion of pectinolytic enzymes, penetration of cellophane membranes, vegetative hyphal fusion, or adhesion to tomato roots (data not shown). Together, these results suggest that Fpr1 is dispensable for vegetative growth, different stress responses, and virulence of F. oxysporum on the plant host.
Fpr1 Is Essential for Efficient Dissemination and Virulence in a Mouse Model
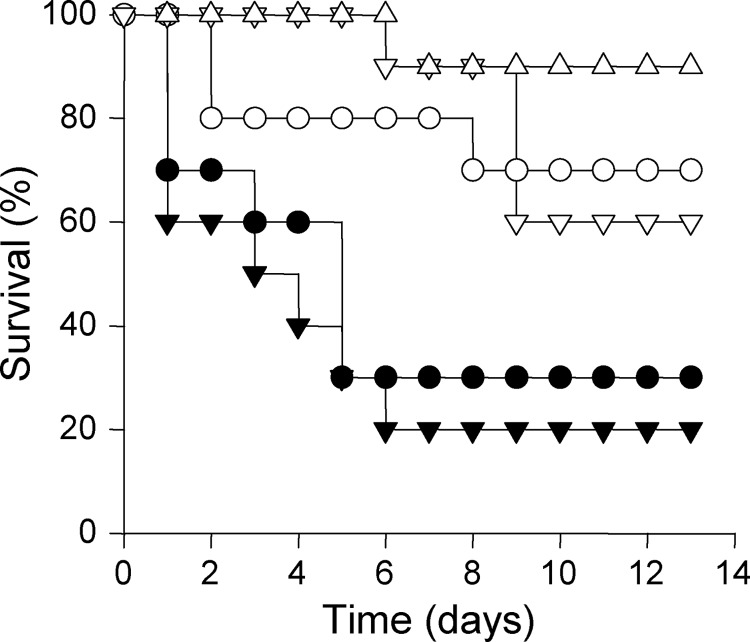
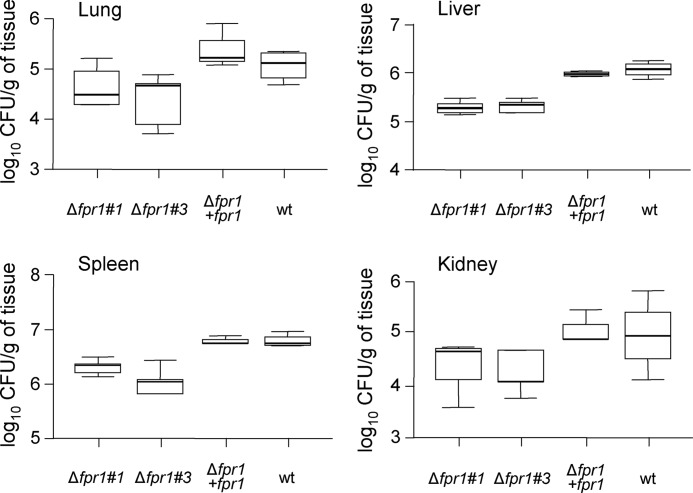
Mortality rates of immunodepressed mice infected with the two independent fungal mutants Δfpr1#1 and Δfpr1#3 were significantly lower (p < 0.05) than in mice infected with the wild-type strain (Fig. 6). Complementation of the Δfpr1#1 mutant with the native fpr1 allele restored virulence to wild-type levels, but introduction of the fpr1H170A,E177A allele into the same mutant failed to do so. Fungal tissue burden in lung, liver, spleen, and kidney of surviving mice sacrificed 7 days after challenge was significantly (p < 0.05) lower in mice infected with the Δfpr1#1 and Δfpr1#3 mutants than in those infected with the wild-type or the Δfpr1#1+fpr1 strain (Fig. 7). These results demonstrate that Fpr1 function is required for full virulence of F. oxysporum on a mammalian host.
FIGURE 6.
Fpr1 function is required for virulence of F. oxysporum on immunodepressed mice. Groups of ten mice were infected by lateral tail vein injection with 2 × 107 microconidia of the following strains: wild type strain 4287 (▾), knockout mutants Δfpr1#1 (○) and Δfpr1#3 (▿ and △), and the Δfpr1#1 mutant complemented either with a wild-type fpr1 allele (●) or the fpr1H170A,E177A allele (▿). Percent survival was plotted for 14 days. The data shown are from one representative experiment.
FIGURE 7.
Fungal burden is reduced in mice infected with Δfpr1 strains. Five surviving mice infected with the indicated strains were sacrificed on day 7 post-infection, and homogenates obtained from the indicated organs were cultured quantitatively on PDA medium. CFU, Colony Forming Units.
Expression of fpr1 Is Induced during Fungal Growth in Human Blood and in a Mammalian Host
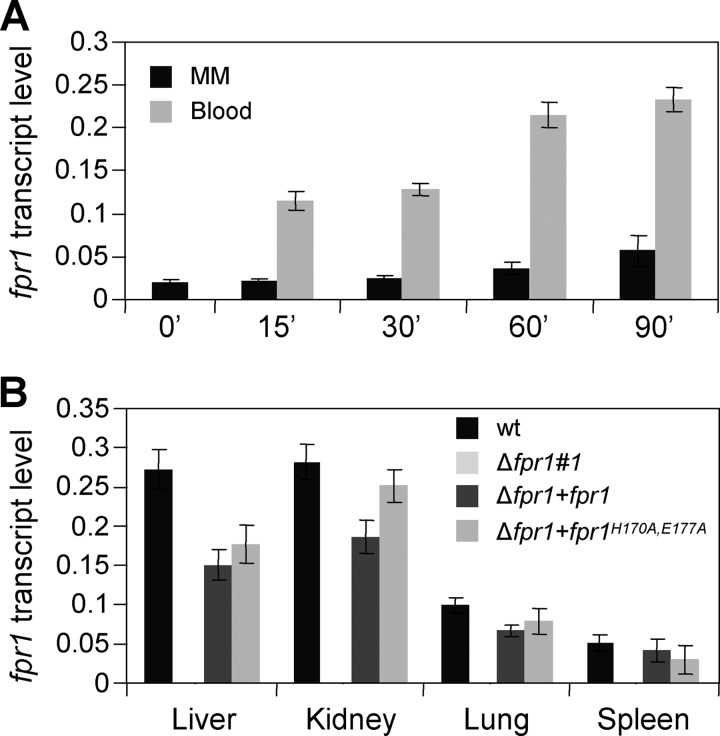
Transfer of F. oxysporum germlings to whole human blood resulted in rapid activation of fpr1 expression (Fig. 8A). Within 60 min, transcript levels increased ∼4-fold compared with mycelia grown in MM. Induced transcript levels of fpr1 were also observed in kidney and liver of immunodepressed mice 3 days after inoculation, whereas lower levels were detected in lung and spleen (Fig. 8B). As expected, no fpr1 transcript was detected in organs of mice inoculated with the Δfpr1#1 mutant, confirming the absence of background PCR amplification. Thus, expression of fpr1 is induced during growth of F. oxysporum in human blood and within the mammalian host.
FIGURE 8.
Expression of fpr1 in F. oxysporum is up-regulated during growth in human blood and infection of mice. Quantitative real-time RT-PCR of cDNA extracted from the wild-type strain germinated 12 h in MM and then transferred for the indicated time periods (min) to MM or human blood (A) or from different organs of immunodepressed mice 3 days after inoculation with the indicated strains (B). Transcript levels of fpr1 are normalized to act1. Error bars represent mean ± S.E. from two independent infection experiments with three technical replicates each.
Fpr1 Is Part of a PR-1-like Protein Family in F. oxysporum
A BLASTp search of the F. oxysporum genome database with the Fpr1 amino acid sequence detected, besides Fpr1 (FOXG_09795), five additional predicted proteins with a PR-1-like domain, all containing a putative N-terminal secretion signal: FOXG_06245, FOXG_10300, FOXG_12428, FOXG_12292, and FOXG_14109 (supplemental Fig. 6). FOXG_12428 and FOXG_14109 are identical, suggesting that they originated from a recent gene duplication event. The putative catalytic tetrad His-Glu-Glu-His was present in all members except FOXG_06245. Interestingly, FOXG_10300 lacks the intermediate region located between the signal peptide and the SCP domain that is present in the other PR-1-like proteins and contains multiple predicted N-glycosylation sites in the amino terminal part as well as a predicted glycosylphosphatidyl-inositol (GPI) motif in the carboxy terminal region.
The presence of transcripts of the PR-1-like genes was determined in different F. oxysporum strains (supplemental Fig. 7). FOXG_06245 produced similar results in all strains and conditions studied, whereas FOXG_12428/FOXG_14109 showed a stronger signal in the Δfpr1 knockout mutants. No transcripts of FOXG_10300 and FOXG_12292 were detected under any of the conditions studied. Thus, F. oxysporum contains multiple PR-1-like genes that are differentially regulated at the transcriptional level.
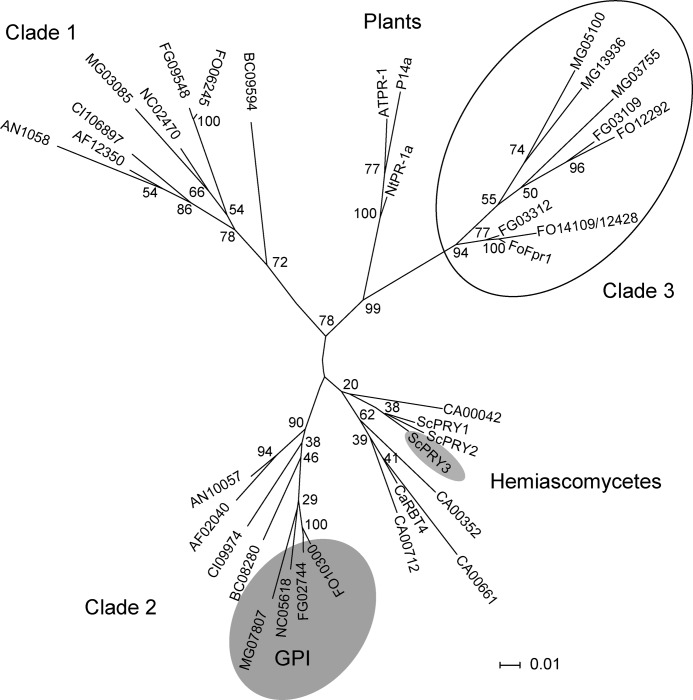
Fpr1 Belongs to a Clade of PR-1-like Proteins That Has Expanded in Plant Pathogenic Sordariomycetes
The presence of PR-1-like proteins in a number of sequenced ascomycete genomes was examined on the Web server of the Broad Institute using the BLASTp algorithm. In addition, we included the plant PR-1 proteins P14a from tomato (32), PR1a from tobacco (3), and AtPR-1 from Arabidopsis (33) in the analysis. The phylogram obtained from sequence alignment is depicted in Fig. 9. Most Euascomycetes, including the saprophytes Neurospora crassa and Aspergillus nidulans, the human pathogens Aspergillus fumigatus and Coccidioides immitis, and the plant pathogen Botrytis cinerea, contain two PR-1-like proteins that cluster into well separated clades (designated clades 1 and 2). The basidiomycetes Ustilago maydis and Coprinus cinereus and the Zygomycete Rhizopus oryzae also have two PR-1-like proteins (results not shown). Only clade 2 members from Sordariomycetes, including FOXG_10300, share a predicted GPI motif in the carboxy terminal region (shaded in gray in Fig. 8). Candida albicans shows a remarkable increase in the number of PR-1-like proteins with five members, all of which cluster in a Hemiascomycete-specific clade (Fig. 8).
FIGURE 9.
Fpr1 is part of a gene family that has expanded in plant pathogenic Sordariomycetes. Phylogram of PR-1-like proteins in fungi. Fpr1 from F. oxysporum (FOXG_09795); Rbt4 from C. albicans (XP_718792); Pry1, Pry2, and Pry3 from S. cerevisiae (SGD_YJL078C); P14a from Solanum lycopersicum (P04284); PR1a from Nicotiana tabacum (CAA29022.1); AtPR-1 from Arabidopsis thaliana (P33154); and Sc7 from Schizophyllum commune (P35794). For the remaining proteins, gene ID numbers are given preceded by a species prefix: AF, A. fumigatus; AN, A. nidulans; AT, A. thaliana; BC, B. cinerea; CA, C. albicans; CI, C. immitis; CN, Cryptococcus neoformans; FG, F. graminearum; FO, F. oxysporum; MG, M. grisea; NC, N. crassa; RO, R. oryzae; SC, S. cerevisiae; and UM, U. maydis. Bootstrap values obtained from 1000 replicates are indicated at the nodes. Scale bar = relative length of each branch. The subfamily expanded in the pathogenic Sordariomycetes Fusarium and Magnaporthe is circled. Proteins containing a predicted GPI sequence are embedded in a gray area. Subclades corresponding to PR-1 proteins from Hemiascomycetes and plants are indicated.
A striking expansion of PR-1-like proteins was detected in the plant pathogenic Sordariomycetes F. oxysporum, Fusarium graminearum, and Magnaporthe grisea. These species contain additional PR-1-like proteins clustering into a separate clade, designated clade 3 (circled in Fig. 8). Fpr1 falls within a Fusarium-specific subclade of clade 3. Interestingly, clade 3 appears closer to the plant PR-1 proteins than to the highly conserved fungal clades 1 and 2 (Fig. 8). The Fusarium PR-1-like proteins in clade 3 contain a non-canonical cysteine between the conserved glutamine and valine residues of the CRISP-1 domain (red arrowheads in supplemental Fig. 6). Moreover, the members of clade 3 lack some of the cysteine residues predicted to engage in the formation of two intramolecular disulfide bridges, which are invariably conserved in members of clades 1 and 2 (supplemental Fig. 6). These results suggest that Fpr1 belongs to a subfamily of PR-1-like proteins that has expanded in plant pathogenic Sordariomycetes.
DISCUSSION
Distinct Cellular Pathways Regulate Expression and Posttranslational Processing of Fpr1
Levels of fpr1 transcript and of secreted Fpr1 protein were dramatically reduced in a mutant lacking the MAPK Fmk1, suggesting that Fmk1 controls fpr1 expression through an unknown mechanism. Interestingly, fpr1 transcript levels were decreased further in a double mutant lacking both Fmk1 and the Gβ subunit Fgb1. Fgb1 was shown previously to function in a cAMP/PKA pathway distinct from the Fmk1 cascade (16). The combinatorial control of fpr1 by these two signaling pathways is reminiscent of the flo11 gene from Saccharomyces cerevisiae, which encodes a cell surface protein required for pseudohyphal formation. Expression of flo11 requires coordinated activation by the Kss1 MAPK cascade and the cAMP/protein kinase A pathway (34), which are orthologous, respectively, to the Fmk1 and the Fgb1-regulated pathways of F. oxysporum.
A DNA array analysis of C. albicans mutants lacking either Cph1 or Efg1, two transcription factors functioning downstream of the MAPK and cAMP/protein kinase A pathway, respectively, identified RBT4 as a hypha-specific gene showing the strongest combinatorial control by the two pathways (35). Similar to Fpr1, RBT4 is a member of a PR-1-like gene family, which has expanded in C. albicans (see Fig. 8) and is required for full virulence on a mammalian host (36). Thus, both the regulatory pathways mediating expression and their role during infection appear to be conserved between two PR-1-like genes from Fusarium and Candida.
Western blot analysis revealed the presence of multiple secreted forms of Fpr1. The band with the highest molecular weight migrated at an apparent mass of 40 kDa, although MALDI-MS analysis confirmed the expected molecular mass of 30 kDa. We speculate that the Ser-Pro rich region of Fpr1 could account in part for its non-canonical electrophoretic behavior because conformational restrictions of the proline may prevent the protein from adopting the expected rod-like shape in the presence of SDS. Furthermore, acidic proteins such as Fpr1 tend to show reduced binding affinity for SDS, possibly accounting for migration of Fpr1 at a higher apparent molecular mass.
Proteolytic processing of full-length form Fpr1 into multiple smaller forms was detected in the wild-type strain but not in a mutant lacking Fgb1. In a previous study, a Δfgb1 mutant displayed a marked decrease in extracellular protease activity (37). Thus, Fgb1 may to control activity of Fpr1-processing protease(s), whose identity remains unknown. Our inhibitor studies indicate the possible involvement of multiple proteolytic enzymes in this process.
The biological relevance of Fpr1 cleavage is currently unknown. Proteolytic processing and activation, either by fungal or host proteases, has been reported in a number of secreted effectors from plant pathogens such as the cysteine rich proteins Avr4 from Cladosporium fulvum (38) and Six1 from F. oxysporum (39). Processing of Fpr1 observed in this study is carried out by fungal proteases. It remains to be determined whether processing is required for the unknown biochemical activity of Fpr1 and whether the cleaved protein undergoes further processing within the plant or mammalian host.
Genetic Evidence Supports a Functional Role of the Predicted Active Site of Fpr1
To date, the biological role of PR-1-like proteins remains largely enigmatic. Although they have been associated with fundamental biological processes such as reproduction, immune response, or cancer (5, 25), these functions were inferred indirectly on the basis of gene expression, localization in specific cell types (e.g. glioma or sperm cells), or in response to certain stimuli (e.g. pathogen attack) rather than by firm genetic evidence.
A solvent-exposed spatial cluster of two histidine and two glutamic acid residues is highly conserved among PR-1-like proteins and was proposed as the putative active site of this protein family (6). Because these four residues are also present in Fpr1, we tested their functional role by changing His-170 and Glu-177 into alanines. In contrast to the wild-type allele, the fpr1H170A,E177A allele failed to restore virulence of F. oxysporum on mice when expressed in a Δfpr1 background. This provides the first genetic proof for a functional role of the predicted catalytic site in a PR-1-like protein.
Under non-denaturing conditions, a fraction of recombinant Fpr1 was consistently present as a homodimer. In the human PR-1-like protein GAPR-1, around 10% of total protein was present in the dimeric form (40). Moreover, both in GAPR-1 and Fpr1, the conserved histidine and glutamate residues from the putative active site are not essential for dimerization. Circular dichroism spectra of wild-type Fpr1 and Fpr1H170A,E177A showed that the percentage of secondary structural elements was unchanged by these mutations. These results support an essential role of the predicted active site in the biological function of Fpr1 but argue against its implication in protein structure.
Evidence for an Expansion of PR-1-like Proteins in Plant Pathogenic Sordariomycetes
The evolution of fungal pathogenicity is thought to be associated with the expansion of certain gene families (41, 42). In our survey of ascomycete genomes, we detected two cases of expansion of PR-1-like genes in pathogenic ascomycetes. The first concerns the presence of five PR-1-like genes in the human pathogen C. albicans, all of which cluster within a Hemiascomycete-specific group together with the PR-1-like proteins from S. cerevisiae and PRY1, 2, and 3. One protein from this group, RBT4, was shown previously to contribute to virulence of C. albicans on mammals (36), whereas the role of the remaining members is unknown. The second case of expansion of PR-1-like proteins was found in clade 3, which is unique to the Sordariomycete plant pathogens Fusarium spp. and M. grisea. F. graminearum has two members within this clade, M. grisea three, and F. oxysporum four, including two exact gene duplications. Fpr1 also falls within this clade whose evolutionary origin is unclear. In our phylogenetic analysis, clade 3 was placed closer to plant PR-1 proteins than to the highly conserved fungal clades 1 and 2. Although it is tempting to speculate that clade 3 may have originated from a horizontal gene transfer of a PR-1-like gene from plants to a common pathogenic ancestor of the Fusarium and Magnaporthe, the phylogenetic tree did not resolve with sufficient bootstrap support among deeper branches to conclusively support the hypothesis of horizontal gene transfer.
Regardless of their evolutionary origin, members of clade 3 all lack several conserved cysteine residues involved in the formation of two disulfide bonds that are present in virtually all PR-1-like proteins (5), contributing to the high thermal, pH, and proteolytic stability of this family of secreted proteins (30). The only known CAP superfamily protein lacking disulfide bonds is the human GAPR-1 protein, which is also the only PR-1-like protein with a reported intracellular localization (43). However, in contrast to GAPR-1, the members of clade 3 are predicted to be secreted proteins. The absence of disulfide bonds should render them more accessible to proteolytic cleavage, as supported experimentally for Fpr1 in this study.
What could be the biological significance of the lack of disulfide bridges in the members of clade 3? Secreted proteins from plant pathogens are often detected by the host as microbe-associated molecular patterns, which trigger a strong immune response (44). One way for pathogens to evade detection is to reducing the half-life of the secreted microbe-associated molecular pattern proteins by making them more sensitive to degradation by host proteases. For example, the extracellular AVR4 protein from the pathogen C. fulvum has four disulfide bonds, making it highly resistant to plant proteases, and induces a robust defense response in the resistant tomato genotype Cf-4 (38). Naturally occurring strains of C. fulvum, which are virulent on Cf-4 tomatoes, circumvent host recognition by producing AVR4 versions that lack cysteine residues and, consequently, two of the disulfide bonds and are thus readily degraded by plant proteases (45). The absence of conserved disulfide bonds in PR-1-like proteins of clade 3 could reduce their stability in the presence of host proteases, allowing the fungus to evade detection by the plant immune system.
Role of Fpr1 in Virulence
Microbial pathogens of plants and mammals secrete an array of effector molecules that promote virulence either by directly targeting host cells or by suppressing defense responses (46–50). Two lines of evidence support an active role of Fpr1 protein in virulence of F. oxysporum on mammalian hosts. First, two independent knockout mutants lacking fpr1 caused significantly less mortality on immunodepressed mice than the wild type. Second, Fpr1H170A,E177A, which lacks two conserved residues of the putative active site, did not complement the virulence phenotype, indicating that Fpr1 activity is required for its role in virulence.
Considering the presence of multiple PR-1-like proteins in F. oxysporum, the essential role of Fpr1 in virulence is remarkable and suggests a unique function of this secreted protein during infection. Increased survival of mice infected with the Δfpr1 mutants was concomitant with a reduced fungal burden in different organs. Thus, Fpr1 is required for survival of the pathogen or for its efficient dissemination within the host by either actively promoting fungal invasion or by protecting Fusarium from the host immune system. Several mammalian PR-1-like proteins are preferentially expressed in cells and tissues of the innate or adaptive immune systems (5) or have been associated with tissue invasion by cancer cells (26, 51). Thus, interfering RNAs against RTVP-1, a PR-1-like gene highly expressed in human glioblastomas, decreased proliferation of glioma cells, whereas overexpression of the gene increased their invasiveness (51). Secreted Fpr1 protein could play a similar active role in promoting invasion of the mammalian tissue by the fungal pathogen.
Parasitic nematodes colonize their host through suppression and evasion of the immune system, and PR-1-like proteins appear to have an important function in the infection process (52). The dog hookworm Anclystoma caninium produces several PR-1-like proteins, including a neutrophile inhibiting protein, that are secreted in high abundance immediately after the transition of the free living larvae to the parasitic form (53–55), whereas the human parasite Necator americanus contains a family of nine PR-1-like genes that are highly represented within EST libraries and may contribute to immune evasion and inhibition of platelet aggregation (56). The saliva of blood-feeding ticks, flies, and mosquitoes contains PR-1-like proteins that were suggested to suppress the host immune system or to prevent blood clotting (57). Secreted Fpr1 could play an analogous role in allowing F. oxysporum to evade the host immune system and to successfully complete the infection process.
Impaired virulence on mice was the only phenotype resulting from fpr1 deletion in F. oxysporum. Such a highly specific role contrasts with pleiotropic phenotypes of previously reported F. oxysporum mutants affected in virulence on mammalian hosts, such as those lacking the transcription factors PacC or white collar-1 (12, 58) or the Δfmk1Δfgb1 double mutant (37). Pleiotropic genes tend to encode master regulators that control the expression of multiple downstream targets. By contrast, Fpr1 is unlikely to act as a global regulator but rather may function as a secreted effector. The highly specific role of Fpr1 during the fungus-host interaction makes it a promising target for new antifungal therapeutics.
Acknowledgments
We are grateful to Michael W. Rey and Randy Berka, Novozymes Biotech Inc., Davis, CA for making the sequences of the F. oxysporum EST library available. We also thank Isabel Caballero and Esther Martínez for valuable technical assistance.
This research was supported by the Ramon y Cajal Program. This work was also supported by Ministerio de Ciencia e Innovación (MICINN) Grant BIO2010-15505, by MICINN ERA-NET/PathoGenoMics project TRANSPAT Grant BIO2008-04479-E, MICINN/Plant KBBE Grant EUI2009-03942, Junta de Andalucia Grant BIO-3847, Marie Curie ITN ARIADNE Grant FP7-PEOPLE-ITN-237936), and MICINN PhD fellowships (to R. C. P. R. and M. S. L. B.).

This article contains supplemental Figs. 1–7 and Methods.
- CRISP
- cysteine-rich secretory protein
- GPI
- glycosylphosphatidyl-inositol.
REFERENCES
- 1. Sexton A. C., Howlett B. J. (2006) Parallels in fungal pathogenesis on plant and animal hosts. Eukaryot. Cell 5, 1941–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nürnberger T., Brunner F., Kemmerling B., Piater L. (2004) Innate immunity in plants and animals. Striking similarities and obvious differences. Immunol. Rev. 198, 249–266 [DOI] [PubMed] [Google Scholar]
- 3. Cornelissen B. J., Hooft van Huijsduijnen R. A., Van Loon L. C., Bol J. F. (1986) Molecular characterization of messenger RNAs for “pathogenesis-related” proteins la, lb and lc, induced by TMV infection of tobacco. EMBO J. 5, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Loon L. C., Rep M., Pieterse C. M. (2006) Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 [DOI] [PubMed] [Google Scholar]
- 5. Gibbs G. M., Roelants K., O'Bryan M. K. (2008) The CAP superfamily. Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins. Roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897 [DOI] [PubMed] [Google Scholar]
- 6. Szyperski T., Fernández C., Mumenthaler C., Wüthrich K. (1998) Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Natl. Acad. Sci. U.S.A. 95, 2262–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Armstrong G. M., Armstrong J. K. (1981) in Fusarium: Diseases, Biology and Taxonomy (Cook R., ed.), pp. 391–399, Penn State University Press, University Park, PA [Google Scholar]
- 8. Nucci M., Anaissie E. (2007) Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guarro J., Gené J. (1995) Opportunistic fusarial infections in humans. Eur. J. Clin. Microbiol. Infect. Dis. 14, 741–754 [DOI] [PubMed] [Google Scholar]
- 10. Azor M., Cano J., Gené J., Guarro J. (2009) High genetic diversity and poor in vitro response to antifungals of clinical strains of Fusarium oxysporum. J. Antimicrob. Chemother. 63, 1152–1155 [DOI] [PubMed] [Google Scholar]
- 11. O'Donnell K., Sutton D. A., Rinaldi M. G., Magnon K. C., Cox P. A., Revankar S. G., Sanche S., Geiser D. M., Juba J. H., van Burik J. A., Padhye A., Anaissie E. J., Francesconi A., Walsh T. J., Robinson J. S. (2004) Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses. Evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 42, 5109–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ortoneda M., Guarro J., Madrid M. P., Caracuel Z., Roncero M. I., Mayayo E., Di Pietro A. (2004) Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72, 1760–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Navarro-Velasco G. Y., Prados-Rosales R. C., Ortiz-Urquiza A., Quesada-Moraga E., Di Pietro A. (2011) Galleria mellonella as model host for the trans-kingdom pathogen Fusarium oxysporum. Fungal Genet. Biol. 48, 1124–1129 [DOI] [PubMed] [Google Scholar]
- 14. Di Pietro A., Roncero M. I. (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 11, 91–98 [DOI] [PubMed] [Google Scholar]
- 15. Di Pietro A., García-MacEira F. I., Méglecz E., Roncero M. I. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152 [PubMed] [Google Scholar]
- 16. Delgado-Jarana J., Martínez-Rocha A. L., Roldán-Rodriguez R., Roncero M. I., Di Pietro A. (2005) Fusarium oxysporum G-protein β subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet. Biol. 42, 61–72 [DOI] [PubMed] [Google Scholar]
- 17. Chomczynski P., Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 18. Raeder U., Broda P. (1985) Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–20 [Google Scholar]
- 19. Hazen G. G., Hause J. A., Hubicki J. A. (1965) An automated system for the quantitative determination of proteolytic enzymes using azocasein. Ann. N.Y. Acad. Sci. 130, 761–768 [DOI] [PubMed] [Google Scholar]
- 20. Carr S., Aebersold R., Baldwin M., Burlingame A., Clauser K., Nesvizhskii A. (2004) The need for guidelines in publication of peptide and protein identification data. Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol. Cell Proteomics 3, 531–533 [DOI] [PubMed] [Google Scholar]
- 21. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guindon S., Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 23. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides. SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 24. Eisenhaber B., Schneider G., Wildpaner M., Eisenhaber F. (2004) A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337, 243–253 [DOI] [PubMed] [Google Scholar]
- 25. van Loon L., van Strien E. (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97 [Google Scholar]
- 26. Murphy E. V., Zhang Y., Zhu W., Biggs J. (1995) The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene 159, 131–135 [DOI] [PubMed] [Google Scholar]
- 27. Krätzschmar J., Haendler B., Eberspaecher U., Roosterman D., Donner P., Schleuning W. D. (1996) The human cysteine-rich secretory protein (CRISP) family. Primary structure and tissue distribution of CRISP-1, CRISP-2 and CRISP-3. Eur. J. Biochem. 236, 827–836 [DOI] [PubMed] [Google Scholar]
- 28. Lu G., Villalba M., Coscia M. R., Hoffman D. R., King T. P. (1993) Sequence analysis and antigenic cross-reactivity of a venom allergen, antigen 5, from hornets, wasps, and yellow jackets. J. Immunol. 150, 2823–2830 [PubMed] [Google Scholar]
- 29. Morrissette J., Krätzschmar J., Haendler B., el-Hayek R., Mochca-Morales J., Martin B. M., Patel J. R., Moss R. L., Schleuning W. D., Coronado R. (1995) Primary structure and properties of helothermine, a peptide toxin that blocks ryanodine receptors. Biophys. J. 68, 2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernández C., Szyperski T., Bruyère T., Ramage P., Mösinger E., Wüthrich K. (1997) NMR solution structure of the pathogenesis-related protein P14a. J. Mol. Biol. 266, 576–593 [DOI] [PubMed] [Google Scholar]
- 31. Milne T. J., Abbenante G., Tyndall J. D., Halliday J., Lewis R. J. (2003) Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 278, 31105–31110 [DOI] [PubMed] [Google Scholar]
- 32. Niderman T., Genetet I., Bruyère T., Gees R., Stintzi A., Legrand M., Fritig B., Mösinger E. (1995) Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 108, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., Chandler D., Slusarenko A., Ward E., Ryals J. (1992) Acquired resistance in Arabidopsis. Plant Cell 4, 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rupp S., Summers E., Lo H. J., Madhani H., Fink G. (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18, 1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lane S., Birse C., Zhou S., Matson R., Liu H. (2001) DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276, 48988–48996 [DOI] [PubMed] [Google Scholar]
- 36. Braun B. R., Head W. S., Wang M. X., Johnson A. D. (2000) Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prados-Rosales R. C., Serena C., Delgado-Jarana J., Guarro J., Di Pietro A. (2006) Distinct signalling pathways coordinately contribute to virulence of Fusarium oxysporum on mammalian hosts. Microbes Infect 8, 2825–2831 [DOI] [PubMed] [Google Scholar]
- 38. Joosten M. H., Cozijnsen T. J., De Wit P. J. (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367, 384–386 [DOI] [PubMed] [Google Scholar]
- 39. Rep M., van der Does H. C., Meijer M., van Wijk R., Houterman P. M., Dekker H. L., de Koster C. G., Cornelissen B. J. (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383 [DOI] [PubMed] [Google Scholar]
- 40. Serrano R. L., Kuhn A., Hendricks A., Helms J. B., Sinning I., Groves M. R. (2004) Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism. J. Mol. Biol. 339, 173–183 [DOI] [PubMed] [Google Scholar]
- 41. Soanes D. M., Alam I., Cornell M., Wong H. M., Hedeler C., Paton N. W., Rattray M., Hubbard S. J., Oliver S. G., Talbot N. J. (2008) Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One 3, e2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tunlid A., Talbot N. J. (2002) Genomics of parasitic and symbiotic fungi. Curr. Opin. Microbiol. 5, 513–519 [DOI] [PubMed] [Google Scholar]
- 43. Eberle H. B., Serrano R. L., Füllekrug J., Schlosser A., Lehmann W. D., Lottspeich F., Kaloyanova D., Wieland F. T., Helms J. B. (2002) Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J. Cell Sci. 115, 827–838 [DOI] [PubMed] [Google Scholar]
- 44. Boller T., Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 45. van den Burg H. A., Westerink N., Francoijs K. J., Roth R., Woestenenk E., Boeren S., de Wit P. J., Joosten M. H., Vervoort J. (2003) Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278, 27340–27346 [DOI] [PubMed] [Google Scholar]
- 46. Birch P. R., Boevink P. C., Gilroy E. M., Hein I., Pritchard L., Whisson S. C. (2008) Oomycete RXLR effectors. Delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379 [DOI] [PubMed] [Google Scholar]
- 47. Ellis J. G., Dodds P. N., Lawrence G. J. (2007) The role of secreted proteins in diseases of plants caused by rust, powdery mildew and smut fungi. Curr. Opin. Microbiol. 10, 326–331 [DOI] [PubMed] [Google Scholar]
- 48. Galán J. E. (2009) Common themes in the design and function of bacterial effectors. Cell Host Microbe 5, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naglik J., Albrecht A., Bader O., Hube B. (2004) Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 6, 915–926 [DOI] [PubMed] [Google Scholar]
- 50. Rep M. (2005) Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27 [DOI] [PubMed] [Google Scholar]
- 51. Rosenzweig T., Ziv-Av A., Xiang C., Lu W., Cazacu S., Taler D., Miller C. G., Reich R., Shoshan Y., Anikster Y., Kazimirsky G., Sarid R., Brodie C. (2006) Related to testes-specific, vespid, and pathogenesis protein-1 (RTVP-1) is overexpressed in gliomas and regulates the growth, survival, and invasion of glioma cells. Cancer Res. 66, 4139–4148 [DOI] [PubMed] [Google Scholar]
- 52. Hawdon J. M., Hotez P. J. (1996) Hookworm. Developmental biology of the infectious process. Curr. Opin. Genet. Dev. 6, 618–623 [DOI] [PubMed] [Google Scholar]
- 53. Del Valle A., Jones B. F., Harrison L. M., Chadderdon R. C., Cappello M. (2003) Isolation and molecular cloning of a secreted hookworm platelet inhibitor from adult Ancylostoma caninum. Mol. Biochem. Parasitol. 129, 167–177 [DOI] [PubMed] [Google Scholar]
- 54. Hawdon J. M., Jones B. F., Hoffman D. R., Hotez P. J. (1996) Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J. Biol. Chem. 271, 6672–6678 [DOI] [PubMed] [Google Scholar]
- 55. Moyle M., Foster D. L., McGrath D. E., Brown S. M., Laroche Y., De Meutter J., Stanssens P., Bogowitz C. A., Fried V. A., Ely J. A. (1994) A hookworm glycoprotein that inhibits neutrophil function is a ligand of the integrin CD11b/CD18. J. Biol. Chem. 269, 10008–10015 [PubMed] [Google Scholar]
- 56. Daub J., Loukas A., Pritchard D. I., Blaxter M. (2000) A survey of genes expressed in adults of the human hookworm, Necator americanus. Parasitology 120, 171–184 [DOI] [PubMed] [Google Scholar]
- 57. Ribeiro J. M., Francischetti I. M. (2003) Role of arthropod saliva in blood feeding. Sialome and post-sialome perspectives. Annu. Rev. Entomol. 48, 73–88 [DOI] [PubMed] [Google Scholar]
- 58. Ruiz-Roldán M. C., Garre V., Guarro J., Mariné M., Roncero M. I. (2008) Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot Cell 7, 1227–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]