Background: SMM is activated by RLC phosphorylation in the lever arm.
Results: Modifying RLC-ELC interaction hampers the ability of phosphorylation to activate motor functions.
Conclusion: A major consequence of phosphorylation is to stabilize RLC-ELC interactions and associated conformations of the lever arm elbow.
Significance: Learning how this myosin is regulated furthers the understanding of activation and relaxation of smooth muscle contraction.
Keywords: Actin, ATPases, Myosin, Protein Phosphorylation, Smooth Muscle, Structural Biology, Essential Light Chain, Lever Arm, Regulatory Light Chain, Smooth Muscle Myosin
Abstract
We examined the regulatory importance of interactions between regulatory light chain (RLC), essential light chain (ELC), and adjacent heavy chain (HC) in the regulatory domain of smooth muscle heavy meromyosin. After mutating the HC, RLC, and/or ELC to disrupt their predicted interactions (using scallop myosin coordinates), we measured basal ATPase, Vmax, and KATPase of actin-activated ATPase, actin-sliding velocities, rigor binding to actin, and kinetics of ATP binding and ADP release. If unphosphorylated, all mutants were similar to wild type showing turned-off behaviors. In contrast, if phosphorylated, mutation of RLC residues smM129Q and smG130C in the F-G helix linker, which interact with the ELC (Ca2+ binding in scallop), was sufficient to abolish motility and diminish ATPase activity, without altering other parameters. ELC mutations within this interacting ELC loop (smR20M and smK25A) were normal, but smM129Q/G130C-R20M or -K25A showed a partially recovered phenotype suggesting that interaction between the RLC and ELC is important. A molecular dynamics study suggested that breaking the RLC/ELC interface leads to increased flexibility at the interface and ELC-binding site of the HC. We hypothesize that this leads to hampered activation by allowing a pre-existing equilibrium between activated and inhibited structural distributions (Vileno, B., Chamoun, J., Liang, H., Brewer, P., Haldeman, B. D., Facemyer, K. C., Salzameda, B., Song, L., Li, H. C., Cremo, C. R., and Fajer, P. G. (2011) Broad disorder and the allosteric mechanism of myosin II regulation by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 108, 8218–8223) to be biased strongly toward the inhibited distribution even when the RLC is phosphorylated. We propose that an important structural function of RLC phosphorylation is to promote or assist in the maintenance of an intact RLC/ELC interface. If the RLC/ELC interface is broken, the off-state structures are no longer destabilized by phosphorylation.
Introduction
For all muscles, contraction and relaxation are regulated by changes in the intracellular Ca2+ concentration. In striated muscles (skeletal and cardiac), Ca2+ binding to the tropomyosin-troponin complex on thin filaments activates the actin-activated myosin II ATPase and thus contraction. In contrast, in molluscan and vertebrate smooth muscle (and non-muscle cells), the contractile state is triggered by direct Ca2+ binding to the myosin II ELC2 or by phosphorylation of the N-terminal domain of the RLC by the Ca2+/calmodulin-dependent MLCK, respectively. For all myosins, the generally accepted mechanism explaining how these motors generate movement is that ATP-induced structural changes at the active site cause the RD to rotate, while the motor domain is bound to actin (1). The RD is comprised of a long helix of the HC to which the RLC and ELC bind in an antiparallel manner (Fig. 1A). The light chains stabilize the HC helix so that it can serve as a rigid “lever arm” that functions to amplify small ATP-induced conformational changes in the motor domain into larger conformational changes that produce the working stroke of myosin. The RD is thus named because it is the structural site for Ca2+ binding and phosphorylation in the regulated myosin II isoforms.
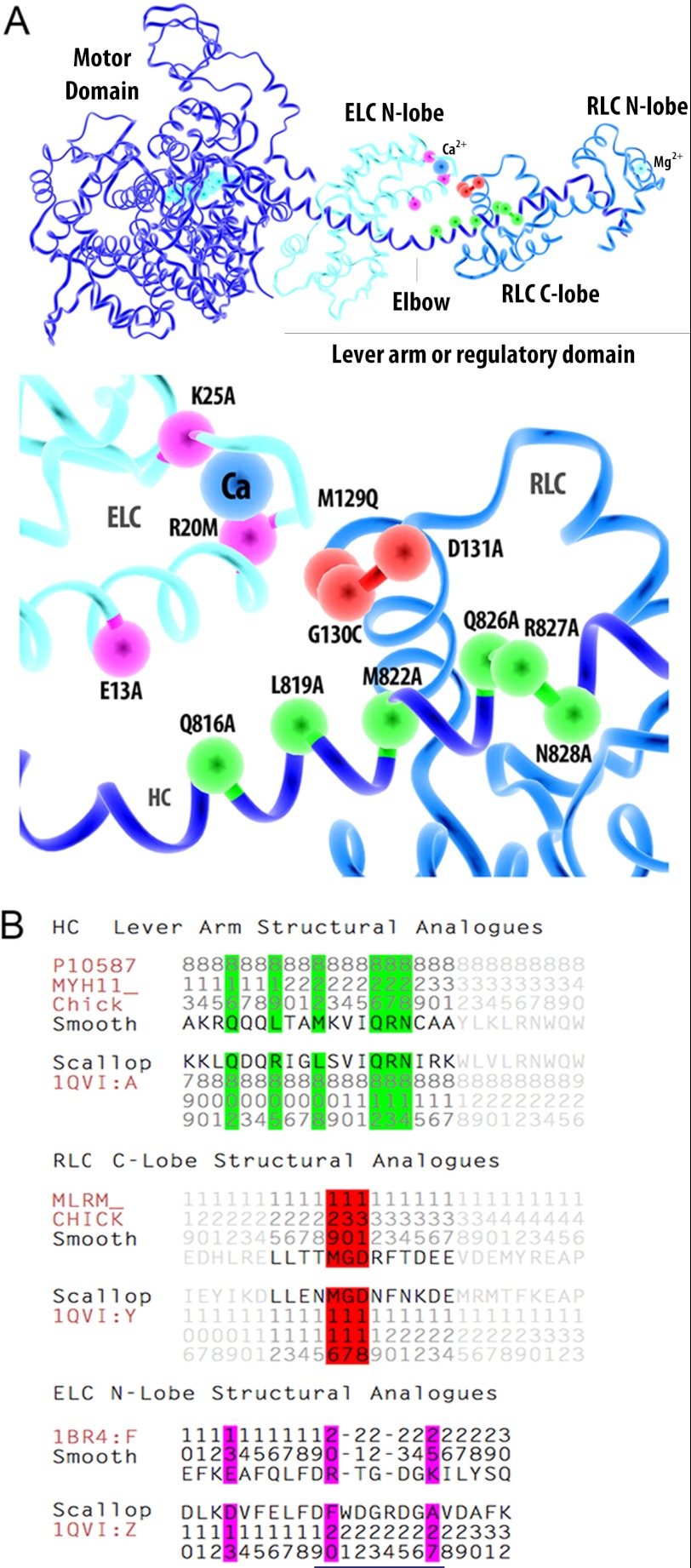
FIGURE 1.
Homologous positions of the amino acids mutated in SMM depicted on the scallop myosin structure. A, panel, ribbon rendering of the scallop myosin subfragment 1 or head domain (PDB code 1QVI). The HC is shown in dark blue; ELC is in cyan (also ATP in motor domain); the RLC is in light blue; the Ca2+ ion bound to the ELC loop I in the N-lobe is shown as a blue ball, and Mg2+ ion bound to the RLC N-lobe is shown in cyan. The RLC, ELC, and HC to which they bind constitute the lever arm or RD. The position of the bend in the HC (elbow) is evident in this view between the RLC and ELC. Bottom panel, the position of the Ca2+ ion bound to an EF hand of the ELC is shown as a blue ball and the other colored balls indicate positions of the mutated amino acids (ball colors coordinate with B). The amino acid numbers are for chicken SMM. B, alignment of the SMM sequence with the scallop sequence for the heavy chain (MYH11), RLC, and ELC regions of interest. Amino acid numbers read from top to bottom. Colors of the letter/number shading correspond to the colors of the balls in A.
We are interested specifically in the structural aspects of the allosteric activation of SMM by phosphorylation. Phosphorylation of Ser-19 on the SMM RLC accelerates the rate of phosphate release from the active site (2) when myosin is bound to actin, and the rate of this step in the cycle is likely limited by a preceding isomerization from a weak binding to strong binding acto-myosin·ADP·Pi state. The result is an increase in the steady-state actin-activated ATPase and an increase in the velocity of actin sliding in an in vitro motility assay. Coupling of the ATPase to movement is mediated by the RLC (3).
Clues to the structural mechanisms of myosin regulation were first revealed in the atomic resolution structures of the RD of scallop myosin II (PDB codes 1SCM and 1WDC), which contain both the ELC (with Ca2+) and the RLC along with the respective portion of the HC to which the light chains bind (4, 5). Similar structures for SMM have not been forthcoming. In scallop myosin, Ca2+ coordinates to the EF hand of domain I residing in the ELC N-lobe (Fig. 1A). The residues of this loop are sc19DFWGDRDGA27, with the three Asp residues involved in Ca2+ coordination (“sc” indicates scallop and “sm” indicates smooth; Fig. 1B, underlined). This Ca2+ binding does not occur in SMM but is required in scallop myosin for activation of both motor and enzymatic activities (6). However, Ca2+ does not bind to the isolated scallop ELC (7). This is because of critical linkages between the ELC Ca2+-binding loop and the only region of the RLC that abuts the ELC, i.e. the linker between the RLC F and G helices in the C-lobe of the RLC (4, 5). This linker is highlighted in red in Fig. 1B. Specifically, scG117 of the scallop RLC (smG130) makes key hydrogen bonds to scF20 and scR24 in the Ca2+-binding loop of the scallop ELC (4, 5, 8). The functional importance of RLC scG117 was demonstrated by mutation to either Cys or Ala, with both mutations giving a phenotype with drastic weakening of RLC binding to the ELC·HC complex, and the partially reconstituted myosin showed neither Ca2+-sensitive ATPase nor specific Ca2+ binding (9).
Further crystallographic studies at higher resolution (PDB code 1WDC) further revealed that residues Asp-118 and Asn-119 of the scallop isoform of RLC (scallop numbering) also make hydrogen bonds to ELC scR24, and apolar interactions between RLC scM116 and ELC scW21 are also involved in the RLC-ELC interaction site (8). Finally, the HC residues scQ812 and scR816 also hydrogen bond to the F-G helix linker of the RLC, further stabilizing the interaction. In summary, the atomic resolution structures for scallop myosin with the Ca2+-binding loop occupied with Ca2+ suggest that local interactions at the interface of the RLC and ELC along with the nearby HC may be important to maintaining the activated state or on-state.
More recently, a structure of the scallop RD (without the motor domain) in the absence of Ca2+ has been obtained (10), where the molecule was modified in the Ca2+-binding loop to prevent Ca2+ binding. Compared with the Ca2+-bound structures, this structure shows the following: 1) the Ca2+-binding loop is more flexible, especially at ELC scR24; 2) the key main-chain hydrogen bond between RLC scG117 and ELC scR24 has been broken; and 3) two hydrogen bonds between the side chains of RLC scD118 and ELC scR24 have also been broken. These data suggest that the off-state of scallop myosin does not require these RLC-ELC interactions. If Ca2+-dependent and phosphorylation-dependent regulation have parallel mechanisms, then we predict that an interaction between the SMM ELC and RLC is critical for activation of SMM by phosphorylation but is not critical to stabilizing the off- or inhibited-state. This prediction is consistent with the fact that the ELC is not required to maintain the off-state of SMM, whereas it is important to maintaining a fully on-state with respect to both ATPase and motility (11, 12).
Here, we address the specific question of whether or not the interaction between the RLC and the ELC in SMM stabilizes the enzymatically and mechanically activated state. Unlike previous studies that used hybrids of subunits or domain substitutions from different isoforms, our approach was to study an intact HMM molecule with mutated RLC, ELC, or HC all from the SMM sequence. HMM is a soluble subfragment of full-length SMM that lacks the C-terminal 2/3rd of the tail domain but retains a high level of regulation by phosphorylation (13). We chose the Ca2+-scallop S1 structure (14), which represents the pre-power stroke state (PDB code 1QVI) to guide our mutations, because it retains a full motor domain containing the converter element that determines the position of the lever arm and interacts with the C-lobe of the ELC. Assuming that this structure approximates the SMM on-state (phosphorylated), we mutated selected amino acids in this region from the ELC, RLC, and nearby HC that were likely involved in a bonding network that we hypothesized to stabilize the appropriate structure for myosin activation. We found that a single mutation of either smM129Q or smG130C, but not smD131A, in the linker between the F and G helices of the RLC was sufficient to completely abolish in vitro motility and diminish the ATPase activity of the P-HMM. In contrast, there was no significant effect on the parameters for the uP-HMM, which remained completely inhibited. Further mutating residues flanking the ELC loop that interacts with this region of the RLC partially recovered both the ATPase activity and motor function. These data provide direct evidence that the native RLC-ELC interaction is important for motor function.
To further understand the mechanism whereby such mutations alter motor function, we used discrete molecular dynamics simulations to compare the flexibility of two scallop S1 structures as follows: one in which Ca2+ is bound to loop I of the ELC with native RLC-ELC interactions intact (PDB code 1QVI), and one in which we substituted in silico the Ca2+-binding loop with the smooth ELC sequence that does not bind to Ca2+ (modified PDB code 1QVI). In the latter structure, we expected the RLC/ELC interface to be broken or no longer functional. The simulations showed that the modified 1QVI structure was more flexible specifically at the RLC/ELC interface and the surrounding HC. These simulations along with the experimental data on smHMM reported herein imply that an important role for phosphorylation is to stabilize the RLC/ELC interface and potentially the bend in the HC of the lever arm dubbed the “elbow.” A specific mechanism for this has been proposed.
EXPERIMENTAL PROCEDURES
Protein Preparations
Chicken smooth muscle HMM was expressed in Sf9 insect cells by co-infection with three viruses to obtain the HC (1106 residues with a C-terminal His tag), full-length RLC, and full-length ELC. The molecular mass = 314,000 daltons and the E280 nm0.1% = 0.61. Details of cloning, expression, and purification have been previously described (15). The RLC mutants M129Q/G130C, M129Q, and G130C were obtained using a site-directed mutagenesis kit (Stratagene). RLC mutants D131A, ELC mutants E13A, R20M, and K25A, and HC mutants Q816A, Q826A, R827M, N828A, and L819A/M822A/Q826A were obtained by multi-PCR. All constructs were confirmed by sequencing. HMMs were thiophosphorylated using ATPγS in the presence of MLCK/Ca2+-CaM as described previously (16). For data collection, each HMM construct was expressed at least twice, and the reported data reflect the average of these data or otherwise as indicated in the figures and tables. All HMM preparations were stored on ice and used within 2 weeks of purification. Actin was purified from frozen rabbit skeletal muscle acetone powder (17) and was labeled with pyrene-iodoacetamide (Invitrogen) as described (18).
Steady-state NH4+-EDTA ATPase Assay
The NH4+-EDTA ATPase was measured at 25 °C in a solution containing 50–100 μg/ml HMM, 0.23 m KCl, 6.25 mm ATP, 0.56 m NH4Cl, 38 mm EDTA, and 62.5 mm Tris (pH 8.0). The production of inorganic phosphate was measured by the method of Fiske and SubbaRow (19). Samples were quenched into an aliquot of molybdate reagent (0.75 n H2SO4 and 0.66% ammonium molybdate) at 3 and 10 min. The resulting phosphomolybdate complex was reduced by addition of freshly prepared 10% FeSO4 in 0.15 n H2SO4. After 2 min, the absorbance at 700 nm was measured. A standard curve was prepared using KH2PO4.
Transient Kinetic Assays
All transient kinetic experiments (Tables 1 and 2) were done at 25 °C in 50 mm NaCl, 0.1 mm EGTA, 1 mm MgCl2, 1 mm DTT, and 10 mm MOPS (pH 7.0). Reported concentrations refer to values after mixing (unless otherwise indicated), and HMM concentrations refer to the concentration of heads (two heads per HMM). A Hi-Tech stopped-flow spectrofluorimeter was used for all studies. See Cremo and Geeves (20) for instrument settings and filters used.
TABLE 1.
Summary of kinetic and motility data for HMM constructs
| HMM constructs | Scallop structural homologsa | NH4+-ATPaseb | Basal MgATPasec | Vmaxd | KATPased | Motilitye | NVmaxNmotf |
|---|---|---|---|---|---|---|---|
| s−1 head−1 | s−1 | s−1 | μm | μm s−1 | |||
| WT | 7.6 ± 0.1 | 0.032, 0.032 | 2.7 ± 0.7 | 38 ± 8 | 0.25 ± 0.02 | 4,5 | |
| M129Q (RLC) | Met-116 | 7.1 ± 0.1 | 0.021, 0.018 | 0.35 ± 0.05 | <10 | 0 | 2,3 |
| G130C (RLC) | Gly-117 | 7.1 ± 0.2 | 0.024, 0.025 | 0.38 ± 0.04 | <10 | 0 | 2,3 |
| M129Q/G130C (RLC) | Met-116/Gly-117 | 7.1 ± 0.1 | 0.017, 0.010 | 1.2 ± 0.1 | 10 ± 3 | 0 | 2,2 |
| M129Q/G130C/R20 M (RLC-ELC) | Met-116/Gly-117/Trp-21 | 7.1 ± 0.1 | 0.037, 0.021 | 1.4 ± 0.2 | 45 ± 15 | 0.11 ± 0.00 | 2,2 |
| M129Q/G130C/K25A (RLC-ELC) | Met-116/Gly-117/Ala-27 | 7.1 ± 0.1 | 0.012, 0.012 | 1.8 ± 0.5 | 28 ± 4 | 0.13 ± 0.04 | 2,4 |
| R20M (ELC) | g | 7.1 ± 0.1 | 0.019, 0.012 | 2.8 ± 0.1 | 50 ± 0 | 0.22 ± 0.02 | 2,2 |
| K25A (ELC) | Ala-27 | 7.1 ± 0.1 | 0.030, 0.017 | 2.7 ± 0.3 | 46 ± 2 | 0.21 ± 0.03 | 2,2 |
| D131A (RLC) | Asp-118 | 7.5 ± 0.3 | 0.035, 0.033 | 2.5 ± 0.3 | 37 ± 3 | 0.23 ± 0.00 | 2,2 |
| Q826A (HC) | Gln-812 | 7.5 ± 0.3 | 0.017, 0.011 | 2.5 ± 0.3 | 30 ± 6 | 0.25 ± 0.02 | 3,2 |
| Q826A/D131A (HC-RLC) | Gln-816/Asp-118 | 7.5 ± 0.3 | 0.032, 0.024 | 0.75 ± 0.2 | 13 ± 5 | 0.07 ± 0.02 | 2,2 |
| E13A (ELC) | Asp-13 | 7.5 ± 0.3 | 0.010, 0.026 | 1.2 ± 0.1 | 59 ± 22 | 0.11 ± 0.01 | 2,2 |
| Q816A (HC) | Gln-802 | 7.5 ± 0.3 | 0.024, 0.021 | 2.3 ± 0.2 | 71 ± 15 | 0.16 ± 0.04 | 2,3 |
| Q816A/E13A (HC-ELC) | Gln-802/Asp-13 | 7.5 ± 0.3 | 0.029, 0.029 | 0.97 ± 0.09 | 25 ± 8 | 0.05 ± 0.01 | 2,2 |
| R827M (HC) | Arg-813 | 7.5 ± 0.3 | 0.032, 0.034 | 2.5 ± 0.3 | 52 ± 4 | 0.15 ± 0.03 | 2,2 |
| N828A (HC) | Asn-814 | 7.5 ± 0.3 | 0.031, 0.039 | 2.1 ± 0.03 | 19 ± 4 | 0.22 ± 0.02 | 2,2 |
| L819A/M822A/Q826A (HC) | Arg-805/Leu-808/Gln-812 | ND | 0.011, 0.017 | 2.7 ± 0.1 | 48 ± 8 | 0.24 ± 0.03 | 2,2 |
a Data are based upon PDB code 1QVI (scallop). The HC of the smooth muscle myosin structure (PDB code 1BR1) is not solved beyond sm821, so the structural homologs of 822, 826, 827, and 828 are extrapolated and are therefore estimates.
b Steady-state NH4+-ATPase rates for the uP-state in the absence of actin are shown.
c Basal MgATPase rates for the P-state (absence of actin) are shown, but otherwise are measured under identical conditions for samples used to determine the Vmax (see Footnote d). Two independently determined values are shown. Values for the uP-state ranged between 0.010 and 0.030 for all constructs studied (data not shown).
d To determine Vmax and KATPase, P-HMM was assayed as described (22, 27) by actin-activated single-turnover of 2′-(3′)-O-(N-methylanthraniloyl)-ATP. See under “Experimental Procedures” for definitions of variables, fitting equations, number of determinations, and meaning of the indicated errors.
e Data are for the P-state. No motility was detected for any of the mutants in the uP-state (data not shown).
f NVmax and Nmot refer to the number of times that independent measurements of Vmax and motility were performed on independent samples prepared from different batches of cells.
g RLC homologs are based upon sequence alignment. ELC homologs for Glu-13 and Lys-25 are based upon structural alignment using smooth PDB code 1BR4. Arg-20 in smooth is in a loop that is two amino acids shorter than in scallop (21WDG23). Therefore, there is no unequivocal structural homolog.
TABLE 2.
Summary of other kinetic constants for unphosphorylated proteins
| Protein | K1k+2a | k+3 + k−3b | K+1k+2c | k−ADd |
|---|---|---|---|---|
| m−1 s−1 | s−1 | m−1 s−1 | s−1 | |
| WT HMM | 1.6 × 106 | 51 | 0.46 × 106 | 51 ± 2 |
| M129Q HMM | 1.9 × 106 | 48 | 48 ± 5 | |
| G130C HMM | 1.6 × 106 | 45 | 0.40 × 106 | 45 ± 6 |
| Native cHMMe | 1.0 × 106 | |||
| Native V8S1e | 2.1 × 106f | 50f | 0.47 × 106f | 22f |
| Chicken skeletal S1e | 5.6 × 106 |
a K1k+2 is the apparent second-order rate constant for ATP binding to myosin. See Cremo and Geeves (20) for kinetic schemes with the same nomenclature used here for all kinetic constants.
b k+3 + k−3 defines the maximal rate of ATP binding to myosin.
c K+1k+2 defines the apparent second-order rate constant for ATP binding to acto-myosin.
d k−AD is the rate of ADP release from the acto·myosin complex.
e Native HMM was prepared by chymotryptic digestion of chicken gizzard SMM as described (50). Gizzard SMM was prepared from frozen chicken gizzards (13) obtained from Pel-Freez Biologicals (Rogers, AR). Native V8S1 was prepared by digestion of SMM with Staphylococcus aureus protease as described (13). Chicken skeletal S1 was prepared by papain digestion (51) of fast skeletal muscle myosin purified from chicken pectoralis muscle (52).
f Values were taken from Ref. 20.
The actin-activated MgATPase was measured by the single turnover of 2′-(3′)-O-(N-methylanthraniloyl)-ATP as described previously (15, 16, 21). At least two transients were collected per actin concentration (0, 5, 10, 25, 50, 75, and 100 μm). The two observed rates resulting from a double-exponential fit to each transient were weighted by their respective amplitudes, and the weighted average for each transient was averaged (21). A plot of the actin concentration versus the average turnover rate was prepared at least twice from independently expressed and purified batches of protein. For P-HMM, each of the plots were analyzed separately and fit to the Michaelis-Menten equation, V = (Vmax)[actin])/(KATPase + [actin]) + C, where C is the rate in the absence of actin or basal MgATPase; Vmax is the maximal turnover rate, and KATPase is the concentration of actin at half-maximal activity corresponding to the dissociation constant of HMM for actin in the weakly bound state. The reported errors (Table 1) indicate ½ the range of the values obtained from the independent fits, and the number of determinations is listed. For the uP state, there was little difference in the turnover rates as the actin concentration was increased, and therefore the plots were not fit (data not shown). Table 1 shows the values at zero actin concentration (basal).
The rate of ADP release from the pyrene-actin·uP-HMM·ADP complex was measured by monitoring the increase in pyrene-actin fluorescence following rapid mixing of 0.6 μm HMM, 1 μm pyrene actin, and 100 μm MgADP with 2 mm ATP. At least four transients were averaged, and the data were fit to a single exponential equation (20).
The rate of association of HMM (0.75 μm heads) with ATP was measured by fitting the increase in tryptophan fluorescence to a single exponential (excitation 290 nm and emission was collected through a WG 320 filter (Schott)). The dependence of kobs on ATP concentration was plotted, and the slope of the initial points was fit to a line to determine K1k+2 as described (20). The maximum kobs obtained by fitting to a binding isotherm corresponds to the rate of ATP cleavage (k+3 + k−3) (20). All constructs showed similar amplitudes of increased tryptophan fluorescence as the WT.
The rigor binding of uP-HMM to actin (data not shown) was determined by mixing phalloidin-stabilized pyrene-actin (80 nm, before mixing) and various concentrations of uP-HMM with 80 μm ATP (before mixing), and the amplitude of the observed dissociation reaction was used to estimate the fraction of actin bound to HMM as described for native smHMM (22) using the method of Kurzawa and Geeves (23). Data were similar to our previous data for uP-HMM prepared from native myosin (22). Pyrene fluorescence was excited at 365 nm, and emission was observed through a KV 399 cutoff filter (Schott). At least four transients were averaged for each actin concentration, and the experiment was repeated twice with independent samples.
In Vitro Motility Assays
In vitro motility assays were performed as described in detail previously (15). The velocity of each actin filament was manually collected, and the mean velocity was calculated from the velocities of 20–30 filaments for each of at least two independently expressed and purified protein preparations. Reported values represent the average of all data. Phosphorylated native SMM served as a positive control.
Discrete Molecular Dynamics Simulations for Flexibility Analysis
Discrete molecular dynamics simulations (24) were performed on course grain models (one sphere per α-carbon) of native scallop myosin (PDB code 1QVI) and myosin with a modified Ca2+-binding loop of subdomain I of the ELC (1QVI with the native ELC residues 20–31 replaced with residues 20–29 of the smooth ELC; see Fig. 1B). Simulations were run at 300 K with the following set parameters: α-carbon, α-carbon pairwise cutoff of 8 Å, well amplitudes (Sigma cutoff) of 0.05 Å for consecutive atoms, and (Sigma Go cutoff) of 0.1 Å for nonconsecutive atoms. Data were downloaded from the FlexServ server and examined in Excel. The averages of the changes between one amino acid and all other amino acids outside a 5-Å cutoff were placed into the occupancy record of the PDB file and the colors were plotted using the programs InsightII and Visual Molecular Dynamics.
RESULTS
Fig. 1A (top panel) shows the structure of scallop myosin subfragment 1 or head domain (PDB code 1QVI) containing the motor domain and the lever arm or RD with associated subunits, the RLC and ELC. The bottom panel of Fig. 1A is an expanded view of the ELC-RLC-HC interfacing region showing the amino acid substitutions made in this study using the SMM sequence numbers. Fig. 1B shows the amino acid alignments and numbering of the regions of interest, comparing the chicken SMM to the scallop myosin sequences. All sequence numbers listed in the text represent the SMM numbers as indicated by the prefix “sm” versus “sc” for scallop. Table 1 lists all the mutants prepared for this study. Because the Ca2+-binding loop of domain I of the ELC in the scallop sequence has little homology to the smooth sequence, homology modeling was not an effective approach to predict specific interactions due to uncertainties in sequence alignment. For example, an important residue of the ELC, scR24 (see below), identified from the scallop structures does not exist in the SMM ELC sequence. Nevertheless, we were able to make reasonable judgments for mutant selection based upon the scallop structure.
RLC of All Constructs Can Be Fully Phosphorylated by MLCK
Double-headed HMM constructs were expressed in Sf9 cells by co-infection with three recombinant baculoviruses to express residues 1–1106 of the HC with a C-terminal His tag, the ELC, and the RLC, as we have previously described (15). All HMM constructs were soluble and were expressed at similar levels to the WT construct. To properly interpret the effects of the mutations on functional parameters, we confirmed that all HMM constructs could be completely phosphorylated by MLCK. Fig. 2A shows images of Coomassie-stained urea gels of the constructs before (Up) and after (P) phosphorylation. Note that the RLC subunit of all constructs shifts to the higher mobility mono-phosphorylated RLC (P-RLC) position after treatment with MLCK, indicating complete phosphorylation. Under these conditions only Ser-19 is phosphorylated (15).
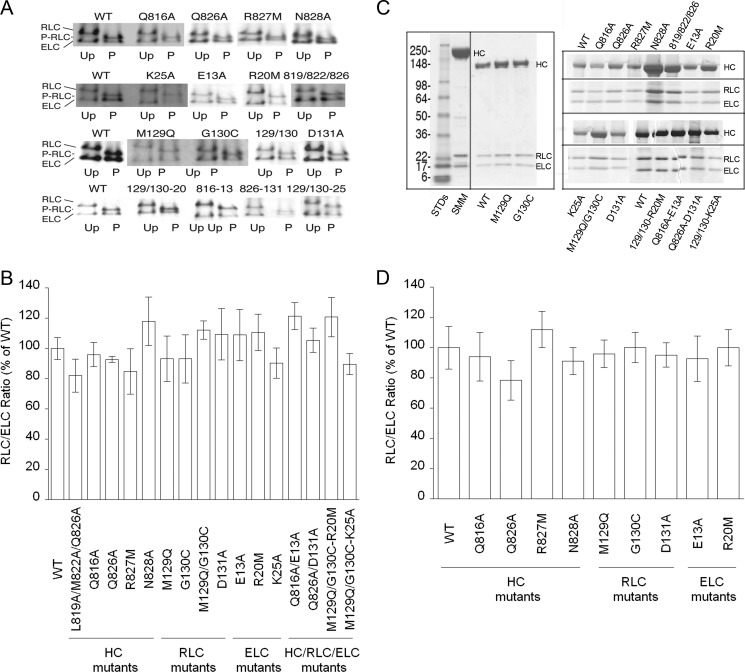
FIGURE 2.
Functional and structural properties of HMM constructs. A, images of representative Coomassie-stained urea gels comparing the mobility of the RLC and ELC before (uP) and after (P) phosphorylation by MLCK. The mutations are indicated above the paired (uP and P) gel images. Row 1, HC mutants; row 2, ELC mutants except for triple HC mutant; row 3, RLC mutants; row 4, double and triple mutants. Note that the RLC increases mobility after phosphorylation (P-RLC) as expected due to an increase in negative charge. B, urea gel images were scanned, and the density of the RLC/ELC was calculated and expressed as % of WT. Error bars are the ½ the range of two measurements on independent samples. C, representative images of Coomassie-stained SDS gels. Left panel, full-length of gel showing three samples compared with native full-length SMM. Right panel, two representative gels showing only HC and light chain regions, with mutant identities shown above and below the respective gels. D, similar to B, except data are from SDS gels. All gels were 10–20% polyacrylamide gradients from GE Healthcare. Urea gels were performed as described (15).
Gel Analysis Shows That the Constructs Have a Normal Subunit Composition
Previous studies in scallop myosin have shown that mutations in the ELC/RLC region that we are interested in can lead to changes in subunit composition (9) by significantly weakening the binding of one of the light chains. To determine whether or not our constructs had a normal subunit composition, we quantified the RLC/ELC ratio from images of stained gels. It is unlikely that any specific mutation(s) will precisely alter both light chain compositions simultaneously. The RLC/ELC is more accurate than the HC/light chain ratio, because the staining for the HC is so much heavier than the light chains, which become barely detectable under conditions where the HC is not oversaturated on the images. Fig. 2B shows the RLC/ELC ratio determined from the urea gels in Fig. 2A, indicating no significant difference from the WT construct. Fig. 2C shows images of representative SDS gels of the constructs. In Fig. 2C, left panel, two critical constructs, M129Q and G130C, are compared with WT HMM and full-length native SMM. In Fig. 2C, right panel, other constructs are compared with the WT, showing only the HC, RLC, and ELC regions of the gels. There were no apparent differences in the HC/light chain ratios upon visual inspection. Fig. 2D shows the results of quantifying the RLC/ELC ratio from the SDS gels in Fig. 2C, along with other similar images (data not shown). The results showed that there were no significant differences in the RLC/ELC ratios relative to the WT. The data in Fig. 2 suggest that all the constructs have a normal subunit composition and that none of the mutations significantly altered the binding of the light chains to the HCs. This allows us to interpret changes in the kinetic and mechanical behaviors of the mutants as bona fide effects of the mutated residues rather than changes in subunit composition.
RLC/ELC Interface and Surrounding HC Amino Acids Are Not Critical for Maintaining the Off-state for uP-HMM
To evaluate the effects of the mutations on phosphorylation-dependent regulation, we measured the actin-activated single-turnover rate of 2′-(3′)-O-(N-methylanthraniloyl)-ATP in the uP state as a function of actin concentration in addition to the in vitro actin sliding velocity (Table 1). The basal MgATPase rates (in the absence of actin) for all mutants in the uP state were low and essentially the same as the WT (0.01–0.02 s−1). The effect of actin on this rate was minimal for all samples, and the fastest rate that we observed at 100 μm actin was 0.035 s−1 (data not shown). This shows that none of the mutations altered the ability of the protein to adopt the Off-state when unphosphorylated. As expected, no moving filaments could be detected in the in vitro motility analysis of any of the uP constructs. Because the tested mutations were designed to disrupt the RLC/ELC interface, these data suggest that the interface between the RLC/ELC and the nearby HC residues are not critical for maintaining the off-state.
Changes in the RLC/ELC Interface Do Not Affect the Rate-limiting Step of the ATPase for P-HMM in the Absence of Actin
Table 1 shows the basal ATPase activities (absence of actin) for all the constructs in the P-state. Note that all values are low and very similar to the WT. This means that the kinetics of the rate-limiting step in the absence of actin is not appreciably affected. This step is thought to be the reverse recovery step (called the power-stroke if myosin was bound to actin) in which the lever arm transitions from an “up” to “down” position (25). In this process, the 50-kDa cleft in the motor domain goes from an open to closed state, although switch I and II proceed from a closed to an open state. The changes in the switch I and II conformations ultimately drive changes in the relay helix and the SH1 helix to rotate the converter domain and thus the lever arm (25, 26), followed by Pi release.
RLC/ELC Interface and Surrounding HC Amino Acids Are Critical for Maintaining the On-state Characteristics of P-HMM
To further characterize the P-state, we calculated Vmax and KATPase by measuring ATPase activity at actin concentrations ranging from 0 to 100 μm and fitting the data to a rectangular hyperbola in a manner similar to our previously published work (15). Also, we measured the velocity of myosin-based actin movement (V) observed in an in vitro motility assay and other kinetic constants. Table 1 summarizes the results for all the mutants in the phosphorylated state, Table 2 shows other kinetic parameters for selected mutants, and Fig. 3 shows a plot of the relationship between the Vmax of the ATPase and the speed of sliding of actin filaments in the in vitro motility assay (V) to visually emphasize the relationships in the data.
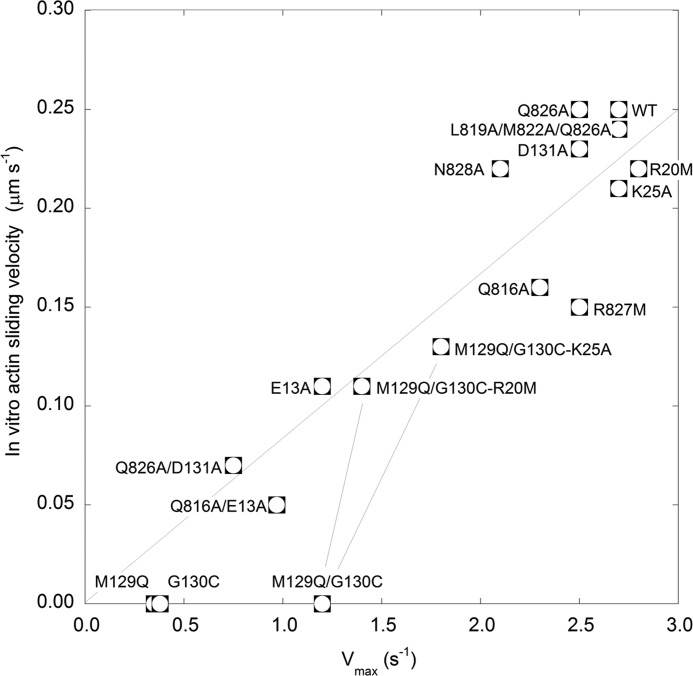
FIGURE 3.
Relationship between actin sliding velocity and Vmax for P-HMM constructs. Actin-sliding velocities and Vmax for P-HMM constructs are plotted with a line drawn between zero and the value for the WT construct. This line is not a fit to the data. The lines between the mutants M129Q/G130C and M129Q/G130C/R20M and M129Q/G130C/K25A merely highlight the partially recovered nature of the triple mutants with respect to both Vmax and motility. Data are from Table 1.
Residues in the RLC That Interact with Loop I of the ELC Are Critical for Activation of the Acto-myosin ATPase and Motility in P-HMM
We characterized the P-state of four constructs with mutations in the linker between the F and G helices of the RLC, M129Q, G130C, M129Q/G130C, and D131A. Table 3 shows that these residues are part of a conserved region of the RLC, but the Gly residue is specific to the regulated myosin RLCs. In 1QVI, these three residues participate in specific interactions with the ELC and nearby HC and therefore may be important for maintaining the On-state, because 1QVI is a putative On-state structure with Ca2+ bound in the ELC loop. The Asp-131 mutant was found to be similar to WT suggesting that it is not required for activation, even though it hydrogen bonds to two HC residues, smQ826 and smR830, in addition to making several interactions with scR24 (residue not present in the smooth sequence). The single mutants, smM129Q and smG130C were chosen because the QC sequence is present in the skeletal muscle RLC isoforms, and these muscles are not similarly regulated by phosphorylation (Table 3). These two mutants even when phosphorylated showed very low Vmax (∼13% of P-WT) and no motility (Table 1; Fig. 3). The double mutant smM129Q/G130C showed a partially recovered ATPase with a Vmax less than ½ of the WT, but the motility remained undetectable. In contrast, the NH4+-ATPase for these mutants was essentially the same as the WT (Table 1), which had the same activity as measured previously for porcine aortic full-length tissue-purified myosin (27). This activity is a measure of the intrinsic ATPase, which would be expected to be low if the mutation caused a structural disturbance around the ATPase catalytic center due to mis-folding for example. Therefore, lowered Vmax and motility are most likely due to local changes at or near the region of the mutation(s). All three mutations in this RLC region caused a modest decrease in the KATPase, which reflects the affinity for actin in the weakly bound state (presence of ATP).
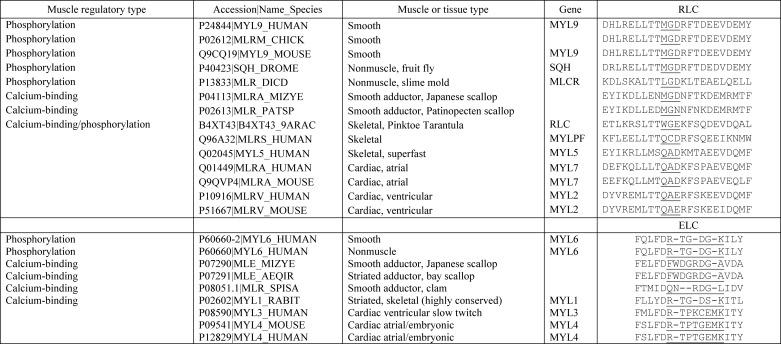
TABLE 3.
Sequence alignment of the RLC-ELC interacting loops for regulated and other myosin II isoforms from several speciesa
a Sequences were aligned using COBALT (NCBI), with minor adjustments to account for structural data, as described in Fig. 1. For each light chain, the loop sequences are underlined.
We also measured the ability of the two single mutants, smM129Q and smG130C, to bind to actin in the absence of ATP (data not shown) by measuring the amplitude of the change in pyrene-actin fluorescence as the acto·HMM complex is dissociated by ATP (22, 23). Both mutants behaved similarly to WT, suggesting that the mutations do not affect the affinity of HMM to actin in rigor. Several other kinetic constants were measured for smM129Q and smG130C (Table 2; see Cremo and Geeves (20) for all kinetic schemes). The apparent second-order rate constant for ATP binding to HMM (K1k+2), the maximal rate of ATP binding to HMM (k+3 + k−3), and the apparent second-order rate constant for ATP binding to acto-HMM (K+1k+2) were all similar to WT and native HMM (purified from chicken gizzard tissue). The first three constants are measures of the interactions of the HMMs or acto-HMMs with ATP, which were all normal. Importantly, for the measurements of K1k+2, ATP binding increased the intrinsic tryptophan fluorescence to the normal extent for all constructs tested. It is generally assumed that this increase of fluorescence intensity mainly reflects the converter swing during the recovery step from the post-stroke or down orientation to the pre-stroke or up orientation (25).
The rate of ADP release from the acto·HMM complex (k−AD) is an important rate constant because is thought to limit the rate of movement of actin in the motility assay under the conditions used here (mm MgATP (28–30)). Velocity in the motility assay then closely corresponds to an unloaded shortening velocity in muscle (31). Although motility was zero, k−AD was similar to the WT value for both M129Q and G130C (Table 2). Using a detachment-limited model, V = d/Ton, where V is velocity; d is the working stroke distance; and Ton is the time in the ATPase cycle where cross-bridges are attached to actin. At high ATP, Ton is inversely proportional to k−AD (28). This suggests that the observed actin sliding velocities for these mutants are slowed by some other factor. It is possible that the mutations result in a smaller d (see below), but this has not been directly measured here. Noncross-bridge-dependent mechanical factors may become experimentally important at slow velocities (32) and therefore cannot be ruled out to explain the lack of moving filaments. For example, if the number of heads binding to actin is below a saturating value, maximal velocity will not be reached. We tested for this by applying the HMM to the coverslip surface at higher concentrations, but this did not recover a detectable motility. Alternatively, interactions of actin with the surface or with a small number of residual “dead heads” (even though we used actin standard selection protocols) could impose a sufficient load to HMMs on the surface to further slow any load-dependent transitions of the active heads.
The main effects of the mutations at the RLC/ELC interface in the P-state were to significantly lower the Vmax of the acto-HMM MgATPase activity and to slow the velocity of actin sliding in the motility assay to undetectable levels. In the 1QVI scallop structure, smG130 makes a backbone-backbone H-bond with smR20 in the ELC Ca2+-binding loop, in addition to smG22 on the ELC. smM129 has no obvious interactions in 1QVI. Our data show that both RLC residues, Met-129 and Gly-130, at the interface between the RLC and ELC are important for triggering actin-induced phosphate release and the strong binding to actin, which are required to maintain the on-state of SMM. Therefore, it appears that the major functional defect occurs at a step in which acto-HMM progresses through conformational changes that contribute to the weak to strong binding transition. Other steps in the cycle, including ATP interactions with HMM and acto-HMM, the rate of ADP release from the acto·HMM complex, and rigor binding of actin to HMM, were not appreciably affected.
The above data suggest that specific RLC-ELC interactions are required to adopt the normal fully activated HMM molecules with respect to both enzymatic and mechanical activity. To test this directly, we attempted to disrupt these interactions by also mutating two ELC residues smR20M and smK25A that are part of the ELC Ca2+-binding loop in hopes of finding recovery-phenotype mutants from the M129Q/G130C background. In 1QVI, smR20M is within the ELC loop (Fig. 1B, underlined) and makes a backbone-backbone interaction with smG130C of the RLC. Therefore, we predicted that smR20M would alone have a normal phenotype but might allow for changes in the RLC/ELC interface that could recover the activity of the smM129Q/G130C mutant. Similarly, we predicted a similar effect for smK25A because it makes only a backbone interaction with scD23, which is smT21. Neither smR20M nor smK25A directly interact with the HC, so as predicted we did not observe any changes in binding affinity to the HC (Fig. 2). These two ELC residues when mutated alone showed a relatively normal phenotype (Table 1; Fig. 3). Interestingly, the two triple mutants, M129Q/G130C/R20M and M129Q/G130C/K25A, had a “recovery” phenotype, where both the Vmax and the motility were restored to much higher levels (Fig. 3). The KATPase also increased toward the WT level. These data strengthen the argument that the RLC-ELC interaction in this region is important for normal Vmax and motility in P-HMM. It cannot be ruled out that the recovery phenotypes are due to repairing other interactions such as RLC-HC interactions in the RLC M129Q/G130C mutant (see below). The specific nature of these interactions will be revealed in future mutagenesis studies as appropriate structural data become available.
Importance of RLC-HC Interactions to Maintaining the Activated State
It has been suggested that HC interactions with the RLC linker containing smM129 and smG130 are important to maintain the linker's structure in scallop myosin (5). The side chain amide nitrogen of smQ826 in the first part of the IQ motif of the HC makes a hydrogen bond with the backbone NH of the donor smD131 on the RLC. The D131A mutation had no effect alone, suggesting that a change in side chain alone is not sufficient to disrupt this interaction. However, the smQ826A mutation alone also had no effect, which would not be expected if this is an important interaction. However, our rotamer analysis of the D131 side chain suggests that it could become the acceptor for the NH backbone of Q826A, thus making a new non-native interaction that might stabilize the RLC-HC interaction sufficiently. This would explain the normal phenotype of smQ826A. To test for this, we made the double mutant smQ826A/D131A, which showed significantly reduced ATPase and motility. In this mutant, the above-mentioned non-native interaction would not be possible. These data suggest that RLC-HC interactions contribute to the structural requirements for a fully activated HMM in the phosphorylated state.
Effect of HC and ELC Mutations Near the Elbow of the HC
To test the idea that the RLC/ELC interface may be important to controlling the bend in the elbow of the HC (Fig. 1A), we mutated residues close to the elbow that do not directly participate in the RLC/ELC interface, ELC-HC, or RLC-HC interactions in 1QVI (Fig. 1). The side chain of smQ816 faces toward the ELC, and the effects of mutation to an Ala were modest, but both the Vmax and the motility were lower than WT. Interestingly, mutation of the closest ELC residue to smQ816, smE13A, had a more obvious inhibitory effect. This could be simply because it is an H-donor for the backbone carbonyl of smL17 within the same helix that is connected to the Ca2+ binding loop of the ELC. The double mutant smQ816A/E13A was even more inhibited with respect to both motility and Vmax. These data suggest that mutations in the elbow region alone are sufficient to give a similar phenotype as the RLC/ELC interface mutations. The elbow may need to be in a specific conformation to allow for a stable RLC/ELC interface. Or it may be that the need for an RLC/ELC interface is to stabilize the proper elbow conformation for activation. More work is needed to reveal more specific mechanistic information.
Lack of an Effect of Other HC Mutants Not Interacting with the RLC-ELC Interfacial Region
The N-terminal part of the IQ motif for the RLC is defined by residues sm825–828 (825IQRNCAAYLKL) roughly following the consensus sequence IQXXXRGXXXR. Above, we have described the importance of smQ826, which has a direct interaction with RLC Asp-131. Because the side chains of smR827 and smN828 face away from the RLC and they are far from the elbow region, we predicted that these residues could be mutated without large changes in the RLC/ELC interface. Table 1 shows that smN828A was normal. smR827A showed a normal Vmax but unexpectedly low motility. This is perhaps due to its proximity to smD97 on the RLC (scD84) with which it forms a salt bridge in the 1QVI structure. Interestingly, at the time we selected this mutant, we did not anticipate the importance of the smD97 interaction with smK163 in the H helix. smK163 is conserved only in regulated myosins, and the H helix is known to be critical to regulation in SMM (3). We propose that smR827-Asp-97-Lys-163 interactions could be altered in the R827A mutant, biasing the structure toward the inhibited state. This would be consistent with the finding that the equivalent residue in scallop myosin, scK149, forms a salt bridge with scT83 in the Ca2+ -in structure, but the interaction is broken in the Ca2+-out structure (10). We also prepared the triple mutant smL819A/M822A/Q826A. The nonconserved Leu-819 and Met-822 do not appear to make specific interactions with the RLC/ELC interface, and as expected, this alanine triple mutant had a normal phenotype. These data provide further support to the idea that the RLC/ELC interfacial area has a specific critical function with regard to the kinetic and mechanical activation of SMM in the phosphorylated state.
Computational Flexibility Analysis, Possible Structural Mechanism for Effects of Altered RLC/ELC Interface
Based upon structural considerations, Houdusse and Cohen (5) proposed a model in which an initial flexibility in the RD is generated upon the loss of Ca2+ from the ELC of the Ca2+-regulated myosins, and this may allow for further interactions to stabilize the Off-state in the context of the whole myosin molecule, perhaps by motor domain-motor domain or head-tail interactions predicted from cryo-EM data (33, 34). The idea that Ca2+ binding to the ELC stiffens the RD is supported directly by experimental data showing that the Ca2+-free complex of the isolated RD of scallop myosin is more flexible than the Ca2+-bound complex by two measures. The Ca2+-free complex showed greater mobility of the protein structure around tryptophan residues and a smaller Stokes radius (an apparently less asymmetric structure) presumably due to increased flexibility (35). With regard to smooth muscle, it has been shown that rigor stiffness is significantly increased by phosphorylation of the smooth muscle RLC in studies of α-toxin-permeabilized smooth muscle (36). Although the region of change in stiffness on the myosin molecule was not identified, it was suggested that phosphorylation may stiffen the lever arm.
Our interest was to try to verify and further understand the specific structural consequences of the increased flexibility of the RD upon loss of Ca2+ binding and to further relate this to our results. We used discrete molecular dynamics simulations to compare the flexibility of two S1 structures, one in which the native RLC-ELC interactions were intact (PDB code 1QVI), and one in which we expected them to be broken or no longer functional (modified 1QVI) based upon direct experimental evidence of function. We used S1 structural coordinates here instead of isolated RD coordinates so that the RD interactions with the converter and motor domain remained intact. Our computational comparison experiment was guided by direct experimental data showing that the source of the ELC must be from a Ca2+-binding (i.e. molluscan) myosin for scallop myosin to bind Ca2+ (9, 37, 38). Therefore, we compared the flexibility of the native scallop structure (1QVI) to the flexibility of the native structure in which the ELC Ca2+-binding loop from the smELC (not able to bind Ca2+) was inserted by model building (modified 1QVI). This modified 1QVI structure represents a molecule that can no longer bind to Ca2+ and therefore may approximate some attributes of the apo-Ca2+ structural cousin of 1QVI. Note that the experiment could not be performed with coordinates from SMM because they are not available. Fig. 4 depicts the results of the computational flexibility analysis where the colors represent the difference in flexibility between the two structures. In Fig. 4, dark blue indicates regions where the modified 1QVI structure is less flexible than 1QVI and red regions are where the modified 1QVI structure is more flexible. Interestingly, a notable aspect of the difference map is that much of the ELC, the HC to which the ELC binds, and the RLC/ELC interfacial region, were found to be more flexible (Fig. 4, red) after the in silico modification of the ELC Ca2+-binding loop. With the exception of a region of the upper 50-kDa domain (Fig. 4, top left), the remainder of the molecule either did not change in flexibility or became less flexible. The observed increase in flexibility of the RLC/ELC interface is consistent with temperature factors of the scallop RD (without the motor domain) in the absence of Ca2+ (10). This result highlights the potential importance of the ELC/RLC interface and interacting HC in maintenance of RD flexibility.
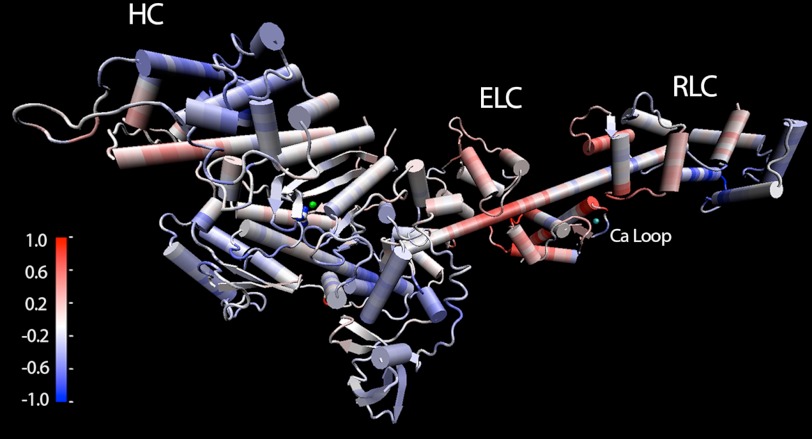
FIGURE 4.
Differences in flexibility between 1QVI and 1QVI in which the ELC Ca2+-binding loop from the smELC was inserted by model building (modified 1QVI). The scale plotted in kcal mol−1 Å−2 denotes flexibility differences. Red color shows areas in which the modified 1QVI is more flexible than the unmodified 1QVI, and blue color shows areas in which the modified 1QVI is less flexible than the unmodified 1QVI. The green Mg2+ and blue vanadate ions are visible at the active site in the motor domain, and the Ca2+ ion in the Ca2+-binding loop of the ELC is in cyan. Note the prominent increase in flexibility at the ELC/RLC interface and the region of the HC to which the ELC binds. The catalytic site remains largely unchanged.
DISCUSSION
We have expressed 16 smHMM constructs in which the interface between the two light chains and the surrounding HC has been modified by mutagenesis. Without exception, the mutant constructs behaved similarly to WT by the following measures. 1) The RLC was fully phosphorylated by MLCK. 2) The subunit composition was normal, suggesting that the mutations did not drastically weaken the binding of either the ELC or the RLC to the HC. 3) The kinetics of the basal and actin-activated ATPase and the mechanical behavior in an in vitro motility assay in the uP state were essentially the same as the fully inhibited WT HMM. 4) The ATPase for P-HMM in the absence of actin was also fully inhibited.
Our most interesting finding is that for constructs in which we deliberately tried to disrupt the interaction between the RLC and ELC, phosphorylation no longer activated the ability of the HMMs to move actin in an in vitro motility assay and also the Vmax of the actin-activated ATPase was significantly diminished to ∼10% of the P-WT construct. This behavior was associated with the M129Q and G130C mutations of the RLC, but D131A was normal. Therefore, M129Q and G130C are important for stabilizing structural interactions that are required for activation of function in the presence of actin. In an effort to further disrupt native interactions, we prepared the double mutant M129Q/G130C. Surprisingly, we found that this mutant had a higher Vmax than either of the single mutants. However, the function of the double mutant was still significantly inhibited because the motility remained undetectable.
Our hypothesis was that an intra-head interaction between the RLC and ELC is important for full activation of the molecule. To test for this, we sought ELC residues near the interface that are part of the ELC Ca2+-binding loop. smR20M and smK25A define the N- and C-terminal ends of the ELC loop but when mutated alone did not cause a loss of function. In contrast, smR20M and smK25A in the M129Q/G130C background showed a recovery phenotype in which both the actin-activated ATPase and the motility were partially activated upon phosphorylation. These data suggest that the interaction between the RLC and the ELC in this region is important to allow for activation upon RLC phosphorylation. It is informative to compare our results to previous work on scallop myosin (9). They showed that the structural analog of smM129Q (scM116Q) had a normal phenotype in contrast to our study. Scallop myosin containing the structural analog of smG130C (scG117C) was not activated by Ca2+, suggesting its phenotype was similar to what we found for SMM. However, the mutant also bound very poorly to Ca2+ and bound poorly to the HC making conclusions about its specific role difficult. A similar experiment, but in the background of the skeletal RLC, showed that the scG117C mutation marginally restored Ca2+-dependent regulation along with HC and Ca2+ binding. Therefore, their study in scallop myosin and our study in SMM both point to an important role for this highly conserved Gly residue. Interestingly, this Gly residue appears to be conserved only in the RLCs of regulated myosins (molluscan, smooth and nonmuscle) and is absent in skeletal and cardiac RLCs (Table 3).
Also we found that the HC mutant Q826A, which interacts with Asp-131, also may stabilize the RLC/ELC interface, and mutations that could affect the conformation of the elbow of the RD also showed an inhibited phenotype. Other mutations of HC residues in this area that were predicted not to stabilize the RLC/ELC interfacial region were found to be normal suggesting that the effects of the M129Q and G130C mutations were specific.
Our data suggest that the reduced level of actin activation of the ATPase observed for our RLC/ELC/HC interface mutants is primarily due to a decreased Vmax rather than a Km effect (Table 1). Thus, this reduction probably results from a rate change in a step that occurs after actin is bound. This is most likely due to a decreased rate of phosphate release from the acto·myosin complex or a rate-limiting step preceding it corresponding to the weak to strong actin binding transition (2, 39, 40). Because we have no evidence that, relative to HMM, these mutated HMMs bind more weakly to actin in the presence of ATP, we assume that the flux through the actin-bound states versus the actin-unbound states in the cycle is normal. It is more likely that modifying the RLC/ELC interface is affecting directly the ability of actin to accelerate the lever arm swing associated with the transition from the weak actin-binding to a strong actin-binding state.
To extend our findings, we designed a computational experiment to reveal a structural mechanism to explain the phenotype of the RLC/ELC interface-disrupted mutants. Why do these mutants have such a low actin-activated MgATPase but a normal low basal MgATPase (no actin)? Our computational flexibility analysis compared the putative activated state of scallop myosin (with bound Ca2+) to a computationally modified inactive state (lacking Ca2+) in which the RLC/ELC interface was broken. The modified structure showed increased flexibility specifically at the RLC/ELC interface and at the ELC-binding region of the HC (Fig. 4).
The observed low basal ATPase rates of the RLC/ELC-modified mutants show that the RD is not prevented from going from the down to the up position in the absence of actin, a motion that is thought to accompany the recovery step (41). This explains the slow Pi release rates (low ATPase) in the absence of actin, because the full converter rotation driving the RD motion during the recovery step is coupled to closure of the “back door” or “trap door” for Pi release from the active site (42). Therefore, increased RD flexibility does not hamper the ability of the heads to adopt the post-recovery conformation. Low resolution structures of the smHMM by cryo-EM show that the two motor domains of a single molecule interact in an asymmetric manner in the unphosphorylated state (33, 34). The so-called blocked head interacts with the actin-binding domain of the free head, and the interaction appears to require full converter rotation (42) from the down to up position. Therefore, we conclude that the RLC-ELC-HC interactions probed here are not important for stabilizing the normal inhibited structure characteristic of uP-HMM.
Our recent EPR results in collaboration with the Fajer laboratory show that there is a distribution of structures corresponding to the cryo-EM unphosphorylated inhibited structure (motor domains interacting intramolecularly) (33, 34) co-existing in solution with a distribution of open-head (motor domains not interacting) structures, even in an uP-HMM or uP-myosin sample (43). Phosphorylation strongly stabilizes the open-head structure at the expense of the closed-head structural distribution. The flexibility result and the actin-activated ATPase data presented here can be reconciled by the notion that increased flexibility in the lever arm simply allows this pre-existing rapid equilibrium between the two distributions to be biased strongly toward the inhibited head-head interacting distribution even when the RLC is phosphorylated. This implies that increased flexibility at the RLC/ELC interface promotes the conformation in which the converter of the free head is fully accessible to the actin-binding domain of the blocked head. The inhibition of the actin-activated ATPase in the RLC/ELC interface mutants must be due to the hampered ability of the lever to go down, which is normally promoted by actin binding. Normally, phosphorylation would increase the rate of the “up to down” process, but this is hampered in the RLC/ELC interface-disrupted mutants.
Our results are further informed by recent cryo-EM data on P-HMM at 20 Å resolution (PDB code 3J04 (44)). The intramolecular interactions between the two motor domains seen in the uP state are replaced by similar intermolecular interactions in P-HMM in which the heads are splayed apart. The authors note a large change in the angle of a hinging region in the HC between the RLC and ELC, what we call the “elbow,” from a more bent in the uP state to a less bent conformation in the P-state, for both the blocked and free heads. It is possible that this reflects an important conformational change in the RLC/ELC interface driven by or driving these HC changes. This is supported directly by our data showing that mutation of residues very close to the elbow on both the HC (Q816A) and the ELC (E13A) hamper the ability of the molecule to be activated by phosphorylation (Table 1), although neither of these residues participates directly in the RLC/ELC interface.
Our data suggest that a major structural function of RLC phosphorylation is to stabilize the interface between the RLC and ELC and the associated conformation of the HC elbow. If the interface is already broken, or is incapable of forming, as in our RLC loop mutants, then phosphorylation cannot as effectively induce the required conformational changes to prevent or bias against the inhibited conformational distributions.
Although the chemical effects (ATPase) and mechanical effects (V) need not share the same mechanism, the loss of mechanical function of the most severe mutants of smHMM, M129Q, G130C, and M129Q/G130C, may be partly due to an increased flexibility of the RD. An increase in the flexibility in the ELC/HC and RLC/ELC interface might reduce lever arm stability, and thus reduce the ability to support productive coupling of ATP hydrolysis to mechanical force production. This would likely require that alterations in the ELC further transmit to the relay helix/loop and the SH1-SH2 helix. Also, a more flexible lever arm could lead to a shorter working stroke, but we have no direct evidence for this as yet. It is known that the working stroke distance (d) is a mechanical property that depends upon the effective length of the RD (28), which may amplify small conformational changes arising in the motor domain by acting as a swinging lever arm (45–48). Because the mutations studied here do not appreciably change the length of the lever arm, a smaller d could arise from increasing the flexibility of the lever arm region. There is precedence for this interpretation in a study of a single mutation in the RLC (F102L) of skeletal myosin in which the step size was diminished, without changing the ATPase rate or the step duration (49). F102L does not directly interact with the RLC/ELC interface but is buried and facing toward the HC, potentially making important hydrophobic interactions with the equivalent of smI825, which is in the region of the HC that we are probing.
Acknowledgments
We thank Paul D. Brewer, Olivia Hall, Mike Carter, and Travis Stewart for technical assistance and other members of the Cremo and Baker laboratories for discussions. We especially thank Dr. Andrew Szent-Györgyi. His excellent work motivated and enabled this study.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AR040917 from NIAMS (to C. R. C. and K. C. F.).
- ELC
- essential light chain
- RLC
- regulatory light chain
- HMM
- heavy meromyosin
- uP
- unphosphorylated
- P
- phosphorylated or thiophosphorylated
- SMM
- smooth muscle myosin
- HC
- heavy chain of myosin
- S1
- subfragment 1 of myosin includes the motor and regulatory domains
- RD
- regulatory domain includes the ELC, RLC and the HC to which they bind
- MLCK
- myosin light chain kinase
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- PDB
- Protein Data Bank
- sm
- smooth muscle
- sc
- scallop muscle.
REFERENCES
- 1. Holmes K. C., Geeves M. A. (2000) The structural basis of muscle contraction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sellers J. R. (1985) Mechanism of the phosphorylation-dependent regulation of smooth muscle heavy meromyosin. J. Biol. Chem. 260, 15815–15819 [PubMed] [Google Scholar]
- 3. Trybus K. M., Waller G. S., Chatman T. A. (1994) Coupling of ATPase activity and motility in smooth muscle myosin is mediated by the regulatory light chain. J. Cell Biol. 124, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie X., Harrison D. H., Schlichting I., Sweet R. M., Kalabokis V. N., Szent-Györgyi A. G., Cohen C. (1994) Structure of the regulatory domain of scallop myosin at 2.8 Å resolution. Nature 368, 306–312 [DOI] [PubMed] [Google Scholar]
- 5. Houdusse A., Cohen C. (1996) Structure of the regulatory domain of scallop myosin at 2 Å resolution. Implications for regulation. Structure 4, 21–32 [DOI] [PubMed] [Google Scholar]
- 6. Szent-Györgyi A. G. (2007) Regulation by myosin. How calcium regulates some myosins, past and present. Adv. Exp. Med. Biol. 592, 253–264 [DOI] [PubMed] [Google Scholar]
- 7. Szent-Györgyi A. G., Chantler P. D. (1994) in Myology (Engel A. G., Franzini-Armstrong C., eds) pp. 506–528, McGraw-Hill Inc., New York [Google Scholar]
- 8. Houdusse A., Silver M., Cohen C. (1996) A model of Ca2+-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure 4, 1475–1490 [DOI] [PubMed] [Google Scholar]
- 9. Jancso A., Szent-Györgyi A. G. (1994) Regulation of scallop myosin by the regulatory light chain depends on a single glycine residue. Proc. Natl. Acad. Sci. U.S.A. 91, 8762–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Himmel D. M., Mui S., O'Neall-Hennessey E., Szent-Györgyi A. G., Cohen C. (2009) The on-off switch in regulated myosins. Different triggers but related mechanisms. J. Mol. Biol. 394, 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katoh T., Morita F. (1996) Roles of light chains in the activity and conformation of smooth muscle myosin. J. Biol. Chem. 271, 9992–9996 [DOI] [PubMed] [Google Scholar]
- 12. Trybus K. M. (1994) Regulation of expressed truncated smooth muscle myosins. Role of the essential light chain and tail length. J. Biol. Chem. 269, 20819–20822 [PubMed] [Google Scholar]
- 13. Ikebe M., Hartshorne D. J. (1985) Proteolysis of smooth muscle myosin by Staphylococcus aureus protease. Preparation of heavy meromyosin and subfragment 1 with intact 20,000-dalton light chains. Biochemistry 24, 2380–2387 [DOI] [PubMed] [Google Scholar]
- 14. Gourinath S., Himmel D. M., Brown J. H., Reshetnikova L., Szent-Györgyi A. G., Cohen C. (2003) Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Å resolution. Flexibility and function in the head. Structure 11, 1621–1627 [DOI] [PubMed] [Google Scholar]
- 15. Ni S., Hong F., Brewer P. D., Ikebe M., Onishi H., Baker J. E., Facemyer K. C., Cremo C. R. (2009) Kinetic and motor functions mediated by distinct regions of the regulatory light chain of smooth muscle myosin. Biochim. Biophys. Acta 1794, 1599–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellison P. A., Sellers J. R., Cremo C. R. (2000) Kinetics of smooth muscle heavy meromyosin with one thiophosphorylated head. J. Biol. Chem. 275, 15142–15151 [DOI] [PubMed] [Google Scholar]
- 17. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 18. Criddle A. H., Geeves M. A., Jeffries T. (1985) The use of actin labeled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem. J. 232, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fiske C. H., SubbaRow Y. (1925) The colorimetric determination of phosphorus. J. Biol. Chem. 66, 375–400 [Google Scholar]
- 20. Cremo C. R., Geeves M. A. (1998) Interaction of actin and ADP with the head domain of smooth muscle myosin. Implications for strain-dependent ADP release in smooth muscle. Biochemistry 37, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 21. Cremo C. R., Wang F., Facemyer K., Sellers J. R. (2001) Phosphorylation-dependent regulation is absent in a non-muscle heavy meromyosin construct with one complete head and one head lacking the motor domain. J. Biol. Chem. 276, 41465–41472 [DOI] [PubMed] [Google Scholar]
- 22. Ellison P. A., DePew Z. S., Cremo C. R. (2003) Both heads of tissue-derived smooth muscle heavy meromyosin bind to actin in the presence of ADP. J. Biol. Chem. 278, 4410–4415 [DOI] [PubMed] [Google Scholar]
- 23. Kurzawa S. E., Geeves M. A. (1996) A novel stopped-flow method for measuring the affinity of actin for myosin head fragments using ug quantities of protein. J. Muscle Res. Cell Motil. 17, 669–676 [DOI] [PubMed] [Google Scholar]
- 24. Camps J., Carrillo O., Emperador A., Orellana L., Hospital A., Rueda M., Cicin-Sain D., D'Abramo M., Gelpí J. L., Orozco M. (2009) FlexServ. An integrated tool for the analysis of protein flexibility. Bioinformatics 25, 1709–1710 [DOI] [PubMed] [Google Scholar]
- 25. Gyimesi M., Kintses B., Bodor A., Perczel A., Fischer S., Bagshaw C. R., Málnási-Csizmadia A. (2008) The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J. Biol. Chem. 283, 8153–8163 [DOI] [PubMed] [Google Scholar]
- 26. Fischer S., Windshügel B., Horak D., Holmes K. C., Smith J. C. (2005) Structural mechanism of the recovery stroke in the myosin molecular motor. Proc. Natl. Acad. Sci. U.S.A. 102, 6873–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh T., Konishi K., Yazawa M. (2002) Essential light chain modulates phosphorylation-dependent regulation of smooth muscle myosin. J. Biochem. 131, 641–645 [DOI] [PubMed] [Google Scholar]
- 28. Tyska M. J., Warshaw D. M. (2002) The myosin power stroke. Cell Motil. Cytoskeleton 51, 1–15 [DOI] [PubMed] [Google Scholar]
- 29. Warshaw D. M., Desrosiers J. M., Work S. S., Trybus K. M. (1991) Effects of MgATP, MgADP, and Pi on actin movement by smooth muscle myosin. J. Biol. Chem. 266, 24339–24343 [PubMed] [Google Scholar]
- 30. Howard J. (2001) Mechanics of Motor Proteins and the Cytoskeleton, pp. 213–227, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 31. Siemankowski R. F., Wiseman M. O., White H. D. (1985) ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc. Natl. Acad. Sci. U.S.A. 82, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cuda G., Pate E., Cooke R., Sellers J. R. (1997) In vitro actin filament sliding velocities produced by mixtures of different types of myosin. Biophys. J. 72, 1767–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wendt T., Taylor D., Trybus K. M., Taylor K. (2001) Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. U.S.A. 98, 4361–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lowey S., Trybus K. M. (2010) Common structural motifs for the regulation of divergent class II myosins. J. Biol. Chem. 285, 16403–16407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Málnási-Csizmadia A., Hegyi G., Tolgyesi F., Szent-Györgyi A. G., Nyitray L. (1999) Fluorescence measurements detect changes in scallop myosin regulatory domain. Eur. J. Biochem. 261, 452–458 [DOI] [PubMed] [Google Scholar]
- 36. Khromov A. S., Somlyo A. V., Somlyo A. P. (1998) Thiophosphorylation of myosin light chain increases rigor stiffness of rabbit smooth muscle. J. Physiol. 512, 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwon H., Goodwin E. B., Nyitray L., Berliner E., O'Neall-Hennessey E., Melandri F. D., Szent-Györgyi A. G. (1990) Isolation of the regulatory domain of scallop myosin. Role of the essential light chain in calcium binding. Proc. Natl. Acad. Sci. U.S.A. 87, 4771–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kwon H., Melandri F. D., Szent-Györgyi A. G. (1992) Role of gizzard myosin light chains in calcium binding. J. Muscle Res. Cell Motil. 13, 315–320 [DOI] [PubMed] [Google Scholar]
- 39. Taylor E. W. (1979) Mechanism of actomyosin ATPase and the problem of muscle contraction. CRC Crit. Rev. Biochem. 6, 103–164 [DOI] [PubMed] [Google Scholar]
- 40. Geeves M. A. (1991) The dynamics of actin and myosin association and the cross-bridge model of muscle contraction. Biochem. J. 274, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mesentean S., Koppole S., Smith J. C., Fischer S. (2007) The principal motions involved in the coupling mechanism of the recovery stroke of the myosin motor. J. Mol. Biol. 367, 591–602 [DOI] [PubMed] [Google Scholar]
- 42. Onishi H., Nitanai Y. (2008) Thiol reactivity as a sensor of rotation of the converter in myosin. Biochem. Biophys. Res. Commun. 369, 115–123 [DOI] [PubMed] [Google Scholar]
- 43. Vileno B., Chamoun J., Liang H., Brewer P., Haldeman B. D., Facemyer K. C., Salzameda B., Song L., Li H. C., Cremo C. R., Fajer P. G. (2011) Broad disorder and the allosteric mechanism of myosin II regulation by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 108, 8218–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baumann B. A., Taylor D. W., Huang Z., Tama F., Fagnant P. M., Trybus K. M., Taylor K. A. (2012) Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to Activation. J. Mol. Biol. 415, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uyeda T. Q., Abramson P. D., Spudich J. A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S.A. 93, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Warshaw D. M., Guilford W. H., Freyzon Y., Krementsova E., Palmiter K. A., Tyska M. J., Baker J. E., Trybus K. M. (2000) The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J. Biol. Chem. 275, 37167–37172 [DOI] [PubMed] [Google Scholar]
- 47. VanBuren P., Waller G. S., Harris D. E., Trybus K. M., Warshaw D. M., Lowey S. (1994) The essential light chain is required for full force production by skeletal muscle myosin. Proc. Natl. Acad. Sci. U.S.A. 91, 12403–12407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruff C., Furch M., Brenner B., Manstein D. J., Meyhöfer E. (2001) Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat. Struct. Biol. 8, 226–229 [DOI] [PubMed] [Google Scholar]
- 49. Sherwood J. J., Waller G. S., Warshaw D. M., Lowey S. (2004) A point mutation in the regulatory light chain reduces the step size of skeletal muscle myosin. Proc. Natl. Acad. Sci. U.S.A. 101, 10973–10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seidel J. C. (1980) Fragmentation of gizzard myosin by α-chymotrypsin and papain, the effects on ATPase activity, and the interaction with actin. J. Biol. Chem. 255, 4355–4361 [PubMed] [Google Scholar]
- 51. Lowey S., Slayter H. S., Weeds A. G., Baker H. (1969) Substructure of the myosin molecule. I. Subfragments of myosin by enzymic degradation. J. Mol. Biol. 42, 1–29 [DOI] [PubMed] [Google Scholar]
- 52. Warshaw D. M., Desrosiers J. M., Work S. S., Trybus K. M. (1990) Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 111, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]