Background: Short human kininogen (HK) peptides can be used as therapeutic agents.
Results: The biological activity of the HK peptides is dramatically enhanced following insertion into GST.
Conclusion: The efficacy of short peptides can be enhanced by insertion into GST without affecting GST structure.
Significance: Intra-backbone insertions of small peptides into easily expressed, well characterized soluble proteins may help to create novel protein therapeutics.
Keywords: Anticancer Drug, Molecular Biology, Molecular Modeling, Peptides, Protein Chimeras, Protein Design
Abstract
The study of synthetic peptides corresponding to discrete regions of proteins has facilitated the understanding of protein structure-activity relationships. Short peptides can also be used as powerful therapeutic agents. However, in many instances, small peptides are prone to rapid degradation or aggregation and may lack the conformation required to mimic the functional motifs of the protein. For peptides to function as pharmacologically active agents, efficient production or expression, high solubility, and retention of biological activity through purification and storage steps are required. We report here the design, expression, and functional analysis of eight engineered GST proteins (denoted GSHKTs) in which peptides ranging in size from 8 to 16 amino acids and derived from human high molecular weight kininogen (HK) domain 5 were inserted into GST (between Gly-49 and Leu-50). Peptides derived from HK are known to inhibit cell proliferation, angiogenesis, and tumor metastasis, and the biological activity of the HK peptides was dramatically (>50-fold) enhanced following insertion into GST. GSHKTs are soluble and easily purified from Escherichia coli by affinity chromatography. Functionally, these hybrid proteins cause inhibition of endothelial cell proliferation. Crystallographic analysis of GSHKT10 and GSHKT13 (harboring 10- and 13-residue HK peptides, respectively) showed that the overall GST structure was not perturbed. These results suggest that the therapeutic efficacy of short peptides can be enhanced by insertion into larger proteins that are easily expressed and purified and that GST may potentially be used as such a carrier.
Introduction
Proteins are constructed in many cases from a number of structurally and/or functionally conserved modules that are genetically mobile and used repeatedly in the course of evolution (1, 2). These modules may be sufficiently large so as to constitute an entire protein domain or as small as a short peptide composed of 5–40 amino acids. In some cases, the latter may mimic selected biological activities of the full-length proteins from which they were derived, and some are currently being used clinically (e.g. Fuzeon, inhibitor of HIV-1 cell entry) (3, 4). However, the functional activity of many short peptides is usually substantially lower (50–200-fold) than that of their parental proteins (5–9). In general, this is due to their diminished solubility, stability, and/or enhanced propensity for aggregation (5–11). Short peptides are also quite flexible in solution and do not readily adapt to a specific functional conformation (5, 7–9). Introduction of chemical or structural constraints may reduce the conformational space of such peptides and enhance their biological activity (5–9). In some cases, this has been achieved through cyclization or introduction of intramolecular S–S bridges (5, 8, 9), changes that promote their higher level structural organization (5, 8, 9). However, currently, chemical methods for synthesis of large amounts of such modified peptides are costly, whereas production of recombinant fusion proteins containing these peptides on their N or C termini often does not allow sufficient structural constraint for enhancement of activity and/or solubility. As an alternative approach, short biologically active peptides can be inserted into the backbones of biologically inert (relevant to the targeted process) proteins that otherwise possess the desired properties of high solubility, stability, and ease of purification.
In this study, we present the design and functional characterization of engineered GST proteins carrying 8–16-mer peptide inserts derived from a sequence within domain 5 (D5)4 of human high molecular weight kininogen (HK). HK D5 contains endothelial cell-binding sites and inhibits angiogenesis through its ability to cause apoptosis of proliferating endothelial cells (6, 12, 13) and to inhibit endothelial cell proliferation and migration (14). Moreover, a histidine-glycine-lysine (HGK) motif derived from this domain blocks tumor metastasis (6, 10, 12–16). Although the exact mechanism of HGK peptide action has not been delineated (6, 10, 12–16), this motif nevertheless represents an attractive target for the design of antitumor peptide therapeutics/drugs.
By employing comparative modeling, we designed eight chimeric GST proteins (denoted GSHKTs) in which peptides ranging in size from 8- to 16-mers derived from HK D5 were inserted into Schistosoma japonicum GST (between Gly-49 and Leu-50). We produced all of the chimeric genes by insertional mutagenesis and expressed and purified the engineered proteins to homogeneity from Escherichia coli cells. GSHKTs were further characterized in terms of their thermostability (using differential scanning calorimetry (DSC)) and biological activity (by assessing their ability to inhibit human umbilical vein endothelial cell (HUVEC) proliferation in a dose-dependent manner). We found that although chimeric GSHKTs possessed decreased thermostability, they were able (with the exception of GSHKT8) to inhibit HUVEC proliferation. Specifically, GSHKT16 was 50–100-fold more active than the free ancestor 16-mer peptide (KHGHGHGKHKNKGKKN) alone. No inhibition was observed with the parental GST protein.
We also solved the crystal structures of GSHKT10 and GSHKT13 chimeras (harboring HK peptides of 10 and 13 amino acid residues in length, respectively) at 2.2 Å resolution and found that the overall GST structure was not perturbed, thus validating our design. Our results suggest that GST may potentially be used to express and display a number of short peptides ranging in length from at least 8 to 16 amino acid residues and that the biological activity of these peptides may be substantially enhanced upon insertion into the carrier GST.
EXPERIMENTAL PROCEDURES
S. japonicum GST Redesign
The GST crystal structure available from S. japonicum (Protein Data Bank code 1M99) was used as a starting point for the design. We used four major criteria in designing the chimeric proteins. 1) The inserted HGK peptide(s) must be solvent-accessible. 2) The insertion should introduce little or no side chain steric conflicts with pre-existing atoms. 3) There should be only minimal disruption of hydrogen bonding and hydrophobic interactions. 4) The glutathione-binding site should be largely unaffected. We began our redesign efforts by inserting the largest 16-mer HGK peptide (KHGHGHGKHKNKGKKN, denoted D5-13) and focusing our efforts on loops in GST. Loops were broken at different locations, peptide(s) were inserted, and comparative modeling was accomplished using the 3D-PSSM V2.6.0 algorithm (17) and/or DeepView (Swiss-PdbViewer). Models satisfying the design criteria (see above) were further chosen for experimental verification. Structures were visualized using PyMOL v0.98 (DeLano Scientific, San Carlos, CA) and DeepView 4.01.
Plasmids and Insertional Mutagenesis
The commercially available pGEX-6P-1 vector (GE Healthcare) was used as the source of GST DNA. Insertional mutagenesis was employed to produce hybrid GSHKT proteins. To produce GSHKT16, the pGEX-6P-1 vector was amplified by PCR using PfuTurbo® DNA polymerase (Agilent Technologies, Inc., Santa Clara, CA) and the following primers: 5′-GCGAAACAAAAAGTTTGAATTGGGTAAGCATGGTCATGGCCACGGAAAACATAAAAATAAAGGCAAAAAGAATTTGGAGTTTCCCAATCTTCC-3′ (forward) and 5′-GGAAGATTGGGAAACTCCAAATTCTTTTTGCCTTTATTTTTATGTTTTCCGTGGCCATGACCATGCTTACCCAATTCAAACTTTTTGTTTCGC-3′ (reverse) (with the sequence encoding the NH2-KHGHGHGKHKNKGKKN-COOH peptide in boldface). The methylated parental DNA was digested with DpnI, and the PCR product was transformed into E. coli XL10-Gold cells (Agilent Technologies, Inc.). GSHKT15, GSHKT14, GSHKT13, GSHKT12, GSHKT11, GSHKT10, and GSHKT8 chimeras, containing inserts of HGK peptides of 15 amino acids (NH2-KHGHGHGKHKNKGKK-COOH), 14 amino acids (NH2-KHGHGHGKHKNKGK-COOH), 13 amino acids (NH2-KHGHGHGKHKNKG-COOH), 12 amino acids (NH2-KHGHGHGKHKNK-COOH), 11 amino acids (NH2-KHGHGHGKHKN-COOH), 10 (NH2-KHGHGHGKHK-COOH), and 8 amino acids (NH2-KHGHGHGK-COOH), respectively, were produced following a similar strategy and using primers that were subsequently shorter at the region encoding the inserted peptide. The sequences of the mutagenized plasmids were confirmed by automated sequencing at the DNA Sequencing Core Facility of the Cleveland Clinic.
Protein Expression and Purification
GST and chimeric GSHKT proteins were expressed in E. coli BL21 cells and purified to homogeneity by affinity chromatography, followed by size exclusion chromatography on Superdex-75 (GE Healthcare). Typically, transformed cells were grown in 2 liters of LB culture medium, collected by centrifugation, and disrupted by sonication (three pulses of 15–20 s each) in PBS (137 mm NaCl, 11.9 mm phosphate, and 2.7 mm KCl) at pH 7.4–9.0. Cell debris was removed by centrifugation (4 °C, 16,000 rpm, 30 min; Beckman Ti-65 rotor), and the proteins were further purified on glutathione-Sepharose 4B beads (GE Healthcare). As a rule, the beads were incubated with the protein extract for 1 h at 4 °C, followed by incubation for 30 min at 24 °C with complete rotation. Seven to eight PBS washes were applied before chimeric proteins were eluted with 10 mm reduced glutathione in 50 mm Tris-HCl (pH 8.0). Proteins were further concentrated using Amicon Ultra-15 PLTK Ultracel-PL membrane (30 kDa) centrifugal filter units (Millipore, Billerica, MA) and subjected to gel filtration on a Superdex-75 column using an ÄKTATM purifier HPLC system (GE Healthcare).
CD Spectroscopy
The far-UV CD spectra were acquired on an AVIV Model 202 instrument using a 1-mm path length quartz cell at 25 °C. Protein concentrations were ∼0.25 mg/ml in standard PBS.
DSC
DSC was performed using a MicroCal VP-DSC microcalorimeter with a cell volume of 0.52 ml under a constant pressure of 30 p.s.i. at a scan rate of 1.5 °C/min in PBS. Protein concentrations were ∼0.25–0.5 mg/ml.
Cell Culture and Endothelial Cell Proliferation Assay
HUVECs were isolated from human umbilical cords, and proliferation assays were performed as described previously (12). The effects of recombinant GST, GSHKTs, HK (used as a positive control), and the D5-13 peptide on cell proliferation were determined by including the proteins in the cell cultures for 48 h. In brief, endothelial cells were cultured in gelatin-coated 96-well microplates at a concentration of 3 × 104 cells/ml in Medium 199 containing 2% fetal bovine serum. After incubation for 4 h (to allow cells to adhere and spread), the medium was replaced with fresh Medium 199 containing 10 μm Zn2+, 1% Cosmic calf serum (HyClone, Logan UT), and 10 ng/ml basic FGF (used as a positive control) in the presence or absence of test proteins. The percent inhibition of cell proliferation caused by GST, GSHKT chimeras, HK, and HK peptides was determined using the CellTiter 96® AQueous cell proliferation assay (Promega, Madison, WI).
Crystallization and Structure Determination
GSHKT13 and GSHKT10 crystals were grown using the sitting drop vapor diffusion method at 20 °C. GSHKT13 crystallized from 0.1 m HEPES (pH 7.5), 25% PEG 3350, and either 0.2 m ammonium sulfate or 0.2 m lithium sulfate. GSHKT10 crystals were grown from 0.1 m HEPES (pH 7.5), 200 mm KCl, 35% pentaerythritol propoxylate, and 0.1 m ATP. Crystals were stabilized in artificial mother liquor containing additional PEG 3350 (35%) or pentaerythritol propoxylate (40%) and flash-frozen by dipping in liquid nitrogen. Diffraction data were collected at Advanced Photon Source beamline 19-ID and processed using HKL (18). Both GSHKT10 and GSHKT13 structures were solved by molecular replacement using the wild-type GST structure (Protein Data Bank code 1M99) (19) and the program MOLREP (20). Iterative cycles of model building were carried out in Coot (21) and refinement in REFMAC (22). The GSHKT10 crystal contains two independent protein molecules in the asymmetric unit. Its final refined structure includes residues 2–36 and 50–216 in molecule A, residues 2–35 and 52–216 in molecule B, and 200 water molecules. The refined GSHKT13 crystal structure includes residues 2–39, HGK insert residue Gly-13′, residues 50–216, and 71 water molecules. Data and refinement statistics are provided in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s.d., root mean square deviation.
| Crystal |
||
|---|---|---|
| GSHKT10 | GSHKT13 | |
| Data collection | ||
| Space group | P212121 | P43212 |
| Unit cell dimensions (a, b, c; Å) | 58.43, 92.27, 93.05 | 92.52, 92.52, 58.09 |
| Wavelength (Å) | 0.97931 | 0.97931 |
| Resolution (Å) | 50–2.20 (2.28–2.20) | 50–2.20 (2.28–2.20) |
| Redundancy | 4.6 (3.2) | 9.1 (6.2) |
| Mean I/α (I) | 28.3 (10.0) | 32.2 (5.1) |
| Completeness (%) | 97.9 (99.1) | 96.0 (97.8) |
| Rmerge (%) | 4.9 (14.4) | 5.4 (36.2) |
| Refinement | ||
| Resolution (Å) | 50–2.20 (2.26–2.20) | 50–2.20 (2.26–2.20) |
| No. of reflections | 24,358 | 12,158 |
| Rwork (%) | 19.90 | 22.00 |
| Rfree (%) | 24.80 | 26.89 |
| No. of atoms (protein, water) | 3295, 200 | 1683, 71 |
| r.m.s.d. bond lengths (Å) | 0.013 | 0.015 |
| r.m.s.d. bond angles | 1.39° | 1.36° |
| Average B factor (protein, water; Å2) | 25.24, 30.86 | 40.74, 44.26 |
| Ramachandran plot (favored, allowed, disallowed; %) | 93.1, 6.9, 0.0 | 92.7, 7.3, 0.0 |
Miscellaneous
Molecular cloning was performed following the general procedures described by Sambrook et al. (23). SDS-PAGE was performed according to Laemmli (24).
RESULTS
GSHKT Design
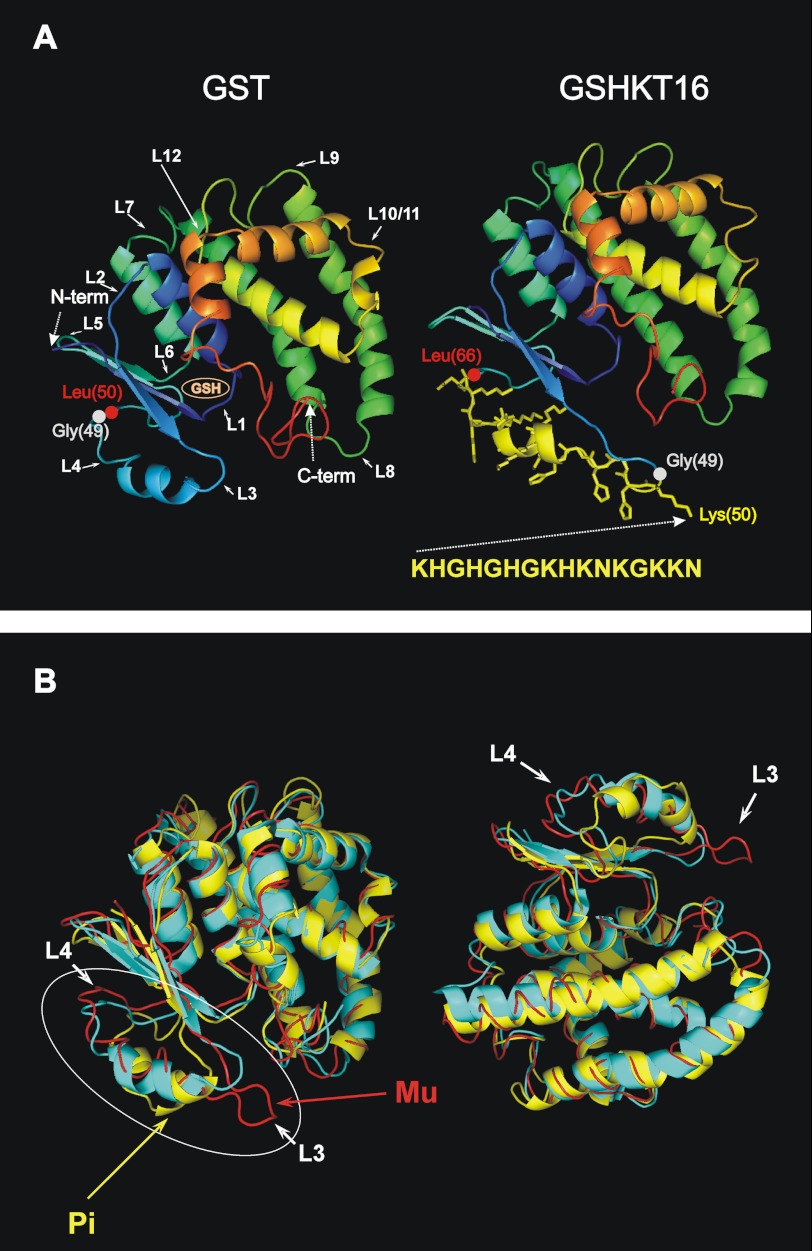
We used GST from S. japonicum for our redesign efforts aimed at producing the chimeric proteins, where the inserted HGK motif-containing peptides would be exposed on the surface and hopefully retain their biological activity and perhaps even possess increased activity due to the imposed structural constraints. We started our redesign by attempting intra-backbone insertion of the largest 16-mer HGK peptide (KHGHGHGKHKNKGKKN, denoted D5-13). One of the challenges in protein redesign is selecting an optimal solution for the enormous amount of sequence and structural possibilities. However, it is generally accepted that loops in proteins can be broken and engineered to have different functions without serious consequences to the overall fold of the protein (25–29). GST is a homodimer composed of two (218-amino acid residue) subunits, each of which is characterized by a modular structure with two spatially distinct domains (19, 30, 31). Domain I consists of a central four-stranded β-sheet flanked on one side by two α-helices and on the other side by a single helix, whereas domain II is entirely α-helical (Fig. 1A, left, with domain I blue). Of the 12 major turns/loops in S. japonicum GST connecting secondary structural elements (Fig. 1A) (19, 30), loop 1 was excluded from modification due to its close proximity to the glutathione-binding site; insertions at this position also introduce steric conflicts. Modifications of loops 2, 5, and 6 were excluded, as they would disrupt the central four-stranded β-sheet fragment in domain 1. Loop 7, which connects domains 1 and 2, was excluded due to our concern that mutations in this region would affect the global folding of the protein and the proper mutual packing of the two domains. Loop 7 is also involved in intersubunit contacts (19, 30). Insertions in loops 11 and 12 would likely disrupt proper packing of domain 2. Insertions in loops 3, 4, and 8–10 seemed feasible. Insertions in loop 4 (Leu-48–Pro-56) seemed at first not to be optimal, as a β-turn containing residues 50–53 is involved in intersubunit interactions and in forming the hydrophobic contact between Phe-52 of one chain and residues 91–94 and 129–133 of the other. However, because the D5-13 peptide (KHGHGHGKHKNKGKKN) is predicted (using a statistical algorithm (32)) to have the propensity to form an α-helix, we assumed that this α-helical region could be accommodated within the short α-helix formed by residues 38–42 of GST. Importantly, this helix is solvent-exposed (19, 31) yet shows the highest degree of structural flexibility and conformational variability between GSTs from different organisms and between different GST classes (e.g. between the helminth enzyme from S. japonicum used in this study (Protein Data Bank code 1M99) and the human Pi class (33) and Μu class (31, 33) enzymes (Protein Data Bank code 10GS and 1HNA, respectively) (Fig. 1B). Thus, we chose loop 4, which connects the domain I β-sheet with a short helix for the insertion. Because the β-turn (amino acids 50–53) is involved in hydrophobic subunit contacts (19, 30), we decided to make insertions prior to this turn, namely between Gly-49 and Leu-50. Comparative modeling revealed that insertion of 16 amino acids of an HGK motif-containing peptide at this site would not significantly perturb the GST structure (Fig. 1A, right).
FIGURE 1.
Comparison of GSHKT16 and GST structures. A, left, three-dimensional structure of S. japonicum GST depicted as a ribbon diagram of the monomer based on the crystal structure (Protein Data Bank code 1M99). Major turn/loop regions are denoted with numbers. Domain I is shown in blue. Right, structural model of the GSHKT16 protein. The side chains of the inserted residues are shown in yellow. B, superimposed three-dimensional structures (ribbon diagrams) of S. japonicum GST (code 1M99; in cyan) and the human Pi class (code 10GS; in yellow) and Mu class (1HNA; in red) enzymes. The most flexible/variable region comprising loops 3 and 4 is circled.
Expression, Purification, and Characterization GSHKT16 Chimera
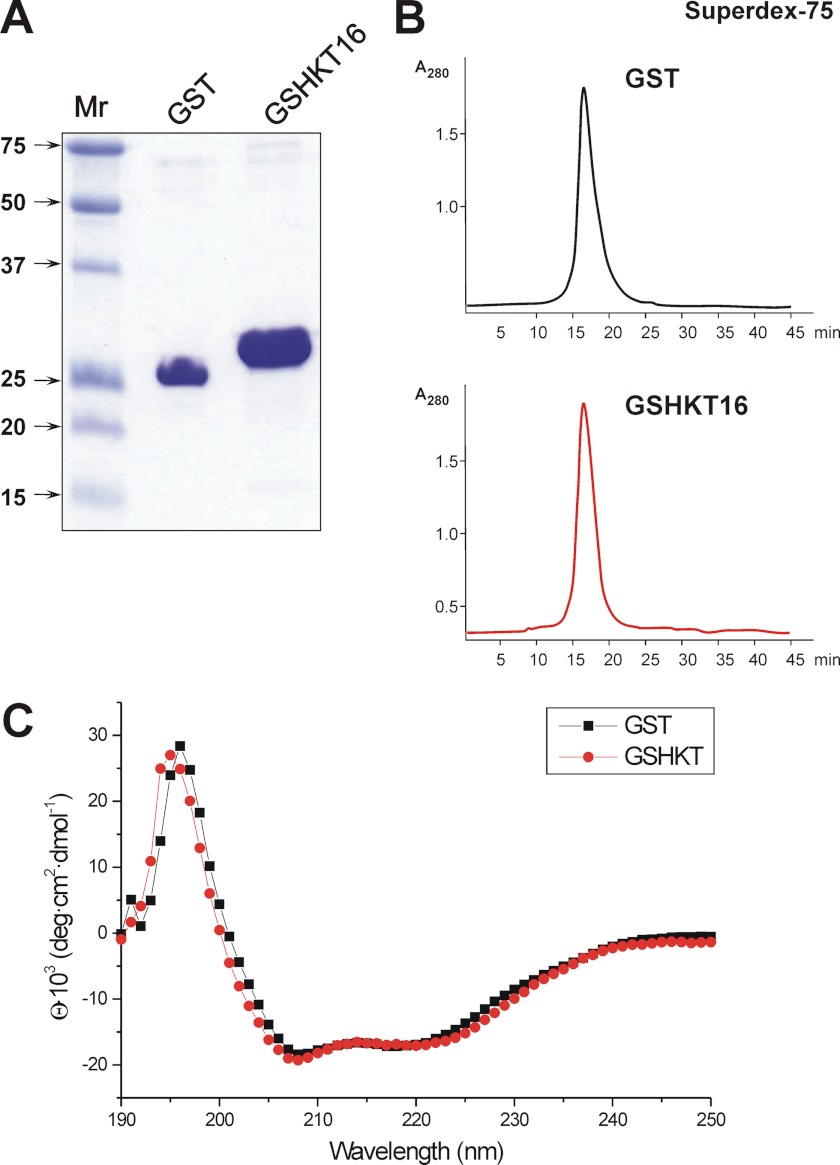
The chimeric GSHKT16 protein was constructed using insertional mutagenesis (as described under “Experimental Procedures”) and produced in E. coli BL21 cells. Recombinant GSHKT16 protein expressed in E. coli was found to be soluble and easily purified by affinity chromatography on glutathione-Sepharose beads (Fig. 2A). GSHKT16 also formed a dimer similarly to GST (Fig. 2B). Furthermore, the far-UV CD spectra obtained from 190 to 250 nm with a bandwidth of 1 nm at room temperature (25 °C) showed no major difference between the structure of GSHKT16 and native GST (Fig. 2C). This suggests that the hybrid protein satisfied the rational design criteria 2–4 as discussed under “Experimental Procedures.”
FIGURE 2.
Characterization of chimeric GSHKT16 protein. A, denaturing gel electrophoresis of the recombinant GST and GSHKT proteins. B, gel filtration of GST (upper) and GSHKT (lower) on Superdex-75. C, far-UV CD spectra of GST (black line) and GSHKT (red line) acquired using a 1-mm path length quartz cell at 25 °C. Protein concentrations were ∼0.25 mg/ml in PBS. deg, degrees.
GSHKT16 Possesses Enhanced Antiproliferative Activity Compared with Native GST and Free D5-13 Peptide
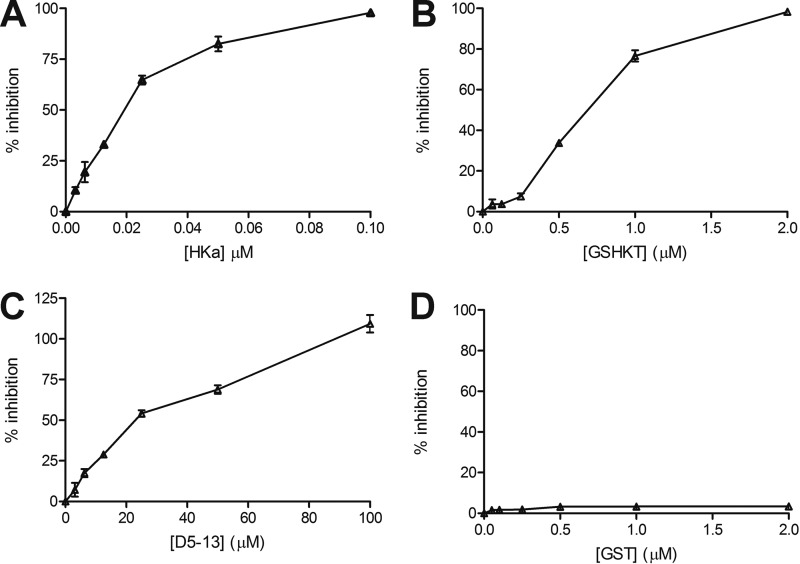
To assess the function of GSHKT16, we measured its ability to inhibit the proliferation of HUVECs. Although not as potent as full-length two-chain HK (Fig. 3A), GSHKT16 efficiently inhibited the proliferation of HUVECs in a dose-dependent manner (Fig. 3B) and was 50–100-fold more active than the free D5-13 peptide (KHGHGHGKHKNKGKKN) (IC50 for inhibition of proliferation of ∼0.6 μm versus ∼30 μm) (Fig. 3, B and C) (12). No inhibition was observed with the parental GST protein (Fig. 3D). These results suggest that D5-13 was indeed exposed on the surface of GST and presented in a conformational state with considerably greater anti-endothelial cell activity compared with the free peptide in solution. This conformational restraint might significantly enhance the interactions of D5-13 with an anti-angiogenic binding site expressed on proliferating endothelial cells (16).
FIGURE 3.
Inhibition of HUVEC proliferation. A, inhibition of HUVEC proliferation by two-chain HK (HKa). B, inhibition of HUVEC proliferation by GSHKT. C, inhibition of HUVEC proliferation by the D5-13 peptide. D, inhibition of HUVEC proliferation by GST.
GSHKT15, GSHKT14, GSHKT13, GSHKT12, GSHKT11, and GSHKT10 Chimeras Exhibit Antiproliferative Activity, whereas GSHKT8 Does Not
Although the chimeric GSHKT16 protein possessed the desired properties, it was prone to aggregation upon freezing and thawing or at high concentrations (>10–15 mg/ml), which precluded our attempts to crystallize it. It is known that an increase in loop length usually results in a decrease in protein stability (34, 35). Therefore, we decided to shorten the size of the insert, aiming to increase the stability of the chimeric protein without substantially decreasing its activity. GSHKT15, GSHKT14, GSHKT13, GSHKT12, GSHKT11, GSHKT10, and GSHKT8 chimeras contained truncated inserts of the D5-13 peptide, namely NH2-KHGHGHGKHKNKGKK-COOH, NH2-KHGHGHGKHKNKGK-COOH, NH2-KHGHGHGKHKNKG-COOH, NH2-KHGHGHGKHKNK-COOH, NH2-KHGHGHGKHKN-COOH, NH2-KHGHGHGKHK-COOH, and NH2-KHGHGHGK-COOH, respectively.
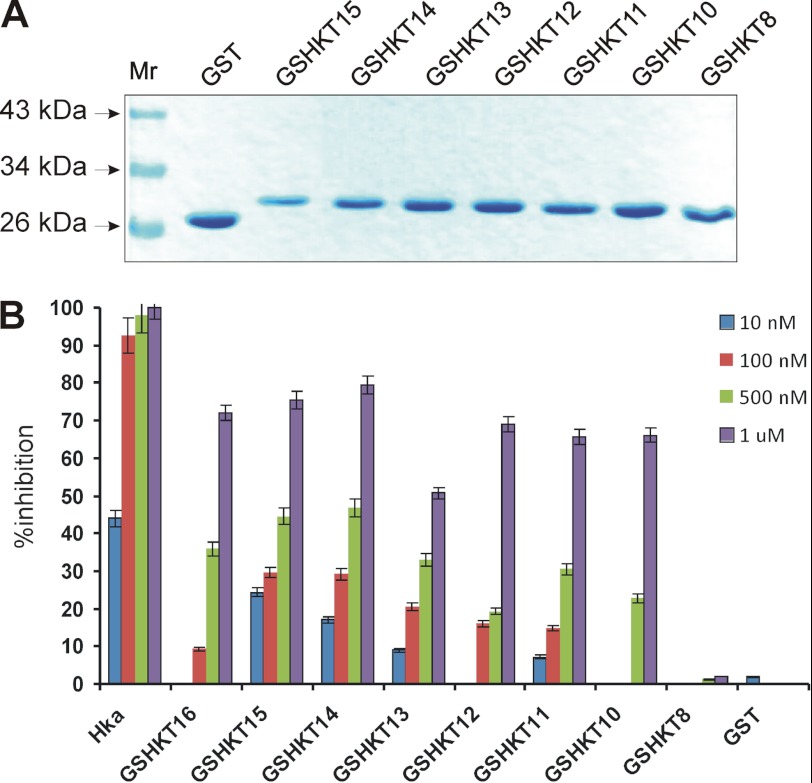
These proteins were produced following the same procedures as described for the GSHKT16 chimera. Recombinant chimeric proteins were expressed in E. coli, found to be soluble, and further purified to homogeneity using the same steps described for GSHKT16 (Fig. 4A). All of the above GSHKT chimeras were dimers (data not shown). GSHKT15, GSHKT14, GSHKT13, GSHKT12, GSHKT11, and GSHKT10 chimeras were able to inhibit the proliferation of HUVECs in a dose-dependent manner, whereas GSHKT8 (similar to GST) revealed no activity (Fig. 4B). We note that the activities of the GSHKT15 and GSHKT14 chimeras were increased in comparison with that of GSHKT16 at lower protein concentrations but were comparable with that of GSHKT16 at higher protein concentrations. Overall, the activity of GSHKT13, GSHKT12, GSHKT11, and GSHKT10 chimeras progressively decreased, which was particularly evident at lower protein concentrations (Fig. 4B).
FIGURE 4.
Chimeric GSHKT15, GSHKT14, GSHKT13, GSHKT12, GSHKT11, GSHKT10, and GSHKT8 proteins and inhibition of HUVEC proliferation. A, denaturing gel electrophoresis. B, inhibition of HUVEC proliferation. Hka, two-chain HK.
GSHKTs Reveal ∼8–10° Lower Phase Transition Temperatures Compared with GST
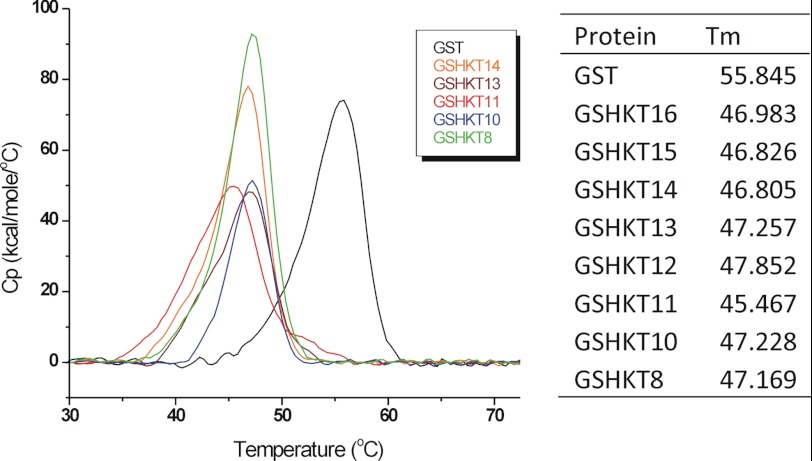
To gain further insight into the stability of the chimeric GSHKT proteins, we subjected them to DSC. The DSC results of GSHKT chimeras in comparison with the ancestor GST protein (36) showed ∼8–10° lower phase transition temperatures for the GSHKT chimeras compared with GST, indicating that the chimeric proteins were less stable than native GST (Fig. 5). Interestingly, the stability of all of the chimeras was quite similar independent of the length of the inserted peptides, although GSHKT10 and GSHKT8 demonstrated slightly greater stability (Fig. 5).
FIGURE 5.
Left, representative DSC profiles of GST and chimeric GSHKT proteins (Cp versus temperature) at a scan rate of 1 °C/min (PBS). Right, exact values of Tm.
Crystal Structures of GSHKT13 and GSHKT10 Confirm That Insertion of Flexible HGK Peptides Is Compatible with Native GST Structure
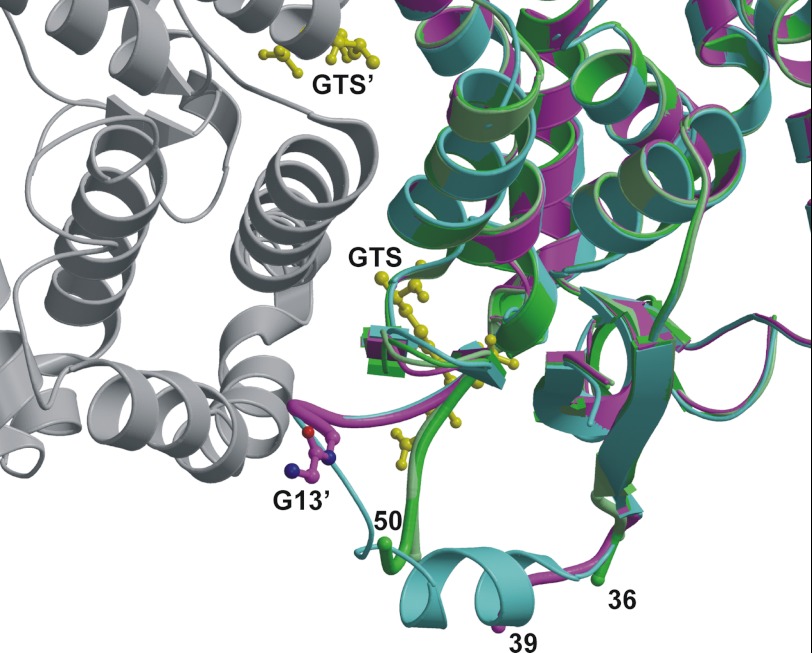
To evaluate the structure of the GSHKT chimeras, crystallization trials were carried out with all eight chimeras. Only GSHKT10 and GSHKT13 crystallized (Table 1), and their structures were determined by molecular replacement. Similar to WT GST, the two chimeras are dimeric: the GSHKT10 crystal contains a dimer in the asymmetric unit, whereas the two halves of the GSHKT13 dimer are related by crystallographic symmetry. The overall protein folds of the two chimeric structures are very similar to that of WT GST (Fig. 6); superposition of Protein Data Bank entry 1M99 onto GSHKT10 and GSHKT13 gave root mean square differences of ∼0.4 Å for ∼200 equivalent Cα atoms. This average coordinate difference is very similar to the average coordinate error of ∼0.3 Å estimated by the Luzzati method. In GSHKT10, the lack of interpretable electron density reveals that residues 37–49, including the 10-residue insert, are disordered. GSHKT10 has the WT conformation up to Asp-36/Arg-35 (in molecules A/B) and resumes the WT conformation at Leu-55. A few loop residues C-terminal to the insert (Leu-50–Asn-54 and Phe-52–Asn-54 in molecules A and B, respectively) deviate from WT conformation such that they shift away from the glutathione-binding site and dimer interface into a space that is occupied by a surface helix in WT GST. In contrast, GSHKT13 follows the WT structure more closely: it has the WT conformation up to Asp-39 and resumes at the insertion site of Leu-50. Interpretable electron density allowed the placement of the C-terminal Gly-13′ residue of the 13-residue insert, extending from Leu-50 (Fig. 6). In both GSHKT10 and GSHKT13, insertion of the HGK peptides caused a dramatic increase in flexibility of a surface helix and loop near the glutathione-binding site. This disordering alters residues found to interact with the glutathione glycyl carboxylate (Trp-41, Lys-45, and Asn-54) (19). Other GST residues that interact with the γ-glutamate or cysteine moieties in glutathione in the WT structure are in similar conformations in both GSHKT10 and GSHKT13.
FIGURE 6.
Superimposed crystal structures of WT and chimeric GSTs. The two molecules of GSHKT10 (dark and light green) and GSHKT13 (purple) are shown as ribbon diagrams superimposed on one-half of the WT S. japonicum GST dimer (cyan and gray) (Protein Data Bank code 1M99). The Gly-13′ residue from the HGK insert in GSHKT13 (purple carbon atoms and bonds) and two glutathione sulfonate (GTS) molecules (yellow) bound to the WT GST dimer are drawn as ball-and-stick structures. Residues 37–49(+insert) and 40–49(+insert) are disordered in GSHKT10 and GSHKT13, respectively. The figure was prepared with MOLSCRIPT (43) and Raster3D (44).
DISCUSSION
The rational design of novel proteins may provide additional insights into protein structure, folding, and function. Since initial attempts, the field of protein design has progressed to the point that it is now possible to consider tertiary and quaternary structures and even functional elements for construction of new proteins (28, 37–39). In this study, we presented the design of engineered GST proteins carrying inserts of the HGK-rich motif derived from a sequence within D5 of HK. HK D5 contains endothelial cell-binding sites and inhibits angiogenesis through its ability to cause apoptosis of proliferating endothelial cells and to inhibit endothelial cell proliferation and migration (6, 10, 12–16). Moreover, HGK peptides were found to block tumor metastasis (6, 10, 12–16). Although the D5 peptide region within the context of intact high molecular weight kininogen or recombinant D5 induces endothelial cell apoptosis with an IC50 in the low nanomolar range, the isolated D5 peptides express at least 2 logs lower activity, presumably due to enhanced flexibility and a failure to maintain the appropriate conformation for meaningful interactions with endothelial cells (6, 10, 12–16). In this study, we demonstrated that constraining flexibility of these peptides through insertion into the GST backbone enhances their activity. Although the activity of the GSHKT proteins is still less than that of two-chain HK D5, presumably due to specific D5 conformational characteristics or additional tertiary interactions, our studies demonstrate that the use of GST to isolate specific peptides in a conformationally restricted form for further study or potential biological applications is worthy of additional efforts.
It has been previously reported that constraining methods (5–9) confer significant biological activity to short peptides (5–9). Cyclization or introduction of intramolecular S–S bridges has been shown to promote structural organization and enhance the biological activity of short peptides derived from, for example, LFA-1 (leukocyte function-associated antigen 1) and ICAM-1 (intercellular adhesion molecule 1) (5). Introduction of chemical constraints and crosslinks has also been shown to enhance the structural organization and biological activity of short peptides derived from FGF and HIV-1 gp41 protein, for example (8, 9).
As an alternative approach, short biologically active peptides can be inserted into the backbones of biologically inert (relevant to the targeted process) proteins that otherwise possess the desired properties of high solubility, stability, and ease of purification. This novel approach may prove useful in defining the functions of small peptides and potentially provide insight into the development of new therapeutic agents.
We used GST from S. japonicum because the protein is well studied and is widely used for affinity purification of fusion proteins expressed in E. coli (26). In addition, several crystallographic structures are available (19, 30, 31), and importantly, GST does not inhibit cell proliferation (6, 16). However, other backbones/carrier proteins also may potentially be used.
It should also be noted that production of HGK motif-containing peptides expressed as fusions with various recombinant proteins has faced numerous problems in the past, and these efforts did not enhance their biological activity (6, 12, 13, 16). For example, production of HK D5 in E. coli as a calmodulin fusion protein resulted in inclusion bodies, requiring renaturation to re-establish biological activity (13). The GSHKT protein generated in the course of these studies proved to be soluble and may also be valuable in further characterizing the biological effects of D5. Moreover, these constructs may be of value in defining the active structure of this intriguing protein domain, which until now has escaped structural characterization.
We note, however, that the chimeric GSHKT proteins are thermodynamically less stable than isolated GST. It has been suggested that loops do not actively dictate the final protein fold (25, 29, 34, 35, 39). However, an increase in loop length usually leads to a decrease in protein stability (34, 35). For example, insertion of a series of glycine linkers (up to 10 residues in length) into a natural two-residue loop in a four-helix bundle protein (Rop) led to a progressive decrease in stability of Rop toward thermal and chemical denaturation (34). Similarly, insertions of short unstructured sequences (four glycines and/or four threonines) into the loops of yeast phosphoglycerate kinase did not affect the enzyme folding but led to a decreased stability of the protein (35). Interestingly, the effect per residue on stability was larger for the first inserted residue than for all of the subsequent ones (35). Our GSHKT10 and GSHKT13 crystal structures indicate that insertions of 10 and 13 residues increase the flexibility of a surface loop significantly, providing an explanation for the observed decrease in thermodynamic stability for these chimeras. In contrast, insertions of 7–13-residue stretches of glycine, alanine, and glutamine repeats had minimal effects on the stability and folding of chymotrypsin inhibitor-2 (39). This suggests that local environment is an important factor and that the presence of certain (e.g. unstructured) regions in proteins does not necessarily have a great impact on energetic penalty (39).
Although a variety of strategies have emerged for rational design of proteins, including manipulation of primary structure, incorporation of chemical and post-translational modifications, and utilization of fusion partners (28, 37, 38, 40–42), successful efforts have often resulted from trial and error (see Ref. 37 for a review). A smaller number of examples exist whereby rational design has achieved the desired result (see Ref. 37 and references therein). For biologically active peptides to function as therapeutic agents, efficient production, high solubility, and retention of biological activity are required. The stability and solubility (but not the activity) of therapeutic peptides can sometimes be enhanced through fusion to a soluble protein with a long biological half-life (e.g. albumin fusion with the anticoagulant proteins/peptides hirudin and barbourin) (see Ref. 40 for a review). Intra-backbone insertions of small peptides into easily expressed, well characterized soluble proteins may provide a unique and different solution to the problem.
Acknowledgments
We thank Drs. Nelson Phillips and Rupinder Singh (Case Western Reserve University School of Medicine) for assistance with the CD measurements. We also thank Drs. Joyce Jentoft, Allison Miketa, and Pamela Hall (Case Western Reserve University School of Medicine) for helpful discussions. X-ray diffraction data were measured at Advanced Photon Source beamline 19-ID at the Argonne National Laboratory, which is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants HL089796 (to K. R. M.), GM68079 (to W. C. M.), and DK075897 and GM61388 (to V. C. Y. and Y. P.). This work was supported by National American Heart Association Grant 0730120N (to A. A. K.).
The atomic coordinates and structure factors (codes 4ECB and 4ECC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- D5
- domain 5
- HK
- human high molecular weight kininogen
- DSC
- differential scanning calorimetry
- HUVEC
- human umbilical vein endothelial cell.
REFERENCES
- 1. Moult J., Melamud E. (2000) From fold to function. Curr. Opin. Struct. Biol. 3, 384–389 [DOI] [PubMed] [Google Scholar]
- 2. Söding J., Lupas A. N. (2003) More than the sum of their parts: on the evolution of proteins from peptides. BioEssays 25, 837–846 [DOI] [PubMed] [Google Scholar]
- 3. Greenberg M., Cammack N., Salgo M., Smiley L. (2004) HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 14, 321–337 [DOI] [PubMed] [Google Scholar]
- 4. Miyamoto F., Kodama E. N. (2012) Novel HIV-1 fusion inhibition peptides: designing the next generation of drugs. Antivir. Chem. Chemother. 22, 151–158 [DOI] [PubMed] [Google Scholar]
- 5. Tibbetts S. A., Seetharama Jois D., Siahaan T. J., Benedict S. H., Chan M. A. (2000) Linear and cyclic LFA-1 and ICAM-1 peptides inhibit T cell adhesion and function. Peptides 21, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 6. Zhang J. C., Claffey K., Sakthivel R., Darzynkiewicz Z., Shaw D. E., Leal J., Wang Y. C., Lu F. M., McCrae K. R. (2000) Two-chain high molecular weight kininogen induces apoptosis and inhibits angiogenesis: partial activity within domain 5. FASEB J. 14, 2589–2600 [DOI] [PubMed] [Google Scholar]
- 7. Church W. B., Inglis A. S., Tseng A., Duell R., Lei P. W., Bryant K. J., Scott K. F. (2001) A novel approach to the design of inhibitors of human secreted phospholipase A2 based on native peptide inhibition. J. Biol. Chem. 276, 33156–33164 [DOI] [PubMed] [Google Scholar]
- 8. Sia S. K., Carr P. A., Cochran A. G., Malashkevich V. N., Kim P. S. (2002) Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. U.S.A. 99, 14664–14669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kiyota S., Franzoni L., Nicastro G., Benedetti A., Oyama S., Jr., Viviani W., Gambarini A. G., Spisni A., Miranda M. T. (2003) Introduction of a chemical constraint in a short peptide derived from human acidic fibroblast growth factor elicits mitogenic structural determinants. J. Med. Chem. 46, 2325–2333 [DOI] [PubMed] [Google Scholar]
- 10. Daly M. E., Makris A., Reed M., Lewis C. E. (2003) Hemostatic regulators of tumor angiogenesis: a source of anti-angiogenic agents for cancer treatment? J. Natl. Cancer Inst. 95, 1660–1673 [DOI] [PubMed] [Google Scholar]
- 11. Fowler S. B., Poon S., Muff R., Chiti F., Dobson C. M., Zurdo J. (2005) Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin. Proc. Natl. Acad. Sci. U.S.A. 102, 10105–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J. C., Qi X., Juarez J., Plunkett M., Donaté F., Sakthivel R., Mazar A. P., McCrae K. R. (2002) Inhibition of angiogenesis by two-chain high molecular weight kininogen (HKa) and kininogen-derived polypeptides. Can. J. Physiol. Pharmacol. 80, 85–90 [DOI] [PubMed] [Google Scholar]
- 13. Zhang J. C., Donate F., Qi X., Ziats N. P., Juarez J. C., Mazar A. P., Pang Y. P., McCrae K. R. (2002) The anti-angiogenic activity of cleaved high molecular weight kininogen is mediated through binding to endothelial tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 99, 12224–12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colman R. W., Jameson B. A., Lin Y., Johnson D., Mousa S. A. (2000) Domain 5 of high molecular weight kininogen (kininostatin) down-regulates endothelial cell proliferation and migration and inhibits angiogenesis. Blood 95, 543–550 [PubMed] [Google Scholar]
- 15. Kamiyama F., Maeda T., Yamane T., Li Y. H., Ogukubo O., Otsuka T., Ueyama H., Takahashi S., Ohkubo I., Matsui N. (2001) Inhibition of vitronectin-mediated haptotaxis and haptoinvasion of MG-63 cells by domain 5 (D5H) of human high molecular weight kininogen and identification of a minimal amino acid sequence. Biochem. Biophys. Res. Commun. 288, 975–980 [DOI] [PubMed] [Google Scholar]
- 16. Kawasaki M., Maeda T., Hanasawa K., Ohkubo I., Tani T. (2003) Effect of His-Gly-Lys motif derived from domain 5 of high molecular weight kininogen on suppression of cancer metastasis both in vitro and in vivo. J. Biol. Chem. 278, 49301–49307 [DOI] [PubMed] [Google Scholar]
- 17. Kelley L. A., MacCallum R., Sternberg M. J. E. (1999) RECOMB 99 Proceedings of the Third Annual Conference on Computational Molecular Biology (Istrail S., Pevzner P., Waterman M., eds) pp. 218–225, Association for Computing Machinery, New York [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Cardoso R. M., Daniels D. S., Bruns C. M., Tainer J. A. (2003) Characterization of the electrophile-binding site and substrate binding mode of the 26-kDa glutathione S-transferase from Schistosoma japonicum. Proteins 51, 137–146 [DOI] [PubMed] [Google Scholar]
- 20. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Fritsch F. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Sibanda B. L., Thornton J. M. (1993) Accommodating sequence changes in β-hairpins in proteins. J. Mol. Biol. 229, 428–447 [DOI] [PubMed] [Google Scholar]
- 26. Thanki N., Zeelen J. P., Mathieu M., Jaenicke R., Abagyan R. A., Wierenga R. K., Schliebs W. (1997) Protein engineering with monomeric triose-phosphate isomerase (monoTIM): the modeling and structure verification of a seven-residue loop. Protein Eng. 10, 159–167 [DOI] [PubMed] [Google Scholar]
- 27. Regan L. (1999) Protein redesign. Curr. Opin. Struct. Biol. 9, 494–499 [DOI] [PubMed] [Google Scholar]
- 28. Pokala N., Handel T. M. (2001) Review: protein design–where we were, where we are, where we're going. J. Struct. Biol. 134, 269–281 [DOI] [PubMed] [Google Scholar]
- 29. Kuhlman B., O'Neill J. W., Kim D. E., Zhang K. Y., Baker D. (2002) Accurate computer-based design of a new backbone conformation in the second turn of protein L. J. Mol. Biol. 315, 471–477 [DOI] [PubMed] [Google Scholar]
- 30. McTigue M. A., Williams D. R., Tainer J. A. (1995) Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonicum and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 246, 21–27 [DOI] [PubMed] [Google Scholar]
- 31. Sheehan D., Meade G., Foley V. M., Dowd C. A. (2001) Structure, function, and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chou P. Y., Fasman G. D. (1974) Prediction of protein conformation. Biochemistry 13, 222–245 [DOI] [PubMed] [Google Scholar]
- 33. Oakley A. J., Lo Bello M., Battistoni A., Ricci G., Rossjohn J., Villar H. O., Parker M. W. (1997) The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution. J. Mol. Biol. 274, 84–100 [DOI] [PubMed] [Google Scholar]
- 34. Nagi A. D., Regan L. (1997) An inverse correlation between loop length and stability in a four-helix bundle protein. Fold. Des. 2, 67–75 [DOI] [PubMed] [Google Scholar]
- 35. Collinet B., Garcia P., Minard P., Desmadril M. (2001) Role of loops in the folding and stability of yeast phosphoglycerate kinase. Eur. J. Biochem. 268, 5107–5118 [DOI] [PubMed] [Google Scholar]
- 36. Brockwell D., Yu L., Cooper S., McCleland S., Cooper A., Attwood D., Gaskell S. J., Barber J. (2001) Physicochemical consequences of the perdeuteration of glutathione S-transferase from S. japonicum. Protein Sci. 10, 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marshall S. A., Lazar G. A., Chirino A. J., Desjarlais J. R. (2003) Rational design and engineering of therapeutic proteins. Drug Discov. Today 8, 212–221 [DOI] [PubMed] [Google Scholar]
- 38. Lazar G. A., Marshall S. A., Plecs J. J., Mayo S. L., Desjarlais J. R. (2003) Designing proteins for therapeutic applications. Curr. Opin. Struct. Biol. 13, 513–518 [DOI] [PubMed] [Google Scholar]
- 39. Ladurner A. G., Fersht A. R. (1997) Glutamine, alanine, or glycine repeats inserted into the loop of a protein have minimal effects on stability and folding rates. J. Mol. Biol. 273, 330–337 [DOI] [PubMed] [Google Scholar]
- 40. Sundaram R., Dakappagari N. K., Kaumaya P. T. (2002) Synthetic peptides as cancer vaccines. Biopolymers 66, 200–216 [DOI] [PubMed] [Google Scholar]
- 41. Mehta N. M., Malootian A., Gilligan J. P. (2003) Calcitonin for osteoporosis and bone pain. Curr. Pharm. Des. 9, 2659–2676 [DOI] [PubMed] [Google Scholar]
- 42. Sheffield W. P. (2001) Modification of clearance of therapeutic and potentially therapeutic proteins. Curr. Drug Targets Cardiovasc. Haematol. Disord. 1, 1–22 [DOI] [PubMed] [Google Scholar]
- 43. Kraulis P. J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 44. Merritt E. A., Bacon D. J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 [DOI] [PubMed] [Google Scholar]