Background: Emerin regulates the expression of a large number of genes.
Results: Emerin binds HDAC3, mediates its nuclear envelope localization, and activates HDAC3 activity.
Conclusion: Decreased HDAC3 activity may contribute to changes in genomic organization seen in emerin-null cells.
Significance: These studies uncovered a putative mechanism for initiating and maintaining repressed genes at the nuclear periphery.
Keywords: Histone Deacetylase, Molecular Cell Biology, Muscular Dystrophy, Nuclear Organization, Nuclear Structure, Emery-Dreifuss Muscular Dystrophy, Emerin, Nuclear Co-repressor, Nuclear Envelope, Nucleoskeleton
Abstract
Organization of the genome is critical for maintaining cell-specific gene expression, ensuring proper cell function. It is well established that the nuclear lamina preferentially associates with repressed chromatin. However, the molecular mechanisms underlying repressive chromatin formation and maintenance at the nuclear lamina remain poorly understood. Here we show that emerin binds directly to HDAC3, the catalytic subunit of the nuclear co-repressor (NCoR) complex, and recruits HDAC3 to the nuclear periphery. Emerin binding stimulated the catalytic activity of HDAC3, and emerin-null cells exhibit increased H4K5 acetylation, which is the preferred target of the NCoR complex. Emerin-null cells exhibit an epigenetic signature similar to that seen in HDAC3-null cells. Emerin-null cells also had significantly less HDAC3 at the nuclear lamina. Collectively, these data support a model whereby emerin facilitates repressive chromatin formation at the nuclear periphery by increasing the catalytic activity of HDAC3.
Introduction
Chromatin architecture is regulated within the nucleus to ensure the coordinated expression of cell type-specific genes. Generally, expressed genes reside within regions of decondensed chromatin called euchromatin. Repressed genes tend to reside in regions of compacted chromatin called heterochromatin, which preferentially localizes to the nuclear periphery. Euchromatin localizes to the nuclear interior. The localization of repressed chromatin to the nuclear lamina predicts that the nuclear lamina may establish or maintain a repressive chromatin environment.
The nuclear envelope is composed of two lipid bilayers: the outer nuclear membrane, which is contiguous with the endoplasmic reticulum, and the inner nuclear membrane (1). The inner nuclear membrane contains >100 integral membrane proteins (2, 3). Emerin is an integral membrane protein of the inner nuclear membrane. Inner nuclear membrane localization of emerin is mediated by binding to A-type lamins, which are nuclear intermediate filament proteins that form a nuclear envelope-associated lattice (4). Together, emerin and lamin A form many stable oligomeric complexes with roles in mRNA transcription and DNA replication (5). The nuclear lamin-based lattice together with lamin-associated inner nuclear membrane proteins defines the nuclear lamina.
Mutations in the genes encoding emerin and lamin A produce diseases with a broad spectrum of both overlapping and distinct phenotypes, including Emery-Dreifuss muscular dystrophy (EDMD).3 These diseases, collectively called laminopathies, include progressive skeletal muscle wasting, life-threatening irregular heart rhythms, contractures of major tendons, abnormal fat deposition, and premature aging (6). Emerin and lamin A are expressed in nearly all differentiated cell types, yet these diseases specifically affect heart, muscle, tendons, and fat. To explain this tissue specificity, emerin and lamin A were variously proposed to have roles in gene expression, cell signaling, or nuclear structure (1, 5, 7).
Growing evidence supports roles for emerin in gene expression and genomic organization as emerin binds directly to many transcription regulators (5, 8–11). Further supporting the role of emerin in gene expression, muscles from EDMD patients and emerin-null mice show increased expression of muscle regeneration pathway components (12, 13). EDMD patient fibroblasts also have significantly reduced heterochromatin both in the nucleoplasm and at the nuclear periphery (14, 15). Emerin associates with chromatin-modifying complexes, including the nuclear co-repressor (NCoR) complex (16), providing a mechanism by which loss of emerin might be linked to decreased heterochromatin formation. Heterochromatin loss and increased gene expression in emerin-null cells suggests repressed chromatin formation and repression of gene expression requires emerin.
The NCoR complex is a histone deacetylase 3- (HDAC3) containing complex that localizes to gene regulatory regions and represses transcription of genes under their control. The NCoR complex is composed of TBL1/TBL1R, GPS2, NCoR, and HDAC3. NCoR has four repression domains and a nuclear receptor interaction domain that mediates binding to unliganded nuclear receptors on DNA (17). The histone-interacting domain (residues 610–690) in NCoR binds to unacetylated H4 N-terminal tails (18). Residues 420–488 of NCoR bind HDAC3, and this domain is called the deacetylase activation domain (DAD); DAD binding to HDAC3 is required for activation of HDAC3 (19). G-protein pathway suppressor 2 (GPS2) binds to NCoR residues 161–225, and TBL1/TBL1R binds NCoR residues 226–312 (19). The N terminus of TBL1/TBL1R binds to G-protein pathway suppressor 2 (GPS2) and stabilizes its interaction with NCoR. TBL1 preferentially binds hypoacetylated H4. Thus, deacetylation of H4 tails by HDAC3 is hypothesized to induce H4 binding by TBL1 and the NCoR histone-interacting domain (HID) and to stabilize NCoR complex binding to chromatin (20).
Recently, genomic regions targeted to the nuclear lamina were repressed, and repression spread from 100 kb to 5 Mbp (21, 22). To examine the features of genomic regions associated with the nuclear lamina, bacterial DNA adenine methyltransferase (DAM) was fused to lamin B1 (DAM-LMNB1) and expressed in fibroblasts (23). Approximately 1,300 genomic regions interacted with DAM-LMNB1. These lamina-associated domains (LADs) were predominantly gene-poor and contained chromatin modifications consistent with transcription repression and chromatin compaction. Similar results were also obtained using DAM-emerin (23).
In Caenorhabditis elegans, chromatin immunoprecipitation (ChIP) of the inner nuclear membrane protein LEM2 coupled to deep sequencing (ChIP-seq) also showed that repressed chromatin preferentially associated with the nuclear lamina (24, 25). LEM2-associated regions were enriched for chromatin modifications consistent with repressed chromatin (e.g. H3K27me3) and lacked transcriptionally active chromatin modifications (e.g. H3K4me3 (26)). LEM2-associated domains also lacked active RNA polymerase II.
Collectively, these studies show that the nuclear lamina is a repressive environment. How the nuclear lamina establishes and maintains this repressive environment remains poorly understood. Here we show that emerin binds directly to HDAC3, a core component of the NCoR complex. Binding of emerin to HDAC3 increases histone deacetylase activity by increasing the Vmax of HDAC3, suggesting that emerin binding to HDAC3 may cause a conformational change in HDAC3 to increase its catalytic activity. Importantly, emerin-null cells exhibit similar epigenetic changes as those seen in HDAC3-null cells, including increased H4K5 acetylation and decreased H3K27 and H3K9 trimethylation.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
Plasmids encoding GFP-HDAC3 and GST-HDAC3 were generous gifts from Ed Seto (Moffitt Cancer Center). The plasmid encoding FLAG-emerin was a generous gift from Glenn Morris (Robert Jones and Agnes Hunt (RJAH) Orthopaedic Hospital). Antibodies against each named protein and the dilution used for Western blotting were: acetylated H4K5 (07-327, Millipore, Inc., 1:1,000), trimethylated H3K27 (07-449, Millipore, 0.4 μg/ml), trimethylated H3K9 (ab4441, Abcam, Inc., 1:500), trimethylated H3K4 (04-745, Millipore, 1:10,000), HDAC3 (05-813, Millipore, 1 μg/ml), H3 (07-690, Millipore, 1:100,000), H4 (05-858, Millipore, 1:50,000), emerin (sc-15378, Santa Cruz Biotechnology, Inc., 1:20,000), GST (sc-138, Santa Cruz Biotechnology, 1:5,000), β-tubulin (sc-9104, Santa Cruz Biotechnology, 1:5,000), lamin B (sc-6216, Santa Cruz Biotechnology, 1:1,000), Myf5 (antibody 69997, Abcam, 1:1,000), and RNA polymerase II (05-952, Millipore, 1:1,000).
Plasmids and Protein Purification
Recombinant GST-HDAC3 was purified from Escherichia coli DH5α. Protein expression was induced by the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside for 4 h at 37 °C with shaking (220 rpm). Bacteria was collected by centrifugation at 15,000 × g and resuspended in modified His Buffer (50 mm HEPES, pH 8, 500 mm NaCl, 2 mm MgCl2, 0.05% Tween 20, 1 mm PMSF, 5 μg/ml each of leupeptin, aprotinin, and pepstatin, 14 μm β-mercaptoethanol). Lysozyme was added to a final concentration of 1 mg/ml and incubated at 4 °C for 30 min followed by sonication. The lysate was centrifuged at 40,000 × g, and the supernatant was incubated with glutathione beads for 1 h at 4 °C. The beads were washed five times with modified His Buffer, and GST-HDAC3 was eluted with 10 mm glutathione in modified His Buffer. Eluted GST-HDAC3 was then dialyzed into PBS containing 14 μm β-mercaptoethanol. Recombinant emerin protein comprising the entire nucleoplasmic domain of emerin (residues 1–222) and lacking the transmembrane domain was expressed in bacteria from pET11c-emerin and purified as described (8).
Cell Culture
C2C12 myoblasts were maintained in growth media (DMEM + 10% fetal bovine serum) at 37 °C and 5% CO2. Wild-type and emerin-null mouse myogenic progenitors were a generous gift from Tatiana Cohen and Terry Partridge (Children's National Medical Center, Washington, D. C.). Myogenic progenitors were grown in proliferative media consisting of DMEM (11995-065, Invitrogen) supplemented with 20% HI-FBS (10082-147, Invitrogen), 2% chick embryo extract (CE6507, Accurate Chemical), 2% l-glutamine (25030-081, Invitrogen), 1% penicillin-streptomycin (15140-122, Invitrogen), and 20 ng/ml γ-Interferon (IF005, Millipore). Cells were grown at 33 °C and 10% CO2. Cells between passages 4 and 10 were used for Western blotting.
Emerin Down-regulation and Western Blotting
Transfection of emerin shRNA and control shRNA plasmids was performed as described previously (27). 1 × 106 cells were resuspended in SDS-PAGE sample buffer, and 50,000 cells were separated by SDS-PAGE, transferred to nitrocellulose, and Western blotted with the indicated antibodies. Densitometry was performed for each protein, and the results were normalized to γ-tubulin (loading control) to determine changes in H3, H4, or HDAC3 expression. Densitometries of acetylated H4K5 (H4K5ac), H3K27me3, H3K4me3, and H3K9me3 were normalized to total H3 or H4. To confirm H4K5ac antibody specificity, 5 μg/ml H4K5ac peptide (12-343, Millipore) or H4K16ac peptide (12-364, Millipore) was incubated with H4K5ac antibodies (1:1,000 dilution) for 4 h at 4 °C and then incubated with nitrocellulose membranes containing whole cell lysate from wild-type myogenic progenitors.
Binding Assays
GST-HDAC3 binding assays were performed by incubating 1.0 μm of recombinant, purified GST-HDAC3 or GST immobilized on glutathione beads with 4.5 μm recombinant, purified wild-type emerin (residues 1–222) or each emerin mutant (residues 1–222). Proteins were incubated 4 h at 4 °C in PBS containing 0.1% Triton X-100 (PBS-T), 2 mm DTT, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 1 mm PMSF for 4 h at 4 °C. The beads were washed five times with PBS-T, and bound proteins were eluted with SDS-PAGE sample buffer, resolved by SDS-PAGE, and Western blotted with antibodies against either emerin (serum 2999; 28) or GST (sc-138, Santa Cruz Biotechnology). The affinity of emerin for HDAC3 was determined by incubating 0–25 μm wild-type emerin (residues 1–222) with 1.0 μm GST-HDAC3 immobilized on glutathione beads. Immunoprecipitations were performed as described previously (16) using protein G Dynabeads (10007D, Invitrogen).
HDAC Assays
HDAC assays were performed per the manufacturer's instructions (17-356, Millipore). Briefly, 19 μm wild-type emerin (residues 1–222) or emerin S54F (residues 1–222) was incubated with 0.3 μm HDAC3·DAD complex (ab42631, Abcam) in 20 μl of PBS-T for 4 h at 4 °C with rotation. These emerin concentrations were used to ensure that >95% HDAC3·DAD was bound by emerin. 15 μl of each binding reaction was then added to the HDAC assay (final volume 40 μl). HDAC3 activity was measured using the FLUOstar OPTIMA plate reader (BMG Labtech). 0–16 μm wild-type emerin was added to 0.3 μm HDAC3·DAD complex to determine the Km for allosteric activation of HDAC activity by emerin. 9.6 μm wild-type emerin was added to 0.3 μm HDAC3·DAD in the presence of 0–300 μm HDAC substrate to determine how emerin altered HDAC3 enzyme kinetics.
Immunofluorescence Microscopy
C2C12 myoblasts were plated on coverslips and transfected with plasmids encoding GFP-HDAC3 (2 μg) or GFP-HDAC3 (1 μg) and FLAG-emerin (1 μg) using Lipofectamine, per the manufacturer's instructions. After 48 h, the coverslips were removed, washed three times with PBS, and fixed with 3.7% formaldehyde for 15 min. The samples were then permeabilized for 23 min by treatment with 0.2% Triton X-100 in PBS followed by blocking with 3% BSA in PBS, 0.1% Triton X-100 for 1 h. Wild-type and emerin-null myogenic progenitors were prepared for immunofluorescence in the same manner. Primary antibodies against emerin (1:10,000, sc-15378, Santa Cruz Biotechnology), GFP (1:1,000, sc-9996, Santa Cruz Biotechnology), FLAG (1:250, F-1804, Sigma-Aldrich), lamin B (1:500, sc-6216, Santa Cruz Biotechnology, Inc), acetylated H4K5 (07-327, Millipore, 1:500), trimethylated H3K4 (04-745, Millipore, 1:250), and trimethylated H3K27 (07-449, Millipore, 1:500) were incubated with the cells overnight at 4 °C, washed three times with PBS, and incubated with either Alexa Fluor 488-conjugated goat anti-rabbit or Alexa Fluor 594-conjugated goat anti-mouse secondary antibodies (1:500 dilution) for 1 h at 22 °C. The cells were rinsed in PBS three times, incubated with 250 nm DAPI for 5 min, and mounted onto slides containing one drop of ProLong Gold (Invitrogen) or VECTASHIELD (Vector Laboratories, Inc.) antifade reagent. All antibodies were diluted into 3% BSA in PBS, 0.1% Triton X-100. Slides were viewed on a Zeiss Axioskop microscope, and images were acquired using a QImaging Retiga EXI camera controlled by iVision (BioVision) software running on an iMac followed by deconvolution through Huygens (SVI, Inc.) software running on Linux. Co-localization analysis was performed using the JACoP plug-in for ImageJ.
Confocal Microscopy
Confocal imaging was performed on a TCS SP5-II laser-scanning confocal system (Leica Microsystems, GmbH). Images of randomly selected nuclei were acquired in 12-bit grayscale and 1,024 × 1,024-pixel resolution with 30-nm voxel size, using an HCX PL APO CS 63.0× 1.40 NA objective and 8× zoom. Comparison of intensity was performed by ensuring identical illumination and acquisition settings on both wild-type and emerin-null samples.
Subcellular Fractionation
5 × 106 wild-type or emerin-null myogenic progenitors were washed in 1× transport buffer (TB; 20 mm HEPES, pH 7.4, 110 mm potassium acetate, 2 mm magnesium acetate, 0.5 mm EGTA) and suspended in 1 ml of ice-cold hypotonic lysis buffer (5 mm HEPES, pH 7.4, 10 mm potassium acetate, 2 mm magnesium acetate, 1 mm EGTA) containing 10 μg/ml pepstatin A, leupeptin and aprotinin, 1 mm PMSF and 14 μm β-mercaptoethanol. The cells were incubated for 10 min on ice with gentle agitation every 2 min until they swelled. Myogenic progenitors were disrupted with a Dounce homogenizer 40–50 times on ice until 90–95% of nuclei stained with Trypan Blue. 100 μl of 10× TB was then added to the cells, and they were centrifuged at 400 × g for 5 min to pellet the nuclei. Cytosol was removed, and nuclei were washed two times with TB. The nuclei were then treated with TB containing 0.2% Triton X-100 for 20 min on ice, swirling every 5 min. The nuclei were then centrifuged at 400 × g for 5 min to collect insoluble nuclear material. The soluble nuclear extract was removed, and the insoluble material (nuclear lamina) was washed two times with TB. 1.25 × 104 cell equivalents of each fraction were separated by SDS-PAGE, transferred to nitrocellulose, and blotted with the indicated antibodies.
RESULTS
Emerin Binds Directly to HDAC3
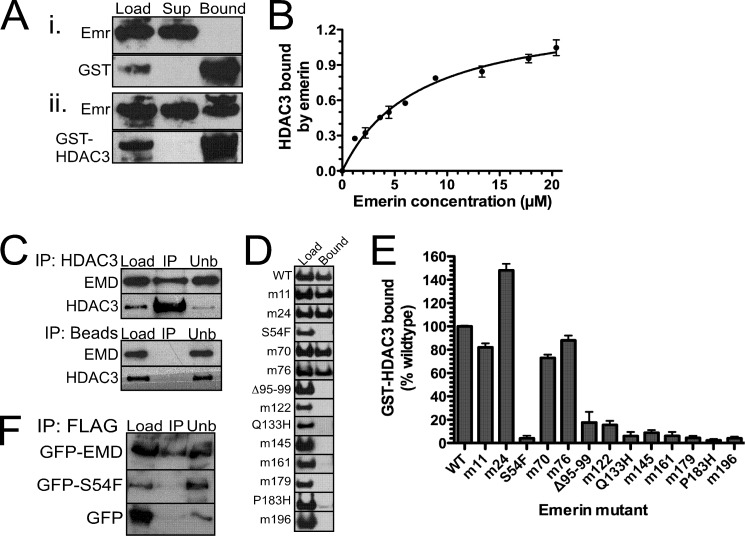
We previously showed that emerin interacted with the NCoR complex in HeLa cells (16). To test whether emerin binds directly to HDAC3, 4.5 μm recombinant, purified emerin (residues 1–222) was incubated with 1 μm GST or 1 μm GST-HDAC3 immobilized on glutathione beads. Emerin specifically bound GST-HDAC3 (Fig. 1A). We next incubated increasing amounts (0–25 μm) of recombinant emerin protein (residues 1–222) with 1 μm GST-HDAC3 to determine the affinity of emerin for HDAC3 (Fig. 1B). The affinity of emerin for HDAC3 was 7.3 ± 1.0 μm (n = 3), which is comparable with the affinities of known HDAC3-binding proteins.
FIGURE 1.
Emerin binds directly to HDAC3 with high affinity. A, 4.5 μm recombinant, purified emerin (Emr; 1–222) was incubated with 1 μm recombinant GST (panel i) or GST-HDAC3 (panel ii) immobilized on glutathione beads. Binding was detected by Western blotting with the indicated antibodies. Sup, supernatant. B, increasing concentrations of emerin (residues 1–222) were incubated with 1.0 μm immobilized GST-HDAC3 to determine the affinity constant. C, emerin binds HDAC3 in vivo. HDAC3 antibodies immobilized on magnetic beads or magnetic beads alone were incubated with C2C12 lysates. Load, bound (IP), and unbound (unb) fractions were separated by SDS-PAGE and Western blotted for emerin (EMD) or HDAC3. D, GST-HDAC3 binds the C terminus of emerin. 4.5 μm wild-type emerin (1–222) or each indicated emerin mutant (1–222) was incubated with immobilized GST-HDAC3, and bound fractions were separated by SDS-PAGE and Western blotted with antibodies against emerin and HDAC3. E, densitometry of HDAC3 binding of each emerin mutant. The amount of GST-HDAC3 binding to each emerin mutant was normalized to wild-type emerin binding. F, emerin S54F mutant fails to bind HDAC3 in vivo. HeLa cells were transfected with FLAG-HDAC3 and GFP, GFP-emerin, or GFP-emerin S54F (GFP-S54F). Immunoprecipitation assays were done using FLAG antibodies. Error bars in B and E indicate S.D.
Co-immunoprecipitation assays were performed to test whether emerin binds HDAC3 in vivo. C2C12 lysates were incubated with HDAC3 antibodies immobilized on Dynabeads or Dynabeads alone. Endogenous emerin is efficiently pulled down by HDAC3 antibodies, but not Dynabeads alone (Fig. 1C).
1.0 μm GST-HDAC3 was incubated with 4.5 μm wild-type emerin (residues 1–222) or each of 15 emerin mutants (residues 1–222) to map the HDAC3-binding domain in emerin (Fig. 1, D and E). The HDAC3-binding domain of emerin mapped to residues 95–202. Most EDMD-causing emerin mutations are nonsense mutations resulting in loss of emerin protein. However, there are four EDMD-causing emerin mutants that are expressed at wild-type levels and localize correctly to the nuclear envelope (28, 29). These are S54F, Δ95–99, Q133H, and P183H. Interestingly, HDAC3 failed to bind all four of these EDMD-causing emerin mutant proteins (n = 4; Fig. 1, D and E). HDAC3 is the first emerin-binding protein disrupted by all EDMD-causing mutants, suggesting that the interaction between emerin and HDAC3 may be important for the EDMD disease mechanism.
GFP-emerin or GFP-S54F was co-expressed with FLAG-HDAC3 in HeLa cells to test whether emerin mutant S54F bound HDAC3 in vivo. GFP-emerin was efficiently pulled down with FLAG antibodies (Fig. 1F). FLAG antibodies failed to pull down GFP-S54F. Thus, emerin mutant S54F is disrupted for binding HDAC3 in vitro and in vivo.
Emerin Binding to HDAC3 Recruits HDAC3 to the Nuclear Periphery
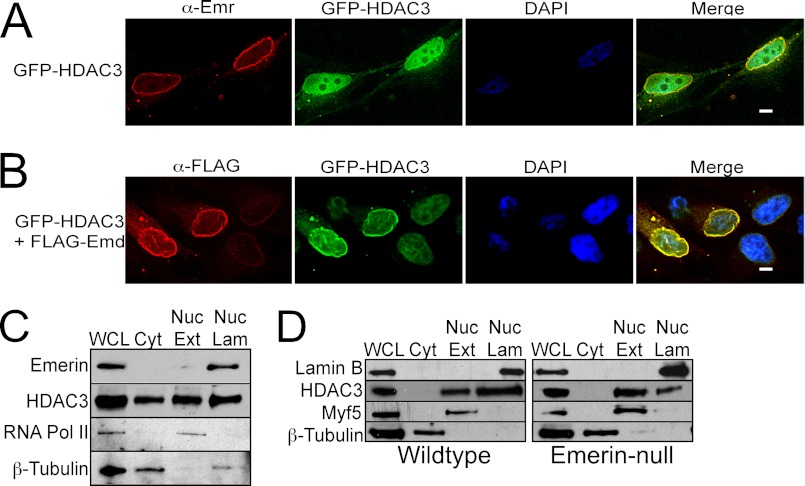
It was previously shown that increased expression of LAP2β, an inner nuclear membrane protein functionally related to emerin, caused increased HDAC3 localization to the nuclear envelope (30). GFP-HDAC3 was expressed in C2C12 cells in the absence or presence of FLAG-emerin (FLAG-Emd) to test whether emerin also recruits HDAC3 to the nuclear envelope. Subnuclear localization was monitored by immunofluorescence microscopy and deconvolution. GFP-HDAC3 was predominantly nucleoplasmic with a fraction localized at the nuclear envelope, where it co-localized with endogenous emerin in the absence of exogenous FLAG-Emd (Fig. 2A). Importantly, expression of FLAG-Emd significantly increased the amount of GFP-HDAC3 localized to the nuclear envelope (Fig. 2B); GFP-HDAC3 was predominantly nucleoplasmic in cells expressing only GFP-HDAC3 (Fig. 2B). Quantification of co-localization of GFP-HDAC3 with endogenous or exogenous emerin in the deconvolved images was analyzed using the JACoP plug-in for ImageJ. 36% of GFP-HDAC3 co-localized with endogenous emerin, whereas 74% co-localized with FLAG-Emd.
FIGURE 2.
Overexpression of emerin recruits HDAC3 to the nuclear envelope. A, deconvolved images of C2C12 myoblasts transfected with plasmids encoding GFP-HDAC3. Localization was monitored 48 h after transfection. Emerin antibodies (α-Emr) were used to mark the nuclear lamina. DAPI was used to stain nuclei. GFP-HDAC3 is predominantly nuclear with a fraction of GFP-HDAC3 at the nuclear periphery overlapping with emerin (Merge). B, deconvolved images of C2C12 myoblasts transfected with plasmids encoding GFP-HDAC3 and FLAG-emerin (FLAG-Emd) 48 h after transfection. More GFP-HDAC3 localized to the nuclear periphery in cells expressing FLAG-Emd (Merge), where it co-localizes with FLAG-Emd. DAPI was used to stain nuclei. C and D, subcellular fractionation confirms that a fraction of endogenous HDAC3 associates with the nuclear lamina. Equal cell equivalents were separated by SDS-PAGE and Western blotted with the indicated antibodies. Less HDAC3 associates with the nuclear lamina in emerin-null myogenic progenitors. Wild-type or emerin-null myogenic progenitors were subjected to subcellular fractionation. Equal cell equivalents were separated by SDS-PAGE and Western blotted with the indicated antibodies. WCL, whole cell lysate; Cyt, cytosol; Nuc Ext, nuclear extract; Nuc Lam, nuclear lamina.
Subcellular fractionation was used to biochemically confirm that endogenous HDAC3 localizes to the nuclear envelope. Importantly, the fractionation of C2C12 myoblasts generated clean cytosolic extract, soluble nuclear extract, and nuclear lamina extract, as determined by Western blotting with emerin, RNA polymerase II, Myf5, and β-tubulin antibodies (Fig. 2C). Emerin is present only in the nuclear lamina fraction, RNA polymerase II and Myf5 are enriched in the soluble nuclear extract, and β-tubulin is highly enriched in the cytosolic fraction (Fig. 2, C and D). HDAC3 was found in the soluble nuclear extract (40.8%) and cytosolic extract (17.9%), as predicted (n = 3; Fig. 2C). Confirming the localization studies above, 41.3% of HDAC3 was detected in the nuclear lamina fraction. Thus, a significant fraction of HDAC3 is found at the nuclear lamina.
Wild-type emerin and emerin-null myogenic progenitors were subjected to subcellular fractionation to test whether nuclear lamina localization of HDAC3 was emerin-dependent. 20.55 ± 7.65% of HDAC3 was found in the soluble nuclear extract, and 65.00 ± 0.3% of HDAC3 was found in the nuclear lamina fraction of wild-type myogenic progenitors (n = 2, Fig. 2D). Unlike C2C12 cells, HDAC3 was undetectable in the cytosolic fractions. There was a dramatic shift in HDAC3 localization in emerin-null myogenic progenitors, as 62.1 ± 0.5 and 28.4 ± 6.5% of HDAC3 were present in the soluble nuclear extract and nuclear lamina, respectively (n = 2, Fig. 2D). These changes in HDAC3 localization were significant (p < 0.033).
Emerin Stimulates HDAC3 Activity
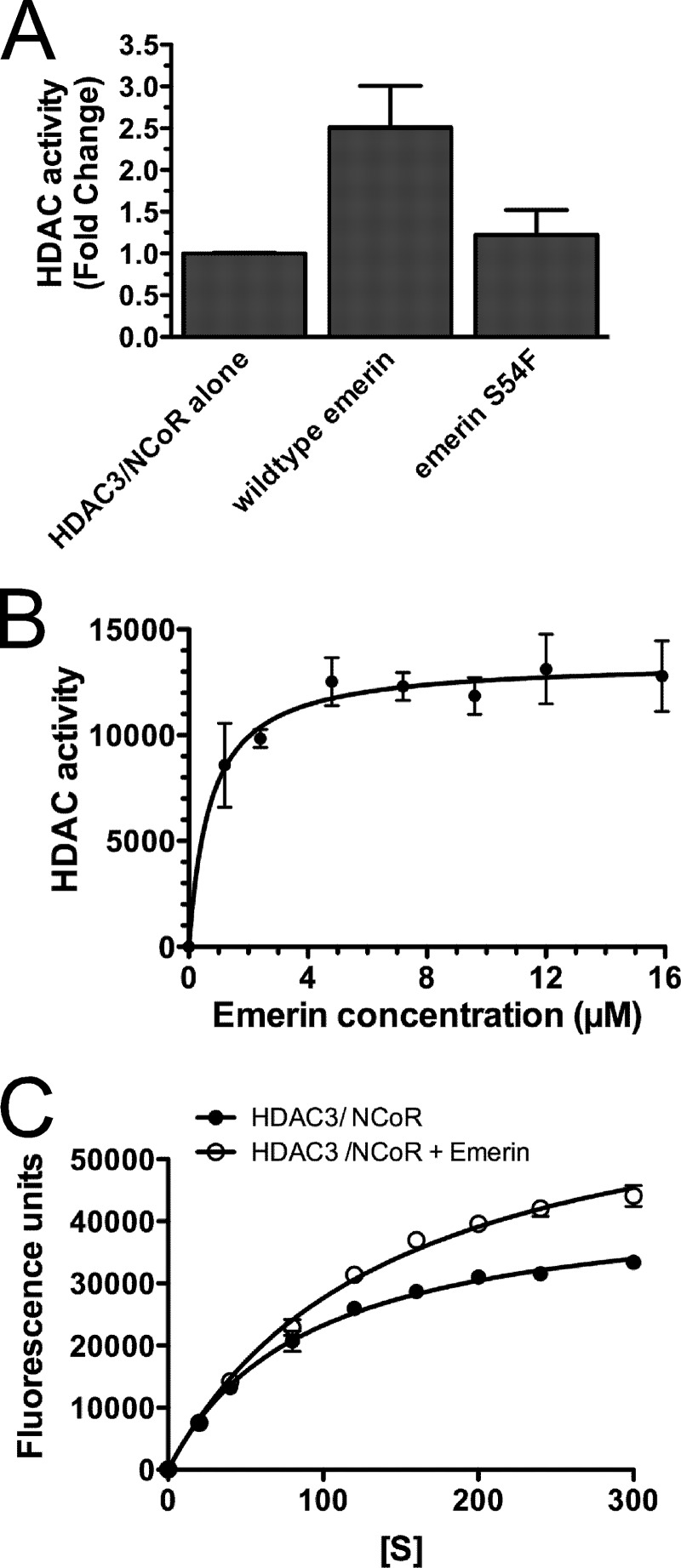
Because LAP2β bound HDAC3 and LAP2β expression increased H4 deacetylation, we tested whether emerin binding to HDAC3 stimulated HDAC3 activity in vitro. For this analysis, we performed histone deacetylase (HDAC) assays (Millipore) in the presence or absence of wild-type emerin (residues 1–222) or an emerin mutant that fails to bind HDAC3 (S54F; residues 1–222). HDAC3 catalytic activity requires binding to the NCoR DAD (31). Thus, HDAC3·NCoR-DAD (HDAC3·DAD) complexes were used for these studies. 0.3 μm HDAC3·DAD complexes were incubated with 19 μm wild-type emerin or emerin S54F for 4 h at 4 °C. Emerin-HDAC3·DAD complexes were then added to the HDAC assay substrate and incubated for 30 min at 30 °C, and HDAC assays were performed per the manufacturer's instructions (Millipore). Wild-type emerin stimulated HDAC activity 2.7-fold (n = 4; Fig. 3A). Emerin S54F failed to stimulate HDAC3 activity (Fig. 3A), demonstrating that emerin binding to HDAC3 is required for activation.
FIGURE 3.
Emerin stimulates HDAC3 activity. A, HDAC3·NCoR-DAD complexes (HDAC3·DAD) were incubated alone or in the presence of wild-type emerin (residues 1–222) or emerin S54F mutant (residues 1–222) for 4 h. HDAC activity was then measured using HDAC assays (Millipore). Wild-type emerin, but not S54F, stimulates HDAC3 activity 2.7-fold, showing that emerin binding to HDAC3 is required for HDAC activation. B, increasing concentrations of emerin were added to HDAC3·DAD and incubated for 4 h prior to performing HDAC assays. HDAC3 activation by emerin had a Km = 0.7 μm. C, HDAC3·DAD complexes were incubated alone or in the presence of 9.6 μm wild-type emerin for 4 h and incubated with increasing concentrations of substrate. The Vmax of HDAC3 increased 1.8-fold and the Km of HDAC3 increased 1.7-fold. Error bars in all panels indicate S.D.
0.3 μm HDAC3·DAD was incubated with increasing concentrations (0–16 μm) of wild-type emerin (residues 1–222) to determine the Km for emerin activation of HDAC3 activity. The Km for emerin activation of HDAC3·DAD was determined to be 0.7 μm (n = 3; Fig. 3B). 0.3 μm HDAC3·DAD was then incubated with increasing substrate concentrations (0–300 μm) in the presence (9.6 μm) or absence of wild-type emerin to test how emerin may regulate the catalytic activity of HDAC3. Interestingly, incubation of HDAC3·DAD with emerin caused a 1.8-fold increase in the Vmax of HDAC3 and a 1.7-fold increase in the Km of HDAC3 (n = 3; Fig. 3C). These results suggest that emerin may stabilize NCoR-DAD binding to HDAC3. Alternatively, emerin may cause a conformational change in HDAC3 that increases the HDAC3 catalytic activity and substrate turnover.
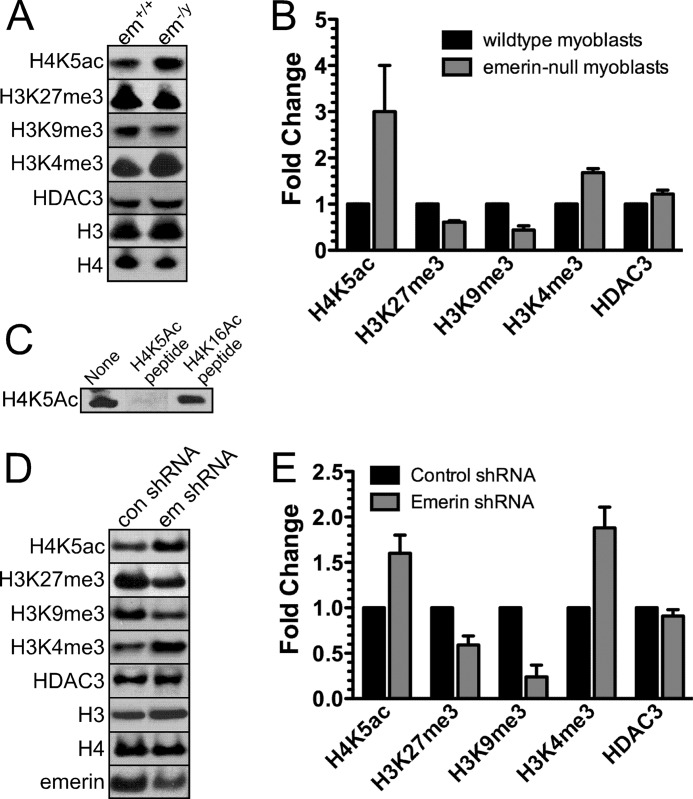
The NCoR complex initially deacetylates H4K5, and repression is enhanced by subsequent deacetylation of adjacent acetyl lysines on the H4 N-terminal tail; initial recruitment of the NCoR complex by H4K5ac is required for stable repression (32). Thus, we tested whether emerin regulates NCoR activity in vivo by examining the levels of H4K5ac in emerin-null mouse myogenic progenitors. Lysates of wild-type and emerin-null mouse myogenic progenitors were separated by SDS-PAGE and Western blotted with antibodies specific to H4K5ac. Levels of H4K5ac were normalized to total H4 protein. Emerin-null myogenic progenitors significantly increased H4K5ac levels (n = 3; Fig. 4, A and B), demonstrating that emerin activates HDAC3 activity in vivo. Levels of the H4K5 acetyltransferase, HAT1 (data not shown), and HDAC3 were unaffected in emerin-null cells (Fig. 4, A and B). Antibody specificity was confirmed by incubation of H4K5ac antibodies with H4K5ac or H4K16ac peptides prior to blotting wild-type myogenic progenitor lysates. Incubation with H4K5ac peptide, but not H4K16ac peptide, blocked antibody binding (Fig. 4C). The abundance of other repressive chromatin marks (H3K27me3 and H3K9me3) were also decreased (n = 3; Fig. 4, A and B). Trimethylation of H3K4 was also up-regulated in emerin-null myogenic progenitors (n = 3; Fig. 4, A and B). Interestingly, these changes in the epigenetic signature of emerin-null cells were similar to the epigenetic changes seen in HDAC3−/− mouse embryonic fibroblasts (33).
FIGURE 4.
Loss of emerin causes epigenetic changes similar to those seen in HDAC3-null cells. A, emerin (em)-null myogenic progenitors were separated by SDS-PAGE and Western blotted with the indicated antibodies. B, densitometry of Western blots from A normalized to wild-type myogenic progenitors. C, H4K5ac antibodies are specific for H4K5ac. H4K5ac antibodies were incubated with H4K5ac or H4K16ac peptides. Wild-type lysates were blotted with these antibody-peptide mixtures to confirm specificity of the H4K5ac antibody. D, HeLa cells transfected with control shRNA (con shRNA) or emerin shRNA (em shRNA) were separated by SDS-PAGE and Western blotted with the indicated antibodies. E, densitometry of Western blots normalized to control shRNA was calculated. Error bars in B and D indicate S.D.
Collectively, these results show that emerin-null myogenic progenitors have a more open chromatin confirmation, which molecularly confirms previous ultrastructural studies (14, 15). To confirm that these epigenetic changes were not cell type-specific, we measured the levels of H4K5ac, H3K27me3, H3K9me3, and H3K4me3 in HeLa cells down-regulated for emerin (Fig. 4, D and E). Emerin levels in emerin-down-regulated cells were 0.5-fold of wild-type cells (Fig. 4, D and E). Emerin-down-regulated cells exhibited increased H4K5ac and H3K4me3 levels and decreased H3K27me3 and H3K9me3 levels (Fig. 4, D and E), demonstrating that these changes were not specific to myogenic progenitors.
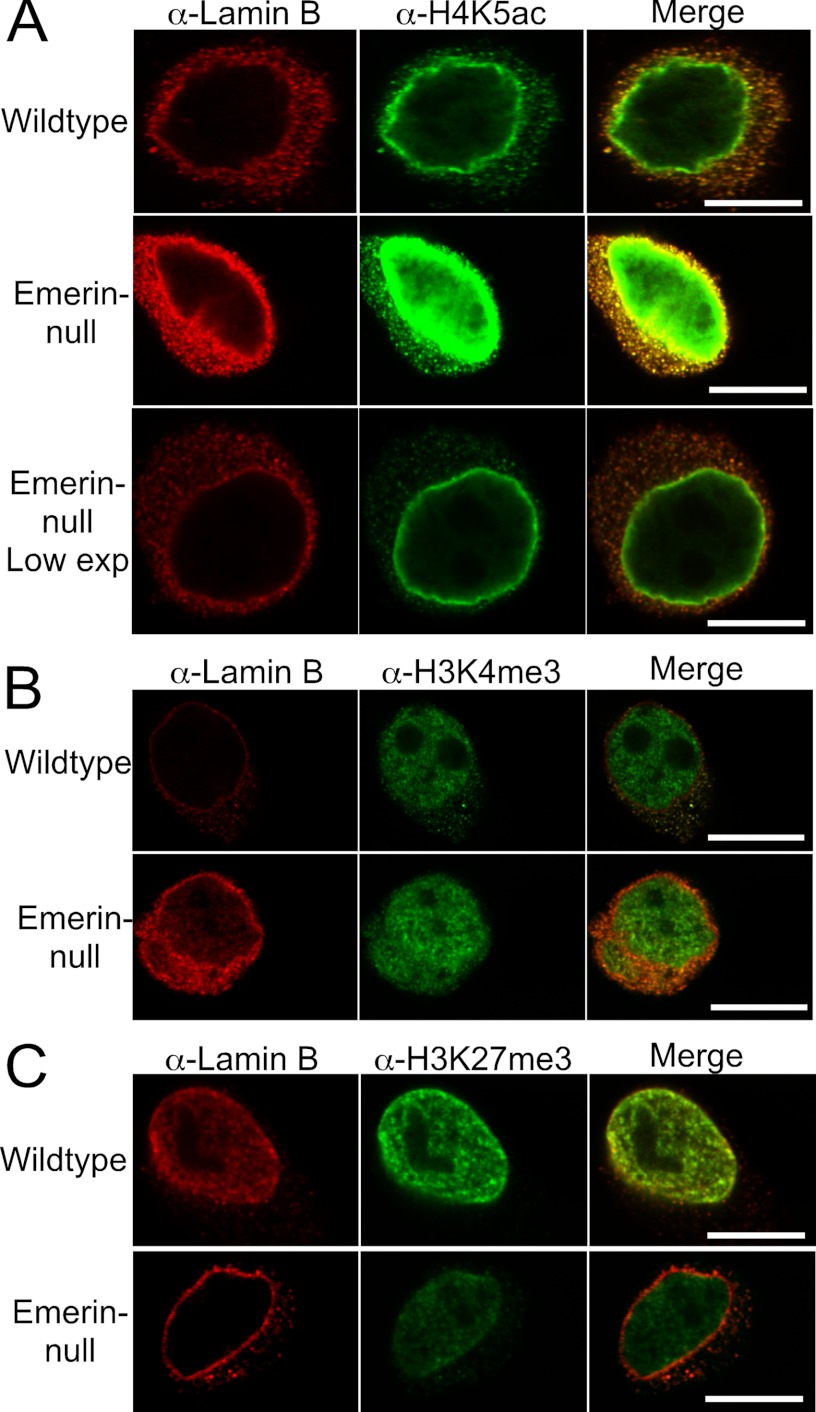
It was unclear from this analysis whether these changes in H4K5ac, H3K4me3, H3K9me3, and H3K27me3 were confined to the nuclear envelope, where emerin interacts with HDAC3, or whether these epigenetic changes occurred throughout the nucleus. Confocal microscopy using antibodies against H4K5ac, H3K4me3, and H3K27me3 was performed to determine the nuclear location of these changes. All images were taken using the same exposure times for comparison of wild-type progenitors with emerin-null myogenic progenitors. H4K5ac predominantly localized to the nuclear envelope in both wild-type and emerin-null myogenic progenitors (Fig. 5A). Nuclear envelope localization of H4K5ac has been seen in other cell types (34, 35). Importantly, significantly more H4K5ac was detected at the nuclear periphery in emerin-null cells (Fig. 5A). A lower exposure is shown in the bottom panel to show that H4K5Ac still preferentially localized to the nuclear envelope in emerin-null cells (Fig. 5A). This is consistent with previous findings showing that trichostatin A treatment causes increased H4K5ac at the nuclear periphery (34, 35), which is hypothesized to be associated with the nuclear pore complex (34) and shows that there is decreased HDAC activity in the absence of emerin. H3K4me3 levels were increased (Fig. 5B) and H3K27me3 levels were decreased (Fig. 5C), as expected. These changes were seen throughout the nucleus and were not confined to the nuclear envelope. Collectively, these results demonstrate that chromatin in emerin-null cells adopts a more open conformation.
FIGURE 5.
Epigenetic changes are not restricted to the nuclear periphery, but occur throughout the nucleus. Confocal microscopy was done on wild-type or emerin-null myogenic progenitors using lamin B antibodies and antibodies against H4K5Ac (A), H3K4me3 (B), or H3K27me3 (C). Exposure times are the same between wild-type and emerin-null cells. The exception is the bottom panel in A, to illustrate that nuclear envelope localization of H4K5Ac is not lost in emerin-null cells. Low exp, low exposure.
DISCUSSION
Here we showed that emerin binds HDAC3 with high affinity and stimulates its activity in vitro. Emerin recruits HDAC3 to the nuclear lamina and is required for localization of HDAC3 to the nuclear lamina. Chromatin adopts a more open chromatin conformation in emerin-null myogenic progenitors, including decreased H3K9me3 and H3K27me3. Importantly, the primary histone target of HDAC3, H4K5ac, was significantly increased in the absence of emerin. This epigenetic signature is similar to that seen in HDAC3-null mouse embryonic fibroblasts (33). Thus, we conclude that emerin is required for optimal HDAC3 activity. We predict that loss of HDAC3 activity contributes to the defective genomic organization seen in cells containing mutations in lamin A (14, 15, 36–38), which mislocalize emerin to the endoplasmic reticulum (39).
Over the last decade, it has become clear that the nuclear lamina is a transcriptionally repressive environment. For example, tethering of genomic regions to the nuclear lamina causes repression of genes 100 kb to 5 Mb from the genomic tethering site (21, 22). Further, electron microscopy shows that chromatin at the nuclear periphery tends to be more condensed (14, 15). The molecular characterization of chromatin at the nuclear lamina confirmed that these LADs are enriched in repressive chromatin marks (H3K27me3 and H3K9me2 (23, 26, 40)), lack active chromatin marks (H3K4me3), and have reduced RNA polymerase II occupancy (23, 26). Further, genes within LADs tend to be transcriptionally silent (23, 26).
Sequence specificity of LAD localization was recently examined, and it was shown that a tandem GAGA sequence was sufficient for localization of lamina-associated sequences (LASs) to the nuclear lamina (41). Localization of LASs was mediated by cKrox, the mammalian ortholog of Drosophila GAGA-associated factor. Nuclear lamina localization of LASs was also dependent on HDAC3 and LAP2β because down-regulation of HDAC3 or LAP2β significantly reduced the association of LASs with the nuclear lamina. However, knockdown of LAP2β failed to completely abolish the interaction of LASs with the nuclear lamina, suggesting that other unidentified LAS-binding proteins may mediate LAS association with the lamina.
LAP2β was previously shown to interact with HDAC3 and regulate the acetylation of H4 (30). Further, inhibition of HDAC3 activity causes LASs to relocalize to the nuclear interior (41), suggesting that HDAC3 activity, presumably at the nuclear lamina, is required for LAS nuclear lamina localization. We found that emerin is also required for optimal HDAC3 localization to the nuclear lamina and localization of repressed genes to the nuclear lamina.4 Collectively, these studies provide compelling evidence that the interaction of HDAC3 with emerin and LAP2β may play an important role in initiating or maintaining repressed chromatin at the nuclear lamina. Our findings that emerin binds HDAC3 and stimulates its catalytic activity provide a potential mechanism for how the association of LASs with HDAC3 at the nuclear lamina may lead to repression of these genomic loci.
Acknowledgments
We thank Katherine Wilson (Johns Hopkins Medical School) for the generous gift of sera 2999 against emerin. We thank Alex Ruthenburg, Harinder Singh, Robert Goldman, and members of the Holaska laboratory for many fruitful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM007197 and T32 HL007381 (to A. J. K.). This work was also supported by the Ellison Medical Foundation (to J. M. H.) and the American Heart Association (to J. M. H.).
J. Demmerle and J. M. Holaska, manuscript in preparation.
- EDMD
- Emery-Dreifuss muscular dystrophy
- HDAC
- histone deacetylase
- HDAC3
- histone deacetylase 3
- NCoR
- nuclear corepressor
- GPS2
- G-protein pathway suppressor 2
- H4K5
- histone 4, lysine 5
- H4K5ac
- histone 4, acetyl lysine 5
- H3K4
- histone 3, lysine 4
- H3K4me3
- histone 3, trimethylated lysine 4
- H3K9
- histone 3, lysine 9
- H3K9me3
- histone 3, trimethylated lysine 9
- H3K9me2
- histone 3, dimethylated lysine 9
- H3K27
- histone 3, lysine 27
- H3K27me3
- histone 3, trimethylated lysine 27
- DAM
- DNA adenine methyltransferase
- LMNB1
- lamin B1
- LAD
- lamina-associated domain
- DAD
- deacetylase activation domain
- TB
- transport buffer
- LAS
- lamina-associated sequence
- Emd
- emerin.
REFERENCES
- 1. Wilson K. L., Berk J. M. (2010) The nuclear envelope at a glance. J. Cell Sci. 123, 1973–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 3. Korfali N., Wilkie G. S., Swanson S. K., Srsen V., Batrakou D. G., Fairley E. A., Malik P., Zuleger N., Goncharevich A., de Las Heras J., Kelly D. A., Kerr A. R., Florens L., Schirmer E. C. (2010) The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol. Cell Proteomics 9, 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vlcek S., Foisner R. (2007) Lamins and lamin-associated proteins in aging and disease. Curr. Opin. Cell Biol. 19, 298–304 [DOI] [PubMed] [Google Scholar]
- 5. Holaska J. M. (2008) Emerin and the nuclear lamina in muscle and cardiac disease. Circ. Res. 103, 16–23 [DOI] [PubMed] [Google Scholar]
- 6. Vlcek S., Foisner R. (2007) A-type lamin networks in light of laminopathic diseases. Biochim. Biophys. Acta 1773, 661–674 [DOI] [PubMed] [Google Scholar]
- 7. Muchir A., Worman H. J. (2007) Emery-Dreifuss muscular dystrophy. Curr. Neurol. Neurosci. Rep. 7, 78–83 [DOI] [PubMed] [Google Scholar]
- 8. Holaska J. M., Lee K. K., Kowalski A. K., Wilson K. L. (2003) Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 278, 6969–6975 [DOI] [PubMed] [Google Scholar]
- 9. Haraguchi T., Holaska J. M., Yamane M., Koujin T., Hashiguchi N., Mori C., Wilson K. L., Hiraoka Y. (2004) Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur. J. Biochem. 271, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 10. Markiewicz E., Tilgner K., Barker N., van de Wetering M., Clevers H., Dorobek M., Hausmanowa-Petrusewicz I., Ramaekers F. C., Broers J. L., Blankesteijn W. M., Salpingidou G., Wilson R. G., Ellis J. A., Hutchison C. J. (2006) The inner nuclear membrane protein emerin regulates β-catenin activity by restricting its accumulation in the nucleus. EMBO J. 25, 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holaska J. M., Rais-Bahrami S., Wilson K. L. (2006) Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum. Mol. Genet. 15, 3459–3472 [DOI] [PubMed] [Google Scholar]
- 12. Bakay M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C., Shneiderman B., Escolar D., Chen Y. W., Winokur S. T., Pachman L. M., Fan C., Mandler R., Nevo Y., Gordon E., Zhu Y., Dong Y., Wang Y., Hoffman E. P. (2006) Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain 129, 996–1013 [DOI] [PubMed] [Google Scholar]
- 13. Melcon G., Kozlov S., Cutler D. A., Sullivan T., Hernandez L., Zhao P., Mitchell S., Nader G., Bakay M., Rottman J. N., Hoffman E. P., Stewart C. L. (2006) Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 15, 637–651 [DOI] [PubMed] [Google Scholar]
- 14. Ognibene A., Sabatelli P., Petrini S., Squarzoni S., Riccio M., Santi S., Villanova M., Palmeri S., Merlini L., Maraldi N. M. (1999) Nuclear changes in a case of X-linked Emery-Dreifuss muscular dystrophy. Muscle Nerve 22, 864–869 [DOI] [PubMed] [Google Scholar]
- 15. Meaburn K. J., Cabuy E., Bonne G., Levy N., Morris G. E., Novelli G., Kill I. R., Bridger J. M. (2007) Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 6, 139–153 [DOI] [PubMed] [Google Scholar]
- 16. Holaska J. M., Wilson K. L. (2007) An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry 46, 8897–8908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoon H. G., Chan D. W., Huang Z. Q., Li J., Fondell J. D., Qin J., Wong J. (2003) Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1, and TBLR1. EMBO J. 22, 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu J., Li Y., Ishizuka T., Guenther M. G., Lazar M. A. (2003) A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 22, 3403–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J., Kalkum M., Chait B. T., Roeder R. G. (2002) The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 [DOI] [PubMed] [Google Scholar]
- 20. Yoon H. G., Choi Y., Cole P. A., Wong J. (2005) Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reddy K. L., Zullo J. M., Bertolino E., Singh H. (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 [DOI] [PubMed] [Google Scholar]
- 22. Finlan L. E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J. R., Bickmore W. A. (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4, e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 24. Huber M. D., Guan T., Gerace L. (2009) Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 29, 5718–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J., Lee K. K., Segura-Totten M., Neufeld E., Wilson K. L., Gruenbaum Y. (2003) MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 100, 4598–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ikegami K., Egelhofer T. A., Strome S., Lieb J. D. (2010) Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11, R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dedeic Z., Cetera M., Cohen T. V., Holaska J. M. (2011) Emerin inhibits Lmo7 binding to the Pax3 and MyoD promoters and expression of myoblast proliferation genes. J. Cell Sci. 124, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 28. Lee K. K., Haraguchi T., Lee R. S., Koujin T., Hiraoka Y., Wilson K. L. (2001) Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114, 4567–4573 [DOI] [PubMed] [Google Scholar]
- 29. Holt I., Clements L., Manilal S., Morris G. E. (2001) How does a g993t mutation in the emerin gene cause Emery-Dreifuss muscular dystrophy? Biochem. Biophys. Res. Commun. 287, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 30. Somech R., Shaklai S., Geller O., Amariglio N., Simon A. J., Rechavi G., Gal-Yam E. N. (2005) The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 118, 4017–4025 [DOI] [PubMed] [Google Scholar]
- 31. Guenther M. G., Barak O., Lazar M. A. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hartman H. B., Yu J., Alenghat T., Ishizuka T., Lazar M. A. (2005) The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep. 6, 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bhaskara S., Knutson S. K., Jiang G., Chandrasekharan M. B., Wilson A. J., Zheng S., Yenamandra A., Locke K., Yuan J. L., Bonine-Summers A. R., Wells C. E., Kaiser J. F., Washington M. K., Zhao Z., Wagner F. F., Sun Z. W., Xia F., Holson E. B., Khabele D., Hiebert S. W. (2010) Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18, 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown C. R., Kennedy C. J., Delmar V. A., Forbes D. J., Silver P. A. (2008) Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22, 627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taddei A., Maison C., Roche D., Almouzni G. (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3, 114–120 [DOI] [PubMed] [Google Scholar]
- 36. Taimen P., Pfleghaar K., Shimi T., Möller D., Ben-Harush K., Erdos M. R., Adam S. A., Herrmann H., Medalia O., Collins F. S., Goldman A. E., Goldman R. D. (2009) A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. U.S.A. 106, 20788–20793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shumaker D. K., Dechat T., Kohlmaier A., Adam S. A., Bozovsky M. R., Erdos M. R., Eriksson M., Goldman A. E., Khuon S., Collins F. S., Jenuwein T., Goldman R. D. (2006) Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U.S.A. 103, 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galiová G., Bártová E., Raska I., Krejcí J., Kozubek S. (2008) Chromatin changes induced by lamin A/C deficiency and the histone deacetylase inhibitor trichostatin A. Eur. J. Cell Biol. 87, 291–303 [DOI] [PubMed] [Google Scholar]
- 39. Vaughan A., Alvarez-Reyes M., Bridger J. M., Broers J. L., Ramaekers F. C., Wehnert M., Morris G. E., Whitfield W. G. F., Hutchison C. J. (2001) Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 114, 2577–2590 [DOI] [PubMed] [Google Scholar]
- 40. Wen B., Wu H., Shinkai Y., Irizarry R. A., Feinberg A. P. (2009) Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zullo J. M., Demarco I. A., Piqué-Regi R., Gaffney D. J., Epstein C. B., Spooner C. J, Luperchio T. R., Bernstein B. E., Pritchard J. K., Reddy K. L., Singh H. (2012) A molecular mechanism for compartmentalization and silencing of chromatin domains at the nuclear lamina. Cell, in press [DOI] [PubMed] [Google Scholar]