Background: The dimeric D. radiodurans single-stranded DNA-binding protein (DrSSB) has poorly defined DNA-binding and cellular mechanisms.
Results: DrSSB binds ssDNA analogously to tetrameric bacterial SSBs and is regulated in response to ionizing radiation.
Conclusion: The DrSSB ssDNA mechanism is conserved with other SSBs, and the protein is dynamically localized during DNA repair.
Significance: The findings suggest a central role for DrSSB in DNA repair.
Keywords: DNA-binding Protein, DNA Damage Response, DNA Recombination, DNA Repair, X-ray Crystallography, Ionizing Radiation Resistance, Single-stranded DNA-binding Protein
Abstract
The single-stranded DNA (ssDNA)-binding protein from the radiation-resistant bacterium Deinococcus radiodurans (DrSSB) functions as a homodimer in which each monomer contains two oligonucleotide-binding (OB) domains. This arrangement is exceedingly rare among bacterial SSBs, which typically form homotetramers of single-OB domain subunits. To better understand how this unusual structure influences the DNA binding and biological functions of DrSSB in D. radiodurans radiation resistance, we have examined the structure of DrSSB in complex with ssDNA and the DNA damage-dependent cellular dynamics of DrSSB. The x-ray crystal structure of the DrSSB-ssDNA complex shows that ssDNA binds to surfaces of DrSSB that are analogous to those mapped in homotetrameric SSBs, although there are distinct contacts in DrSSB that mediate species-specific ssDNA binding. Observations by electron microscopy reveal two salt-dependent ssDNA-binding modes for DrSSB that strongly resemble those of the homotetrameric Escherichia coli SSB, further supporting a shared overall DNA binding mechanism between the two classes of bacterial SSBs. In vivo, DrSSB levels are heavily induced following exposure to ionizing radiation. This accumulation is accompanied by dramatic time-dependent DrSSB cellular dynamics in which a single nucleoid-centric focus of DrSSB is observed within 1 h of irradiation but is dispersed by 3 h after irradiation. These kinetics parallel those of D. radiodurans postirradiation genome reconstitution, suggesting that DrSSB dynamics could play important organizational roles in DNA repair.
Introduction
Deinococccus radiodurans is an extremophilic bacterium that can withstand extraordinary amounts of DNA damage and harsh cycles of desiccation and rehydration (1–5). This remarkable resistance stems in part from a robust DNA repair capacity that can rapidly and accurately restore genome integrity following the introduction of hundreds of double-stranded genomic breaks (DSBs)4 (6). Several novel and classical genome maintenance proteins appear to work together to mediate DNA repair in D. radiodurans. These features have made the bacterium and its highly adapted genome maintenance proteins models for probing the extreme limits of DNA repair mechanisms (7–11).
After exposure to DSB-inducing ionizing radiation, genome reconstitution in D. radiodurans proceeds in two phases (12, 13). During the initial phase (within ∼1 h of ionizing radiation exposure), a process referred to as “extended synthesis-dependent single-stranded DNA annealing” (ESDSA) resects DSB ends, producing 3′ single-stranded DNA (ssDNA) extensions. These ends are then paired with homologous duplex DNA through strand invasion to create templates for DNA synthesis by DNA polymerases. The extended tracts of ssDNA that are produced as intermediates during ESDSA are likely to be bound and protected by cellular ssDNA-binding proteins (SSBs) (12, 13). However, the precise biochemical and cellular roles of D. radiodurans SSB (DrSSB) in this process remain poorly understood. In the second repair phase (∼1–2 h after ionizing radiation), RecA-mediated recombination, which requires removal of SSB and resolution of interlinked chromosomes, completes the repair process. An understanding of the physical and cellular mechanisms that facilitate DrSSB activity is important for appreciating the protein's role in the extreme radio-resistance of D. radiodurans cells.
Bacterial SSBs can be grouped into two classes based on their oligomeric structure and the number of DNA-binding domains in each monomer (14–16). The first group, for which Escherichia coli SSB (EcSSB) serves as a prototype, comprises homotetrameric SSBs in which each subunit contains a single oligonucleotide-binding (OB) domain. This class is present in the vast majority of sequenced bacteria. The second group, which includes SSBs found only in bacteria within the Deinococcus/Thermus (D/T) phylum, is composed of homodimeric SSBs with two OB domains per subunit. Structural analyses of D/T SSBs from D. radiodurans and Thermus aquaticus have revealed an arrangement in which the two OB domains abut one another and are directly connected by a β-hairpin within each monomer (15, 17, 18). Dimerization places the four OB domains in very similar positions to that found in homotetrameric SSBs, which could indicate that both classes of bacterial SSBs bind ssDNA in a conserved manner. However, because the OB domains within D/T SSB monomers are not identical in sequence, the N- and C-terminal OB domains could bind ssDNA differently (15). Structural data mapping the ssDNA binding sites of a number of homotetrameric SSBs are available (19–22), but high resolution structures of D/T SSBs in complex with ssDNA have not been reported.
Bacterial SSBs can bind ssDNA in distinct modes that depend upon monovalent and divalent ion concentrations and protein/DNA ratios (23–29). For example, under low ionic strength and Mg2+ concentrations or high SSB/DNA ratios, EcSSB binds ssDNA in the so-called (SSB)35 mode, which occludes 35 nucleotides (nt)/SSB tetramer. Under higher ionic strength and Mg2+ concentrations or lower SSB/DNA ratios, binding shifts to the (SSB)65 mode, in which 65 nt of ssDNA are bound per SSB tetramer. The (SSB)65 mode is a limited cooperativity binding arrangement in which SSB forms a beaded morphology along ssDNA, as revealed by electron microscopy (27, 30). In contrast, the (SSB)35 mode is a highly cooperative binding mode in which SSB tetramers are clustered on ssDNA. DrSSB ssDNA binding is also salt-dependent, although the site size difference between low and high ionic strength binding modes is less dramatic than that of EcSSB (23, 25). Under low ionic strength conditions, DrSSB binds in an (SSB)45 mode, whereas it binds in an (SSB)50–55 mode under higher ionic strengths. Interestingly, T. aquaticus SSB, a second D/T group homodimeric SSB, has been reported to bind ssDNA with a uniform (SSB)54 binding mode regardless of ionic strength (24). Given the less dramatic differences in salt-dependent DNA-binding site sizes in D/T group SSBs relative to EcSSB, it is unclear whether D/T group SSBs bind ssDNA in binding morphologies similar to those observed with EcSSB.
Here we examine the structural mechanisms of DrSSB ssDNA binding and DNA damage-dependent cellular dynamics of the protein. The x-ray crystal structure of DrSSB bound to ssDNA, the first such complex for a homodimeric D/T group SSB, defines an extended surface on the protein that accommodates ssDNA. Consistent with a common mechanism for ssDNA binding, this surface is strikingly similar to that used by homotetrameric SSBs. A conserved ssDNA binding model is further supported by electron micrographic imaging of DrSSB bound to long ssDNA in which the protein binds in salt-dependent morphologies that parallel those of EcSSB. In vivo, DrSSB protein levels rise significantly upon treatment with ionizing radiation, forming multiple foci in most D. radiodurans cells during the ESDSA phase of DNA repair. These foci accumulate to form one central focus in the nucleoid during the second (RecA-dependent) phase of repair and disperse as genome reconstitution is completed. Taken together, these experiments provide structural and cellular insights that suggest a prominent role for DrSSB in supporting the remarkable radioresistance of D. radiodurans.
EXPERIMENTAL PROCEDURES
Proteins and DNA
EcSSB
E. coli BL21 (DE3) cells transformed with pET21A-EcSSB (which encodes EcSSB) were grown at 37 °C in Luria-Bertani medium supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. Cultures at an A600 nm of ∼0.7 were induced to express EcSSB by the addition of 1 mm isopropyl β-d-thiogalactopyranoside and were harvested by centrifugation after an additional 3.5 h of growth. Cells were lysed by sonication, and protein purification was performed essentially as described (31) with slight modifications. After ammonium sulfate precipitation, SSB was suspended in solubilization buffer (10 mm Tris, pH 8.0, 2.5 mm EDTA, 200 mm NaCl, 10% glycerol), filtered (0.22 μm), and dialyzed at 4 °C against 100× volume dialysis buffer (10 mm Tris, pH 8.0, 2.5 mm EDTA, 100 mm NaCl, 10% glycerol). Samples were heated to 37 °C, diluted 1:1 with dilution buffer (10 mm Tris, pH 8.0, 10% glycerol), filtered (0.22 μm), and loaded to a Q-Sepharose fast flow anion exchange column (GE Healthcare) pre-equilibrated in dilution buffer. The Q-Sepharose fast flow column was washed with 2 column volumes of low salt buffer (10 mm Tris, pH 8.0, 5 mm EDTA, 10% glycerol, 150 mm NaCl), and EcSSB was eluted using a 150–500 mm NaCl gradient over 16 column volumes. Purified fractions were pooled, dialyzed at room temperature for 4–6 h against dialysis buffer, and further purified with 2–3 additional rounds of Q-Sepharose fast flow anion exchange/dialysis to remove all traces of DNA and protein contaminants. Pure EcSSB fractions (>99% as judged by SDS-PAGE) were pooled, adjusted to ∼1 m NaCl, concentrated, and chromatographed through a Sephacryl S100 size exclusion column (GE Healthcare) pre-equilibrated in 10 mm Tris, pH 8.0, 5 mm EDTA, 10% glycerol, 1 m NaCl. EcSSB samples purified with this protocol were free of visible contaminants and nuclease contamination.
DrSSB
E. coli BL21(DE3) cells transformed with pEAW328 (encodes DrSSB (14)) were grown at 30 °C in Luria-Bertani medium supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. Cultures at an A600 nm of ∼0.7 were induced to express DrSSB by the addition of 1 mm isopropyl β-d-thiogalactopyranoside and were harvested by centrifugation after an additional 4 h of growth. Cells were resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10% sucrose, 2.5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF)) supplemented with 1 tablet of EDTA-free protease inhibitor (Roche Applied Science)/50 ml, lysed by sonication, and centrifuged at 4 °C for 15 min at 44,000 × g. Neutralized polyethyleneimine was titrated to 0.4% into the soluble lysate, followed by stirring on ice for 30 min. Precipitated proteins were centrifuged at 4 °C for 15 min at 44,000 × g, washed with 50 ml of lysis buffer supplemented with 1 mm benzamidine, and centrifuged again at 4 °C for 15 min at 44,000 × g. Pellets were homogenized in 50 ml of elution buffer (10 mm Tris-HCl, pH 8.0, 0.4 m NaCl, 5 mm EDTA), and DrSSB was allowed to desorb from the precipitant for an additional 30 min on ice with constant stirring. The slurry was clarified by centrifugation at 4 °C for 15 min at 44,000 × g. The soluble fraction was then precipitated through the slow addition of 0.140 g/ml ammonium sulfate on ice with constant stirring, followed by incubation for 30 min. The ammonium sulfate precipitation was centrifuged at 4 °C for 15 min at 44,000 × g and then solubilized in fresh elution buffer. DrSSB was then reprecipitated with the addition of 0.120 g/ml ammonium sulfate, pelleted, and resuspended in 10 mm Tris-HCl, pH 8.0, 2.5 mm EDTA, 10% glycerol, followed by dialysis against 10 mm Tris-HCl, pH 8.0, 2.5 mm EDTA, 10% glycerol. The dialysate was heated to 37 °C, filtered (0.22 μm), and diluted 1:1 (v/v) with 10 mm Tris-HCl, pH 8.0, 20% glycerol and then loaded onto a heparin fast flow (GE Healthcare) column pre-equilibrated with buffer A (10 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm 2-mercaptoethanol, 1 mm EDTA). DrSSB was eluted with a 0–400 mm NaCl gradient in buffer A and then loaded onto a Q-Sepharose fast flow ion exchange column (GE Healthcare) pre-equilibrated with buffer B (10 mm Tris-HCl, pH 8.0, 5 mm EDTA). DrSSB was eluted with a 0–600 mm NaCl gradient in buffer B. Pooled fractions of DrSSB were dialyzed against buffer A and subjected to 2–3 iterative rounds of purification with anion exchange/dialysis until samples were nuclease-free and homogeneous (as judged by visual inspection using SDS-PAGE). Highly purified samples were pooled, adjusted to ∼1 m NaCl, concentrated, and further purified by size exclusion chromatography on a Sephacryl S100 column (GE Healthcare) equilibrated in buffer B. Purified DrSSB was concentrated, dialyzed against 25 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 5 mm EDTA, 1 mm DTT, 50% glycerol, and stored at −20 °C until needed. Circular single-stranded DNA purified from phage M13mp18 was prepared as described previously (33, 34).
DrSSB-ssDNA Crystal Structure Determination
DrSSB (6.4 mg/ml in 10 mm Tris-HCl, pH 8.0, 150 mm NaCl) was incubated with dT35 at a 1:2 DrSSB (as dimer)/dT35 molar ratio for at least 5 min at room temperature. Crystals of DrSSB-dT35 were grown by hanging drop vapor diffusion by mixing 1:1 (v/v) with mother liquor (40% polyethylene glycol monomethyl ether 2000, 0.1 m sodium acetate, pH 4.6, 0.2 m ammonium sulfate) and equilibration at room temperature. Crystals were cryoprotected with mother liquor supplemented with 25% polyethylene glycol 200 or ethylene glycol and then flash-frozen in liquid nitrogen. Diffraction data were indexed and scaled using HKL2000 (35). Molecular replacement was performed using the previously published DrSSB structure (15) as a search model with Phaser (36). Iterative rounds of model building and structure refinement were carried out using Coot and REFMAC, respectively (37, 38).
Electron Microscopy
Carbon films, mounted on 400-mesh EM grids, were first activated by a brief glow-discharge treatment (39). Activated grids were used immediately for spreading. Samples for EM were prepared by mixing DNA and EcSSB or DrSSB in 10 mm HEPES, pH 7.5, with varying NaCl concentrations (0, 20, 200, and 500 mm). Samples of EcSSB or DrSSB (25 μl of a 0.5 μm stock as tetramer or dimer, respectively) were mixed with 1.1 μl of M13mp18 circular ssDNA (at 4200 μm nucleotide) and allowed to bind for 10 min. Eight μl of 0-, 10-, and 400-fold reaction dilutions were then spread for 3 min to obtain suitable concentrations of SSB-ssDNA complexes for EM. Each grid was then touched to a drop of the respective buffer supplemented with 5% glycerol, followed by floating on a fresh drop of the same for 1 min, and then stained by touching to a drop of 5% uranyl acetate followed by floating on a fresh drop of the prior solution for 30 s. Finally, the grid was washed by touching to a drop of double-distilled water followed by complete immersion (twice) in fresh double-distilled water. Following drying, the samples were rotary-shadowed with platinum. This protocol is designed for visualization of complete reaction mixtures, and no attempt was made to remove unbound material. Although this approach leads to a high background of unbound proteins, it yields results that give a true insight into reaction components. SSB-ssDNA complexes were compared at identical magnifications for each sample. Imaging and photography were carried out with a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 charge-coupled device camera. Digital images of the SSB-ssDNA complexes were taken at 15,000× and 42,000× magnification. Particle dimensions were approximated by manual measurements of width, height, and surface area using the manual elliptical fit in Metamorph (Molecular Dynamics). Complexes analyzed were those from 200–500 mm NaCl conditions. Measurements were done on ∼500 discernible DrSSB and ∼550 EcSSB structures from multiple images.
Cellular DrSSB Protein Analysis
D. radiodurans R1 was grown at 30 °C in 200 ml of 2× TGY medium to an A600 nm of 2 and pelleted. The pellet was resuspended in 0.5 ml of 2× TGY and irradiated on ice in a Mark I 137CsCl irradiator (J.L. Shepherd and Associates) for 430 min at an estimated dose rate of 7.2 Gy/min to a total dose of 3 kGy. The cells were then recovered in 200 ml of 2× TGY medium at 30 °C, and 1-ml aliquots were harvested and pelleted at the times indicated. Each aliquot was resuspended in 10 mm Tris-HCl, pH 8.0, 1 mm EDTA and normalized by the A600 nm of each culture. For each aliquot, an equivalent volume of 6× Laemmli gel loading buffer was added, and the cell extracts were boiled for 5 min before being snap frozen in liquid nitrogen and stored at −80 °C until use.
Purified DrSSB (0.5 pmol) and 5-μl aliquots from each time point were resolved using SDS-PAGE on a 12% gel and electrotransferred onto an Immobilon PVDF membrane (Millipore) using standard procedures. The membrane was blocked with PBS-T (10 mm sodium phosphate, 150 mm NaCl, 0.05% Tween 20, pH 7.5) supplemented with 5% milk and incubated for 2 h at 21 °C with affinity-purified chicken anti-DrSSB polyclonal IgY antibody (GeneTel Laboratories, LLC, Madison, WI). After extensive washes in PBS-T, the membrane was incubated with rabbit anti-chicken IgG coupled with horseradish peroxidase (Sigma), washed again, and developed using the SuperSignal West Pico kit (Thermo Scientific).
Immunofluorescence
Immunofluorescence measurements of D. radiodurans cells were similar to those previously employed (40), with some adaptations. D. radiodurans R1 was grown to exponential phase A600 = 0.08–0.15 (1 A600 ∼5 × 108 cells/ml) in 2× TGY. Cell cultures were pelleted, washed with 1 ml M9 minimal buffer (42.3 mm Na2HPO4, 22.0 mm KH2PO4, 8.56 mm NaCl, 18.7 mm NH4Cl, 1 mm MgSO4, 0.1 mm CaCl2), pelleted again, resuspended in 0.5 ml of M9 minimal buffer, and irradiated with 0, 1, or 3 kGy of ionizing radiation. After irradiation, cells were pelleted, resuspended in 2× TGY, and grown at 30 °C with shaking. One-ml aliquots were removed at the indicated time points and fixed with one-tenth volume of 37% formaldehyde overnight at 4 °C and then pelleted. Fixed cell pellets were washed with PBS and permeabilized with 2 mg/ml lysozyme for 30 min at 37 °C with occasional mixing. Permeabilized cells were pelleted, incubated in 0.5 ml of 0.1% Triton X-100 at room temperature for 5 min, and then washed three times in 1× PBS (137 mm NaCl, 10 mm phosphate, 2.7 mm KCl, pH 7.4) and resuspended in 20 μl of 1× PBS. Samples were applied to poly-l-lysine microscope slides (Electron Microscope Sciences, Hatfield, PA), air-dried, and fixed with 4% formaldehyde for 20 min at 37 °C. The slides were blocked overnight in PBS-T supplemented with 0.1% Tween 20, 2% BSA overnight and incubated in 1:1000 diluted affinity-purified primary anti-DrSSB chicken IgY for 2 h at 37 °C. The slides were rinsed and washed two times in PBST with shaking. Incubation with 1:500 diluted FITC-conjugated goat anti-chicken secondary antibodies (Abcam) in blocking buffer was done at 37 °C for 1 h. DAPI and FM4–64 (Invitrogen) were added to slides, followed by one drop of ProLong antifade (Invitrogen). All slides were examined on a wide field Leica microscope at ×63 with 1.4 numerical aperture and ×1.6 C-mount magnification. Images were captured with a Cooke Sensicam QE fluorescence camera equipped with DAPI, TRITC, and FITC/GFP filters and were analyzed using Slidebook version 4.0.
RESULTS
X-ray Crystal Structure of DrSSB-ssDNA Complex
High resolution structural information describing bacterial SSB-ssDNA complexes has thus far been limited to homotetrameric SSBs (19–22). Because the homodimeric D/T group SSBs have non-identical OB domains present in each monomer, it has been suggested that the DNA binding properties of each domain may differ for this group of SSBs (15). A crystallographic approach was taken to determine the ssDNA-bound structure of DrSSB, the founding member of the D/T group homodimeric SSBs. Crystals of DrSSB in complex with homopolymeric ssDNA (1:2 DrSSB (as dimer)/dT35 molar ratio) were generated, and the structure was determined to 2.4 Å resolution by molecular replacement (Fig. 1 and Table 1). The crystallographic asymmetric unit contained three DrSSB monomers, two of which were present as an apparent dimer, whereas the third formed a dimer with a crystallographic equivalent in an adjacent asymmetric unit (supplemental Fig. 1). Fo − Fc difference electron density identified ssDNA bound to each DrSSB monomer (Fig. 1 and supplemental Fig. 1). Only short ssDNA segments of 1–5 bases were observed, which could be due to destabilization of portions of the DrSSB-ssDNA interface by the crystallization conditions.
FIGURE 1.
Crystal structure of the DrSSB-ssDNA complex. A, ribbon diagram of the DrSSB dimer-ssDNA complex. Visible stretches of ssDNA are colored in yellow (bases) and orange (backbone). Single bases have been omitted for clarity. DrSSB is colored to depict structural features: N-terminal OB fold (salmon), C-terminal OB fold (light blue), β-hairpin linker (cream), and tip of the L45 loop (green). B, SSB65 binding mode of the EcSSB-ssDNA complex (19). C, overlaid DrSSB monomers and bound ssDNA (>2 bases), colored as in A. D, 2Fo − Fc electron density (contoured at 1.0 σ) highlighting a stretch of bound ssDNA.
TABLE 1.
X-ray crystallographic data collection and structure refinement statistics
| Data collection | |
| Space group | C2 |
| Unit cell | |
| a, b, c (Å) | 161.1, 95.7, 65.7 |
| α, β, γ (degrees) | 90, 94.1, 90 |
| Wavelength, Å | 0.97869 |
| Resolution (Å) | 50.0–2.40 (2.49–2.40)a |
| Rsymb | 0.063 (0.272) |
| I/σ(I) | 27.1 (3.1) |
| Completeness (%) | 93.9 (62.6) |
| Redundancy | 3.3 (2.3) |
| Refinement | |
| Resolution (Å) | 50.0–2.40 |
| R/Rfreec | 0.203/0.261 |
| No. of atoms | |
| Protein | 4997 |
| DNA | 575 |
| Waters | 106 |
| Average B factors (Å2) | |
| Protein atoms | 53.6 |
| DNA atoms | 80.1 |
| Waters | 52.1 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.0125 |
| Bond angles (degrees) | 1.664 |
| Ramachandran statistics | |
| Residues in core region (%) | 93.2 |
| Residues in allowed region (%) | 6.2 |
| Residues in generously allowed regions (%) | 0.6 |
| Residues in disallowed regions (%) | 0 |
a Values in parentheses are for the highest resolution shell.
b Rsym = ΣΣj|Ij − 〈I〉|ΣIj, where Ij is the intensity measurement for reflection j, and 〈I〉 is the mean intensity for multiply recorded reflections.
c Rwork/Rfree = Σ‖Fo| − |Fc‖/|Fo|, where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections (5% of the total) were held aside throughout all stages of refinement.
Although the amount of visible ssDNA was limited in the DrSSB-dT35 crystal structure, the observed path for ssDNA binding was quite similar to that of the homotetrameric E. coli SSB (Fig. 1, compare A (left) and B). Also, each of the three DrSSB monomers in the asymmetric unit employs ssDNA-binding surfaces that are similar to those used by other SSBs (Fig. 1C), pointing to a common mechanism of DNA binding by dimeric and tetrameric SSBs. DNA binding induced very few apparent changes in the structure of DrSSB, as evidenced by modest root mean square deviations of atomic positions in the DNA-bound structure compared with the apo-DrSSB structure (15) (root mean square deviation (all atom/main chain) for each monomer is 1.00 Å/0.66 Å, 1.18 Å/0.83 Å, and 1.07 Å/0.71 Å). One notable difference, however, was that the L45 loops of the N-terminal OB folds were ordered in the DNA-bound structure but not in the apo structure (15). Ordering of the L45 region is probably due to its direct participation in DNA binding. The positions of the L45 loops differed somewhat among the three monomers, consistent with mobility in this element (Fig. 1C).
As has been observed with other SSB-ssDNA structures (19–22), DrSSB binds through a combination of apparent base stacking, electrostatic, and polar interactions (Fig. 1 and supplemental Table 1). Several DrSSB residues appear to share conserved ssDNA roles with analogous EcSSB residues. For example, three base-stacking residues in DrSSB (Trp-88, Tyr-53, and Tyr-177) are similarly positioned to base-stacking residues in EcSSB (EcSSB Trp-88 is analogous to DrSSB Trp-88, and EcSSB Trp-54 is analogous to both Tyr-53 from the DrSSB N-terminal OB domain and Tyr-177 from the C-terminal OB domain) (19, 41). There are also some notable species-specific interactions between DrSSB and ssDNA in the complex. DrSSB residues Lys-102 and Lys-174, which project into a cleft formed between the N- and C-terminal OB domains, bind to ssDNA, but the residues in the same positions in EcSSB do not directly bind ssDNA (supplemental Fig. 2). DrSSB residue Glu-84 appears to help position the Lys residues for interaction with DNA. Thus, although the general path of ssDNA on the DrSSB surface and several interactions are conserved with the prototypical EcSSB, there are some distinct interaction surfaces present on DrSSB that could be important in specific formation of DrSSB-ssDNA complexes.
DrSSB-ssDNA Structures Are Salt-dependent
DrSSB and EcSSB bind ssDNA in distinct modes that depend upon solution conditions. DrSSB dimers occlude 45 or 50–55 nt in low or high salt concentrations, respectively, whereas EcSSB tetramers occlude 35 or 65 nt under similar respective conditions (23, 25). For EcSSB, these differences are linked to distinct gross morphological states that can be distinguished by EM (30). To examine whether DrSSB also binds ssDNA in distinct salt-dependent morphologies, DrSSB-M13mp18 ssDNA complexes formed under varying salt conditions were analyzed by EM (Fig. 2). In low NaCl concentrations that favor lower site sizes in both EcSSB and DrSSB, a condensed DrSSB-ssDNA morphology that is strikingly similar to that of EcSSB-ssDNA complexes was observed (Fig. 2A). In these images, the SSB particles merge together in both DrSSB and EcSSB samples to produce condensed, protein-rich structures. However, under higher NaCl concentrations, more disperse discretely beaded structures were found for both DrSSB and EcSSB (Fig. 2, B–D). Interestingly, DrSSB required lower NaCl concentrations to trigger the transition from smooth to beaded morphologies. Structures that appear to be condensed but more loosely associated DrSSB particles were found in 20 mm NaCl (Fig. 2B); these DrSSB particles were easily resolved at 200 and 500 mm NaCl (Fig. 2, C and D). However, EcSSB only formed a beaded structure in the 500 mm NaCl condition (Fig. 2D). These observations were consistent with DrSSB binding in salt-dependent morphologies that resemble those observed with EcSSB but with the transition between the two forms being triggered at lower salt concentrations (∼20 mm NaCl) (25).
FIGURE 2.

Salt-dependent morphologies of DrSSB-ssDNA and EcSSB-ssDNA complexes. DrSSB (left) and EcSSB (right) form distinct salt-dependent morphologies when bound to ssDNA. Salt concentrations are 0 mm NaCl (A), 20 mm NaCl (B), 200 mm NaCl (C), and 500 mm NaCl (D). Images are ×42,000 magnifications with a 100-nm scale bar shown in each. Enlarged images of SSB-ssDNA complexes are inset in each panel.
Individual DrSSB particles (sampled primarily in 200 and 500 mm NaCl conditions) had dimensions that varied from ∼70 × 70 Å to ∼130 × 150 Å. DrSSB particles had mean surface areas of 34,500 ± 7900 Å2 at 200 mm NaCl and 35,800 ± 8500 Å2 at 500 mm NaCl. Given that a dimer of DrSSB has maximum dimensions of ∼45 × 50 Å to ∼50 × 80 Å, depending on orientation, the observed particle size seen in DrSSB EM images fit well to a repeating unit of 1–2 DrSSB dimers/particle. EcSSB beads had similar average surface areas of 46,400 ± 11,800 Å2 and 38,400 ± 11,500 Å2 when measured at 200 and 500 mm NaCl, respectively. Similar fits for EcSSB-ssDNA indicated that ∼1–2 tetramers could account for the dimensions of high salt particles.
DrSSB Protein Levels Are Elevated after Ionizing Radiation
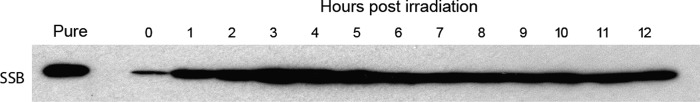
To better understand the roles of SSB in D. radiodurans DNA repair, the effect of a high dose of ionizing radiation (3 kGy) on DrSSB levels was examined using Western blot analysis (Fig. 3). A dramatic accumulation of DrSSB was observed as early as 1 h after ionizing radiation treatment. This accumulation increased to maximal levels by ∼3 h post-ionizing radiation, and elevated levels persisted throughout the 12-h time course. These results are consistent with a recent study in which DrSSB protein levels were shown to increase as early as 0.5 h post-ionizing radiation, reaching a maximum after 2 h and persisting for at least 6 h post-ionizing radiation (42, 43).
FIGURE 3.
Ionizing radiation-dependent accumulation of DrSSB. Western blot analysis of DrSSB protein levels in D. radiodurans cells recovering from treatment with 3 kGy of ionizing radiation. A purified DrSSB protein marker is shown in the left lane.
Ionizing Radiation-dependent SSB Focus Formation in D. radiodurans
The ionizing radiation-dependent accumulation of SSB suggested that SSB could be important in the robust repair mechanisms of D. radiodurans. To further assess the cellular roles of DrSSB, immunofluorescence microscopy was performed on D. radiodurans cells during exponential growth and during recovery from ionizing radiation to map the dynamics of DrSSB under diverse conditions (Fig. 4). D. radiodurans cells were grown to log phase, fixed, and permeated. Permeated cells were then incubated with DrSSB antibodies followed by a FITC-conjugated secondary antibody and visualized using wide field fluorescence microscopy. DNA was visualized with DAPI (blue filter).
FIGURE 4.
Ionizing radiation-dependent cellular dynamics of DrSSB. Shown is time-dependent cellular localization of DrSSB (FITC-conjugated secondary antibody; green filter) and the nucleoid (DAPI; blue filter) following treatment with 0, 1, or 3 kGy of ionizing radiation. DrSSB foci are formed immediately after ionizing radiation treatment (top row). After 1 h of recovery, DrSSB foci condense into a single focus in the center of each nucleoid (middle row). After 3 h of recovery, DrSSB is distributed in the cytosol and excluded from the nucleoid (bottom row). Insets, higher magnification views of the indicated sections. The arrows highlight representative DrSSB foci in each image.
Untreated D. radiodurans cells harvested from log phase had compact nucleoids, which were condensed and toroidal as has been previously observed (Fig. 4) (44). Under these conditions, DrSSB was distributed as multiple foci throughout the cytosol. Some DrSSB foci co-localized with DNA were also observed. These foci could mark sites of DNA replication as has been observed for EcSSB (45).
The nucleoid and SSB localization patterns were monitored after treatment with 1 and 3 kGy of ionizing radiation. Immediately after irradiation, DrSSB was concentrated as foci that appeared to co-localize with the nucleoid (Fig. 4). After 1 h of recovery, SSB localization was altered dramatically, with many of the SSB foci being condensed into a single distinct focus in the center of the nucleoid of most cells (Fig. 4). After an additional 2 h of recovery, the central SSB focus had dissipated in most cells, and the SSB signal was broadly distributed in the cytosol.
The dynamic localization behavior for DrSSB required ionizing radiation. Non-irradiated cultures had SSB foci that were far less intense than those observed in cells from the irradiated samples, consistent with the higher levels of SSB present in irradiated cells (Fig. 3). The foci that were observed in the non-irradiated sample were distributed both within and outside of the D. radiodurans nucleoid. Most notably, the non-irradiated cells entirely lacked the singular SSB focus observed in the irradiated cells, indicating that this feature was a consequence of ionizing radiation treatment.
DISCUSSION
To better understand how DrSSB binds ssDNA and to appreciate its roles in D. radiodurans DNA repair, we have used structural, biochemical, and cellular studies to examine the ssDNA binding mechanism of DrSSB and DNA damage-dependent cellular dynamics. Our crystallographic and EM studies of DrSSB-ssDNA complexes support a ssDNA binding mechanism for DrSSB that is largely conserved with homotetrameric SSBs. Similar surfaces are used for ssDNA binding by representatives from both major classes of SSBs, and comparable salt-dependent SSB-ssDNA complex morphologies are observed for DrSSB and EcSSB. In vivo, DrSSB protein levels were found to rise significantly upon ionizing radiation treatment, accumulating in intense foci within D. radiodurans cells during the ESDSA phase of DNA repair. DrSSB foci condense to form one central focus in the center of the nucleoid that is subsequently dispersed as genomic repair is completed, highlighting SSB dynamics as a potentially important aspect of the DNA repair processes in D. radiodurans cells.
Structure of DrSSB Bound to ssDNA Supports ssDNA Binding Mechanism That Is Conserved with Tetrameric SSBs
Prior to this study, structures of ssDNA-bound bacterial SSBs were only available for homotetrameric SSBs (19–22). Based on differences in the apo structures and DNA binding characteristics of the two classes of SSB, it was possible that homodimeric D/T group SSBs might bind ssDNA in a manner that is distinct from the homotetrameric SSBs found in most bacterial species. In contrast to this idea, the DrSSB-ssDNA complex structure reveals a DrSSB ssDNA-binding surface that is similar to that used in homotetrameric SSBs (Fig. 1). In many instances, ssDNA interactions are mediated by aromatic and basic residues in positions that are conserved in both DrSSB and EcSSB, supporting a model in which diverse bacterial SSBs bind ssDNA using common binding mechanisms (supplemental Table 1). There are some residues that appear to mediate species-specific ssDNA binding in the two proteins as well, but these do not dramatically alter the ssDNA binding paths of the proteins.
EM observations further highlighted similarities between DrSSB and the prototypical tetrameric EcSSB in ssDNA binding. Both SSBs bind ssDNA via a protein-dense “smooth” morphology under low salt conditions and with a beaded morphology at higher salt concentrations. DrSSB required lower salt concentrations than EcSSB for formation of the beaded morphology, transitioning at concentrations of ≤20 mm NaCl. This difference in salt-dependent ssDNA binding could be linked to differential cellular functions of DrSSB and EcSSB.
Cellular Dynamics of DrSSB Imply Role in Genome Reconstitution
The remarkable DNA repair capacity of D. radiodurans has been attributed to a two-phase process, both phases of which are likely to involve SSB. In the first phase, which occurs within ∼1 h of high doses of ionizing radiation, numerous DSBs are processed into ssDNA that can be used to prime DNA synthesis (12, 13). Barring its active exclusion, DrSSB would be expected to coat ssDNA exposed by this process. In the second phase, which occurs during the second half of the 3-h post-ionizing radiation period, the RecA recombinase catalyzes recombinational repair events in which the ssDNA invades homologous dsDNA genomic sequences. DrSSB plays an important role in processes mediated by DrRecA.5
Our observations have revealed intriguing aspects of DrSSB cellular behavior that parallel the timing of DNA repair in D. radiodurans. First, we have found that DrSSB accumulates in cells during the initial phase of genome reconstruction. It is possible that the increase in DrSSB levels is required to preserve newly generated ssDNA ends for their use as primers in ESDSA (13, 46). DNA damage-dependent DrSSB accumulation was also corroborated in a recently published study (42). Second, the cellular localization of SSB changes in an ionizing radiation- and time-dependent manner that parallels the phases of D. radiodurans DNA repair. DrSSB localization was distributed in the cytosol and nucleoid under non-damaging conditions, but, upon ionizing radiation treatment, numerous distinct SSB foci were localized on the nucleoid. These SSB foci subsequently condensed into one focus in the center of the cell as genomic repair progressed. This central focus does not form during the irradiation process, regardless of how long the irradiation proceeds (3 kGy takes 3 times longer to deliver than 1 kGy) but instead appears to require a recovery period to form. Condensation of damaged DNA into a form where its local concentration is increased may facilitate the homology-dependent DNA annealing and recombination processes that are central to the restoration of large chromosomes. This dynamic behavior suggests that SSB could be a central organizational/structural component in DNA repair in D. radiodurans. It is possible that the DNA damage-dependent condensation and subsequent dispersal of DrSSB are linked to the different modes of ssDNA binding. We have shown that these modes are modulated by ionic strength (Fig. 2), but other cues could trigger these transitions in vivo, including DrSSB concentrations, co-localization with other proteins, and/or post-translational modification. We and others have noted that DrSSB levels vary in a DNA damage-dependent manner, and it has been shown that the phosphorylation state of DrSSB is sensitive to genomic stress (42). Future studies will be required to determine whether these features play a role in the cellular DNA repair activities and observed dynamics of DrSSB.
Interestingly, localization dynamics that are very similar to those of DrSSB have been observed for an alternative D. radiodurans SSB (DdrB) that has important roles in DNA repair (47–49). Although DdrB binds ssDNA, it lacks the canonical OB DNA-binding fold shared among most SSBs. However, DdrB does contain a C-terminal tail that is nearly identical with that of DrSSB; this element mediates SSB interactions with numerous genome maintenance enzymes (32) and could help to orchestrate the remarkably efficient DNA repair process in D. radiodurans. Taken together with our findings, it appears likely that coordinated ssDNA binding mediated by SSB and DdrB and subsequent processing by DNA repair factors are important facets of the remarkably robust repair mechanisms of D. radiodurans.
Acknowledgment
We thank Dr. Matthew Brown (Louisiana State University) for assistance with immunofluorescence.
This work was supported, in whole or in part, by National Institutes of Health Grants GM068061 (to J. L. K.) and GM32335 (to M. M. C.).

This article contains supplemental Table 1 and Figs. 1 and 2.
The atomic coordinates and structure factors (code 3UDG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. V. Ngo and M. M. Cox, unpublished results.
- DSB
- double-stranded genomic break
- SSB
- ssDNA-binding protein
- DrSSB
- D. radiodurans SSB
- EcSSB
- E. coli SSB
- OB
- oligonucleotide-binding
- ESDSA
- extended synthesis-dependent single-strand DNA annealing
- D/T
- Deinococcus/Thermus
- nt
- nucleotide(s)
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Bauermeister A., Moeller R., Reitz G., Sommer S., Rettberg P. (2011) Effect of relative humidity on Deinococcus radiodurans' resistance to prolonged desiccation, heat, ionizing, germicidal, and environmentally relevant UV radiation. Microb. Ecol. 61, 715–722 [DOI] [PubMed] [Google Scholar]
- 2. Moseley B. E., Mattingly A. (1971) Repair of irradiation transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J. Bacteriol. 105, 976–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kitayama S. (1982) Adaptive repair of cross-links in DNA of Micrococcus radiodurans. Biochim. Biophys. Acta 697, 381–384 [DOI] [PubMed] [Google Scholar]
- 4. Blasius M., Sommer S., Hübscher U. (2008) Deinococcus radiodurans. What belongs to the survival kit? Crit. Rev. Biochem. Mol. Biol. 43, 221–238 [DOI] [PubMed] [Google Scholar]
- 5. Mattimore V., Battista J. R. (1996) Radioresistance of Deinococcus radiodurans. Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Battista J. R. (1997) Against all odds. The survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51, 203–224 [DOI] [PubMed] [Google Scholar]
- 7. Tanaka M., Earl A. M., Howell H. A., Park M. J., Eisen J. A., Peterson S. N., Battista J. R. (2004) Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White O., Eisen J. A., Heidelberg J. F., Hickey E. K., Peterson J. D., Dodson R. J., Haft D. H., Gwinn M. L., Nelson W. C., Richardson D. L., Moffat K. S., Qin H., Jiang L., Pamphile W., Crosby M., Shen M., Vamathevan J. J., Lam P., McDonald L., Utterback T., Zalewski C., Makarova K. S., Aravind L., Daly M. J., Minton K. W., Fleischmann R. D., Ketchum K. A., Nelson K. E., Salzberg S., Smith H. O., Venter J. C., Fraser C. M. (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova K. S., Aravind L., Wolf Y. I., Tatusov R. L., Minton K. W., Koonin E. V., Daly M. J. (2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox M. M., Battista J. R. (2005) Deinococcus radiodurans. the consummate survivor. Nat. Rev. Microbiol. 3, 882–892 [DOI] [PubMed] [Google Scholar]
- 11. Slade D., Radman M. (2011) Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 75, 133–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daly M. J., Minton K. W. (1996) An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178, 4461–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D., Petranovic M., Lindner A. B., Radman M. (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569–573 [DOI] [PubMed] [Google Scholar]
- 14. Eggington J. M., Haruta N., Wood E. A., Cox M. M. (2004) The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bernstein D. A., Eggington J. M., Killoran M. P., Misic A. M., Cox M. M., Keck J. L. (2004) Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl. Acad. Sci. U.S.A. 101, 8575–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dabrowski S., Olszewski M., Piatek R., Brillowska-Dabrowska A., Konopa G., Kur J. (2002) Identification and characterization of single-stranded DNA-binding proteins from Thermus thermophilus and Thermus aquaticus. New arrangement of binding domains. Microbiology 148, 3307–3315 [DOI] [PubMed] [Google Scholar]
- 17. Fedorov R., Witte G., Urbanke C., Manstein D. J., Curth U. (2006) 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 34, 6708–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jedrzejczak R., Dauter M., Dauter Z., Olszewski M., Długołecka A., Kur J. (2006) Structure of the single-stranded DNA-binding protein SSB from Thermus aquaticus. Acta Crystallogr. D Biol. Crystallogr. 62, 1407–1412 [DOI] [PubMed] [Google Scholar]
- 19. Raghunathan S., Kozlov A. G., Lohman T. M., Waksman G. (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 7, 648–652 [DOI] [PubMed] [Google Scholar]
- 20. Chan K. W., Lee Y. J., Wang C. H., Huang H., Sun Y. J. (2009) Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J. Mol. Biol. 388, 508–519 [DOI] [PubMed] [Google Scholar]
- 21. Yadav T., Carrasco B., Myers A. R., George N. P., Keck J. L., Alonso J. C. (2012) Genetic recombination in Bacillus subtilis. A division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsumoto T., Morimoto Y., Shibata N., Kinebuchi T., Shimamoto N., Tsukihara T., Yasuoka N. (2000) Roles of functional loops and the C-terminal segment of a single-stranded DNA-binding protein elucidated by x-ray structure analysis. J. Biochem. 127, 329–335 [DOI] [PubMed] [Google Scholar]
- 23. Witte G., Urbanke C., Curth U. (2005) Single-stranded DNA-binding protein of Deinococcus radiodurans. A biophysical characterization. Nucleic Acids Res. 33, 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Witte G., Fedorov R., Curth U. (2008) Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys. J. 94, 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kozlov A. G., Eggington J. M., Cox M. M., Lohman T. M. (2010) Binding of the dimeric Deinococcus radiodurans single-stranded DNA-binding protein to single-stranded DNA. Biochemistry 49, 8266–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dabrowski S., Olszewski M., Piatek R., Kur J. (2002) Novel thermostable ssDNA-binding proteins from Thermus thermophilus and T. aquaticus expression and purification. Protein Expr. Purif. 26, 131–138 [DOI] [PubMed] [Google Scholar]
- 27. Griffith J. D., Harris L. D., Register J., 3rd (1984) Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb. Symp. Quant. Biol. 49, 553–559 [DOI] [PubMed] [Google Scholar]
- 28. Bujalowski W., Lohman T. M. (1986) Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 25, 7799–7802 [DOI] [PubMed] [Google Scholar]
- 29. Lohman T. M., Overman L. B., Datta S. (1986) Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J. Mol. Biol. 187, 603–615 [DOI] [PubMed] [Google Scholar]
- 30. Chrysogelos S., Griffith J. (1982) Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl. Acad. Sci. U.S.A. 79, 5803–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lohman T. M., Green J. M., Beyer R. S. (1986) Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry 25, 21–25 [DOI] [PubMed] [Google Scholar]
- 32. Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haruta N., Yu X., Yang S., Egelman E. H., Cox M. M. (2003) A DNA pairing-enhanced conformation of bacterial RecA proteins. J. Biol. Chem. 278, 52710–52723 [DOI] [PubMed] [Google Scholar]
- 34. Neuendorf S. K., Cox M. M. (1986) Exchange of recA protein between adjacent recA protein-single-stranded DNA complexes. J. Biol. Chem. 261, 8276–8282 [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38. Dodson E. J., Winn M., Ralph A. (1997) Collaborative Computational Project, number 4. Providing programs for protein crystallography. Methods Enzymol. 277, 620–633 [DOI] [PubMed] [Google Scholar]
- 39. Grassucci R. A., Taylor D. J., Frank J. (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2, 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen H. H., de la Tour C. B., Toueille M., Vannier F., Sommer S., Servant P. (2009) The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol. Microbiol. 73, 240–252 [DOI] [PubMed] [Google Scholar]
- 41. Curth U., Greipel J., Urbanke C., Maass G. (1993) Multiple binding modes of the single-stranded DNA binding protein from Escherichia coli as detected by tryptophan fluorescence and site-directed mutagenesis. Biochemistry 32, 2585–2591 [DOI] [PubMed] [Google Scholar]
- 42. Basu B., Apte S. K. (2012) Gamma radiation-induced proteome of Deinococcus radiodurans primarily targets DNA repair and oxidative stress alleviation. Mol. Cell Proteomics 11, M111.011734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ujaoney A. K., Potnis A. A., Kane P., Mukhopadhyaya R., Apte S. K. (2010) Radiation desiccation response motif-like sequences are involved in transcriptional activation of the Deinococcal ssb gene by ionizing radiation but not by desiccation. J. Bacteriol. 192, 5637–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levin-Zaidman S., Englander J., Shimoni E., Sharma A. K., Minton K. W., Minsky A. (2003) Ringlike structure of the Deinococcus radiodurans genome. A key to radioresistance? Science 299, 254–256 [DOI] [PubMed] [Google Scholar]
- 45. Reyes-Lamothe R., Sherratt D. J., Leake M. C. (2010) Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328, 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Slade D., Lindner A. B., Paul G., Radman M. (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136, 1044–1055 [DOI] [PubMed] [Google Scholar]
- 47. Bouthier de la Tour C., Boisnard S., Norais C., Toueille M., Bentchikou E., Vannier F., Cox M. M., Sommer S., Servant P. (2011) The deinococcal DdrB protein is involved in an early step of DNA double strand break repair and in plasmid transformation through its single-strand annealing activity. DNA Repair 10, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugiman-Marangos S., Junop M. S. (2010) The structure of DdrB from Deinococcus. A new fold for single-stranded DNA-binding proteins. Nucleic Acids Res. 38, 3432–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Norais C. A., Chitteni-Pattu S., Wood E. A., Inman R. B., Cox M. M. (2009) DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J. Biol. Chem. 284, 21402–21411 [DOI] [PMC free article] [PubMed] [Google Scholar]