Background: miR-140 is a critical regulator of cartilage development and homeostasis.
Results: The proximal upstream region of miR-140 has in vivo chondrogenic promoter activity and an L-Sox5/Sox6/Sox9 (Sox trio) response element.
Conclusion: We reveal that L-Sox5 and Sox6 control miR-140 expression together with Sox9.
Significance: Uncovering molecular mechanisms of chondrogenesis has implications for cartilage repair and restoration of tissue function.
Keywords: Chondrocytes, Development, Gene Regulation, MicroRNA, Transcription Factors, Sox Trio
Abstract
Sox9 plays a critical role in early chondrocyte initiation and promotion as well as repression of later maturation. Fellow Sox family members L-Sox5 and Sox6 also function as regulators of cartilage development by boosting Sox9 activation of chondrocyte-specific genes such as Col2a1 and Agc1; however, the regulatory mechanism and other target genes are largely unknown. MicroRNAs are a class of short, non-coding RNAs that act as negative regulators of gene expression by promoting target mRNA degradation and/or repressing translation. Analysis of genetically modified mice identified miR-140 as a cartilage-specific microRNA that could be a critical regulator of cartilage development and homeostasis. Recent findings suggest Sox9 promotes miR-140 expression, although the detailed mechanisms are not fully understood. In this study we demonstrate that the proximal upstream region of pri-miR-140 has chondrogenic promoter activity in vivo. We found an L-Sox5/Sox6/Sox9 (Sox trio) response element and detailed binding site in the promoter region. Furthermore, detailed analysis suggests the DNA binding and/or transactivation ability of Sox9 as a homodimer is boosted by L-Sox5 and Sox6. These findings provide new insight into cartilage-specific gene regulation by the Sox trio.
Introduction
Chondrogenesis is executed in multiple steps during endochondral ossification. Mesenchymal cells condense, become immature chondroblasts, transform into prehypertrophic chondrocytes, and finally differentiate into hypertrophic chondrocytes (1). Extracellular matrix genes such as Col2a1 and Agc1 are expressed in the early stages; in the prehypertrophic and hypertrophic chondrocyte stages, however, their expression decreases, whereas Col10a1, Ihh, and osteopontin expression increases (2).
The HMG domain-containing transcription factor Sox9 plays a critical role in chondrogenesis except for hypertrophic chondrocyte formation (2, 3). Research with genetically modified mice further revealed that Sox9 initiates and promotes early chondrogenesis but suppresses the maturation stage (4–6). Sox9 recognizes the heptameric DNA sequence (A/T)(A/T)CAA(T/A)G and regulates chondrocyte-specific genes such as Col2a1, Col11a2, CD-RAP, Agc1, and Hapln1 during cartilage development (7–12). Despite this role, molecular mechanisms have not been fully defined for Sox9 regulation of chondrogenesis.
Two other Sox family proteins play a critical role in cartilage development: L-Sox5, a long product of Sox5, and Sox6. L-Sox5 or Sox6 single knock-out mice exhibit a mild skeletal phenotype, whereas double knock-out mice show significant cartilage defects similar to Sox9 knock-out mice (4, 13). These proteins have a coiled-coil DNA-independent homodimerization domain but no other known functional domain (14). L-Sox5 and Sox6 were found to boost Sox9 activity in regulating Col2a1 expression; however, the regulatory mechanism is largely unknown as well as transcription targets beyond Col2a1 and Agc1 (8, 14).
MicroRNAs (miRNAs)3 are a class of short (20–23 nucleotides), non-coding RNAs generated from primary transcripts (pri-miRNAs) by the action of Drosha and Dicer; they negatively regulate gene expression by promoting mRNA degradation and/or repressing translation through formation of RNA-induced silencing complexes and sequence-specific interaction with primarily 3′ untranslated regions on the target mRNA (15–17). Many miRNAs have tissue- and time-specific expression patterns determined at the pri-miRNA transcription level (18–20). Among known miRNAs, Tuddenham et al. (21) showed cartilage-specific expression of miR-140 in mouse embryos. We previously found that miR-140 expression was reduced in human osteoarthritis cartilage or in response to IL-1 stimulation, and miR-140-deficient mice exhibited short stature and age-related osteoarthritis symptoms (22, 23). These observations suggest that miR-140 plays a critical role in cartilage development and homeostasis. Recent findings indicate that Sox9 promotes miR-140 expression (24, 25), although detailed regulatory mechanisms are not fully understood.
We demonstrate in this study that the proximal upstream region of miR-140 has chondrogenic promoter activity in vivo and that cartilage-specific expression of miR-140 is generated from its specific transcript. We also reveal that L-Sox5 and Sox6 control miR-140 expression together with Sox9 through a response element in the promoter. Furthermore, detailed analysis suggests that the DNA binding and/or transactivation ability of Sox9 in its homodimer form is boosted by L-Sox5 and Sox6. The findings provide new insights into cartilage-specific gene regulation by this Sox trio.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Adenovirus Infection
The human kidney cell line 293T and primary mouse chondrocytes were cultured in DMEM with 10% FBS at 37 °C. Primary chondrocytes were prepared from mouse embryo ribs (E16.5) and digested with collagenase. The 293T cell line was transfected using FuGENE HD transfection reagent (Promega). Sox9-expressing recombinant adenovirus was prepared for mouse chondrocytes using the adenovirus expression vector kit (Takara), and infection was performed according to manufacturer's instructions.
Reverse Transcription and Quantitative PCR
Total RNA was extracted with ISOGEN (Nippon Gene) according to the manufacturer's protocol and reverse-transcribed with SuperScript II (Invitrogen) and oligo(dT). Quantitative gene expression analysis was performed via real-time PCR using TaqMan Universal Master Mix reagents and TaqMan Probes (Applied Biosystems) on an ABI PRISM® 7900HT thermal cycler (Applied Biosystems). Col2a1 and Actb were measured using the mouse TaqMan probes Mm00491889_m1 and Mm00607939_m1, respectively (Applied Biosystems). Data were normalized to Actb gene expression for each experiment. Quantitative miRNA expression analysis was performed using the TaqMan MicroRNA reverse transcription kit and TaqMan MicroRNA assay (Applied Biosystems). miR-140 expression was measured using the TaqMan probe TM001187, and snoRNA202 (TM001232) expression was used as an internal control to normalize differences in each sample.
Rapid Amplification of cDNA Ends (RACE)
Total RNA and mRNA were isolated from chondrocytes with TRIzol (Invitrogen) and OligotexdT30 (Takara). 5′- and 3′-RACE were performed using the GeneRace kit (Invitrogen) with region-specific primers (5′ RACE primer (5′-CGATGCAGAGGGTGCTCCAGTACCCTGTCCGTG-3′), 5′ RACE nested primer (5′-CCGTGGTTCTACCCTGTGGTAGAACAGCATGACGT-3′), 3′ RACE primer (5′-ACCCTATGGTAGGTTACGTCATGCTGTTCTACCACAGGG-3′), and 3′ RACE nested primer (5′-ACGTCATGCTGTTCTACCACAGGGTAGAACCACGG-3′).
RNA in Situ Hybridization
Whole mount and section in situ hybridization was performed as previously described (26). Gene-specific fragments were amplified from mouse chondrocyte cDNA by PCR with primers (pri-miR-140, forward 5′-TGGTGTGTGGTTCTATGCCAGC-3′ and reverse 5′-AGCCTCAAGCCAGAATTCAGG-3′; Sox9, forward 5′-TTGAGACCTTCGACGTCAATGAG-3′ and reverse 5′-TCTGGCCACGAGTGGCC-3′). The Col2a1 probe sequence was described in a previous study (27).
Sox9 Conditional Knock-out Mice
In embryos with Sox9, conditional knock-out mice were generated as described using CK-19 Cre and Col2a1 Cre transgenic mice (4, 28).
Reporter Assay
The pGL4.12 vector (Promega), including indicated cloned genome regions, and the indicated gene expression vector were transfected into 293T cells. The Renilla luciferase reporter pRL-TK (Promega) was co-transfected as a control to evaluate transfection efficiency. Cells were lysed, and luciferase activity was measured with the Dual-GloTM Luciferase Assay System (Promega). Data were normalized to Renilla luciferase activity for each experiment. Mutations were introduced with QuikChange® site-directed mutagenesis (Stratagene) according to the manufacturer's instructions.
Generation of miR140 Promoter-LacZ Transgenic (Tg) Mice
To generate miR-140 promoter-LacZ Tg mice, an upstream region of miR-140 was amplified by PCR from mouse genome DNA using specific transgene primers (forward, 5′-ACTGTTCAGAAGGAGACTACTCTGTC-3′; reverse, 5′-ACCGACCTCTGCTCAGCTC-3′). The amplified fragment was cloned into a vector containing the LacZ gene, and Tg BDF1 mice were generated with pronuclear injection of the transgene. Tg mice were confirmed by PCR analysis of genomic DNA using specific transgene primers (forward, 5′-GGTGCTTTGTGAAGGGAAAG-3′; reverse, 5′-GTTGCACCACAGATGAAACG-3′).
X-Gal Staining
LacZ expression from miR140-LacZ Tg mice was detected by X-gal staining. Whole mount Tg embryos were incubated in fixation solution (1% paraformaldehyde, 0.2% glutaraldehyde, and 0.02% Nonidet P-40 in PBS) for 30 min at room temperature. The embryos were then washed with 1 mm MgCl2 in PBS and stained with staining solution (0.01% sodium deoxycholate, 0.02% Nonidet P-40, 1 mm MgCl2, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, and 0.1% X-gal in PBS) overnight at 37 °C.
Electrophoretic Mobility Shift Assay (EMSA)
DNA-protein binding was assayed with DNA probes 32P-radiolabeled by end-filling with Klenow fragment and luciferase or Sox9 with or without anti-Sox9 antibodies. Reactions were carried out at 25 °C for 30 min in binding buffer (20 mm HEPES (pH 7.9),10% glycerol, 50 mm KCl, 0.05% Nonidet P-40, 0.5 mm EDTA, 0.5 mm DTT, and 1 mm PMSF) and 0.5 μg of poly(dG-dI), a nonspecific competitor. Anti-Sox9 antibody was preincubated with Sox9 for 30 min at 25 °C before the addition of the radiolabeled probes. Binding reactions were separated by PAGE on a 4% gel for 3 h at 100 V. Proteins were synthesized in vitro with the TNT T7 Quick Coupled Transcription/Translation System (Promega) and each expression plasmid.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described (29). Briefly, cells were cross-linked with 1% formaldehyde for 15 min at room temperature before glycine was added for a final concentration of 0.125 m. Chromatin was sheared to ∼200–1000 bp by sonication. The chromatin solution was then incubated with the indicated antibodies bound to Dynabeads® Protein A (Dynal Biotech) at 4 °C. Sox9 (Millipore), Sox5 (Abcam, ab94396), and Sox6 (Abcam, ab30455) antibodies were used, with normal rabbit IgG antibodies (Santa Cruz Biotechnology, Inc.) as negative controls. Immune complexes were eluted from the beads and reverse-cross-linked at 65 °C. Chromatin-immunoprecipitated DNA was purified using the MinElute PCR purification kit (Qiagen) and analyzed with whole cell extract by real-time PCR with specific primers (miR140-PSB, forward 5′-GTATTTGCACAAGGCTGGAC-3′ and reverse 5′-AGACCTGGCTGGCTCCAT3′; miR140-Far Ups, forward 5′-CTATCTACCCGGGCCACCTG-3′ and reverse 5′-GGACCTATGCTGGGACAATC-3′).
RESULTS
Chondrogenic Expression of miR-140 Is Regulated by Sox9
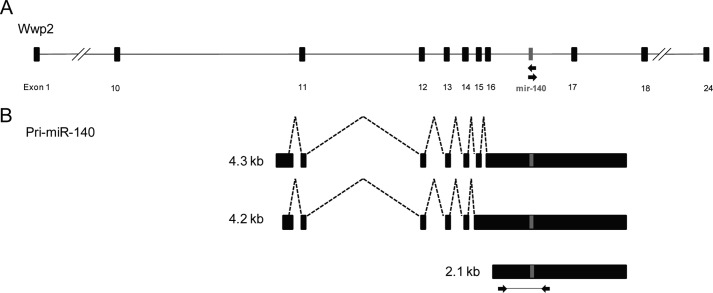
miR-140 is located in intron 16 of the WW domain containing E3 ubiquitin protein ligase 2 (Wwp2 gene) and consists of an N-terminal C2 domain, a C-terminal HECT domain, and four WW domains in the center (Fig. 1A). To determine the transcriptional start site (TSS) and 3′ end of pri-miR-140, we performed 5′- and 3′-RACE using mRNA from cultured mouse chondrocytes. We detected three different transcripts with the miR-140 sequence (Fig. 1B). The shortest was 2.1 kb long, starting inside intron 16. The two longer transcripts were splice variants 4.2 and 4.3 kb long, with the same TSS inside intron 10. All three isoforms had the same 3′ end.
FIGURE 1.
Three isoforms were detected for pri-miR-140. A, miR-140 is located on chromosome 8 and intron 16 of the WW domain containing E3 ubiquitin protein ligase 2 (Wwp2) gene. B, three different-sized transcripts were identified for pri-miR-140 from 5′- and 3′-RACE performed using the primers indicated with arrows in A. The pri-miR-140 probe used for in situ hybridization is indicated with arrows in B.
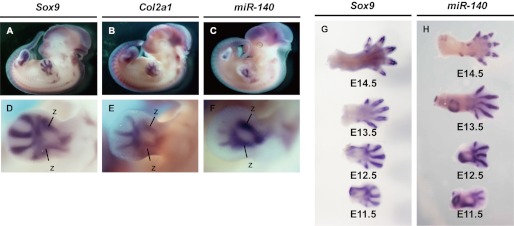
miR-140 expression has been previously detected in the primordia of future bones and across the autopod, zeugopod, and stylopod of E11.5 mouse embryo forelimbs and hind limbs with a mature miR-140 LNA oligonucleotide probe (21). Still, precise temporal and spatial miR-140 expression patterns have not been characterized during chondrogenesis. To explore miR-140 expression further, we performed in situ hybridization using an RNA probe for pri-miR-140 common to all three isoforms (Fig. 1B). The expression pattern in the cartilage of E11.5 embryos was similar to the chondrocyte differentiation markers Sox9 and Col2a1, a target gene of Sox9 (Fig. 2, A–F). Sox9 and miR-140 expressions were also similarly detected in the cartilage of digits from E11.5 to E14.5 (Fig. 2, G and H).
FIGURE 2.
Cartilage-specific expression of pri-miR-140 in mouse embryos. Shown is a comparison of Sox9 (A and D), Col2a1 (B and E), and pri-miR-140 (C and F) expression patterns in mouse whole mount embryos and developing mouse forelimb buds at embryonic stage E11.5. Zeugopod elements are indicated as z. Shown is the Sox9 (G) and Pri-miR-140 (H) expression pattern in the cartilage of developing digits during E11.5 to E14.5.
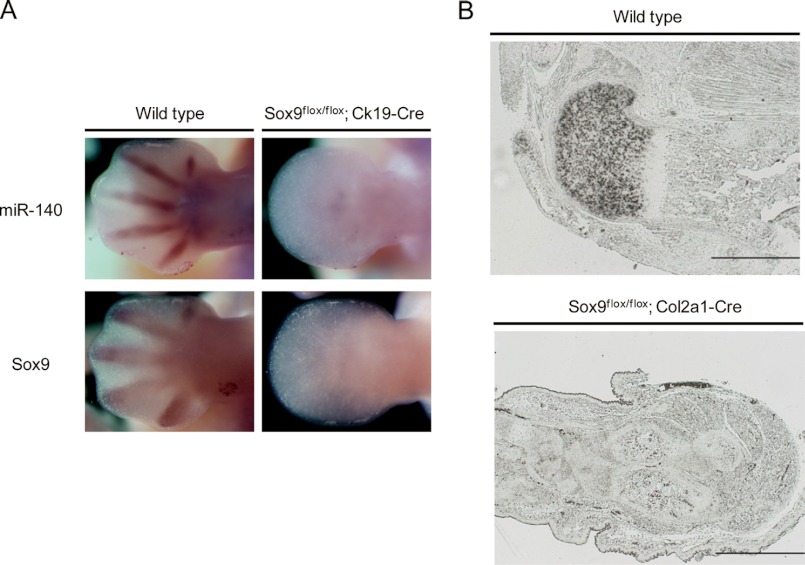
Our in situ hybridization analysis found that miR-140 expression resembled Sox9 and one of its target genes, Col2a1 expression in limb buds, which suggests the possibility that Sox9 regulates miR-140 expression in limb development. To test this hypothesis, we examined the effect of repressing Sox9 expression on miR-140 expression. Sox9flox/flox;Ck19-Cre embryos abolish Sox9 expression before mesenchymal condensation, and chondrogenic cell lineage commitment is profoundly impaired as a consequence (28). In E12.5 wild type and Sox9flox mice without Cre expression, the miR-140 expression pattern overlapped with the Sox9 pattern (Fig. 3A, left). Expression of pri-miR-140 in Sox9 mutant cells at E12.5 was completely absent, however, as was Sox9 expression (Fig. 3A, right). These results suggest that Sox9 is a regulator of miR-140, but there is a possibility that the absence of miR-140 expression in the Sox9flox/flox;Ck19-Cre embryo limb buds was the result of absent chondrogenic lineage cells from impaired chondrogenic mesenchymal condensation. We thus examined Sox9flox/flox;Col2a1-Cre embryos, where Sox9 was inactivated in condensed mesenchymal cells and differentiated chondrocytes through Col2a1-Cre-mediated recombination (4). In E16.5 wild type embryos, pri-miR-140 was expressed in the proliferating chondrocyte zone but absent in the hypertrophic zone (Fig. 3B, top). In contrast, miR-140 was completely absent in condensed chondrogenic mesenchymal cells and differentiated chondrocytes from Sox9flox/flox; Col2a1-Cre embryos (Fig. 3B, bottom). This shows that Sox9 dominates regulation of miR-140 expression in differentiating chondrocytes in vivo.
FIGURE 3.
Pri-miR-140 expression is reduced in Sox9-deficient mouse limb buds and chondrocytes. A, Pri-miR-140 and Sox9 expression patterns in the forelimb buds of wild type and Sox9flox/flox;Ck19-Cre mouse embryos at E12.5 are shown. Expression of both transcripts was drastically reduced in mutant embryos. B, Pri-miR-140 expression was detected in wild type femurs at E16.5 but not E16.5 Sox9flox/flox;Col2a1-Cre femurs.
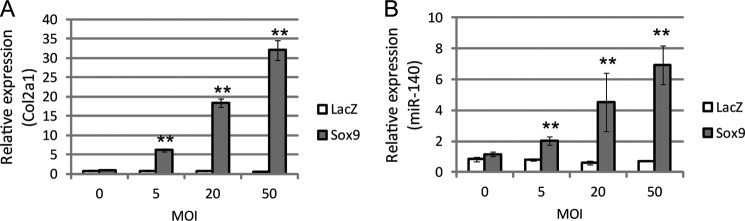
We also studied the effect of Sox9 overexpression on miR-140 expression through quantitative PCR analysis for cultured chondrocytes. Adenovirus infection-mediated Sox9 overexpression increased miR-140 expression as well as Col2a1 expression and correlated with increased multiplicity of infection; adenovirus infection-mediated LacZ overexpression did not affect miR-140 expression (Fig. 4, A and B). Taken together, the results from all these experiments indicate that miR-140 expression is up-regulated by Sox9 in chondrocytes and developing limbs.
FIGURE 4.
miR-140 expression is up-regulated by Sox9 in chondrocytes. LacZ-expressing adenoviruses as a negative control or Sox9-expressing adenoviruses were transduced into cultured mouse chondrocytes with incremental multiplicities of infection (MOI). Col2a1 mRNA (A) and miR-140 (B) expression after Sox9 overexpression were evaluated with quantitative PCR. Data are presented as the mean ± S.D.; n = 3. Statistical differences were calculated using the t test. **, p < 0.01
Upstream Region of miR-140 Is Critical for Its Chondrogenic Expression and Sox9 Regulation
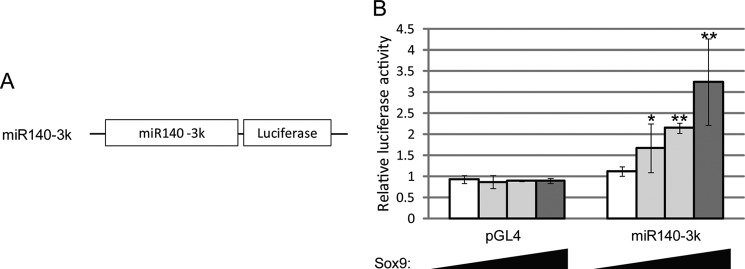
To determine a direct regulatory mechanism for miR-140 expression by Sox9, we constructed a reporter plasmid with a 3-kb region upstream from the miR-140 TSS and a luciferase gene (miR140–3k). With the reporter plasmid and increasing amounts of Sox9-expressing plasmids, luciferase activity in 293T cells increased in a dose-dependent manner (Fig. 5). Sox9 thus regulates miR-140 expression by activating a promoter region 3 kb upstream of its TSS.
FIGURE 5.
miR-140 proximal promoter activity is up-regulated by Sox9. A, shown is a luciferase reporter construct containing a 3-kb proximal upstream region of pri-miR-140. B, 293T cells were co-transfected with pGL4 as a negative control or the miR140–3k reporter plasmid and increasing amounts of Sox9-expressing plasmid (0, 50, 100, and 400 ng). Luciferase activity is presented as the mean ± S.D.; n = 3. Statistical differences were calculated using t test. *, p < 0.05; **, p < 0.01.
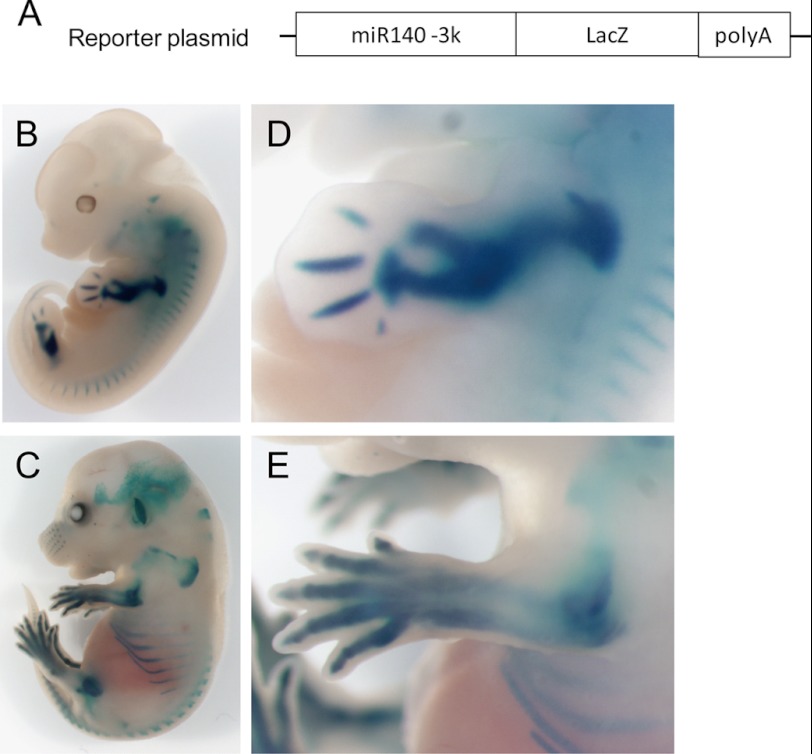
Another reporter plasmid with the 3-kb region upstream of miR-140 and a LacZ gene (Fig. 6A) was prepared and used to generate transgenic (Tg) mice (miR140–3k-LacZ). We generated three lines of transgenic mice, and all of those transgenic E12.5 and E15.5 embryos revealed LacZ expression in forelimbs, hind limbs, and other cartilage tissue such as ribs and vertebrae with an expression pattern similar to miR-140 (Fig. 6, B–E). These in vitro and in vivo results suggest that miR-140 expression in limb development and cartilaginous tissues is regulated by a 3-kb region upstream of its TSS and promoted by Sox9.
FIGURE 6.
The miR-140 proximal promoter has chondrogenic activity. A, shown is an LacZ construct containing a 3-kb upstream region of pri-miR-140 transgene. LacZ expression patterns in miR140–3k-LacZ transgenic (Tg) mouse embryos at E12.5 (B) and E15.5 (C). Shown is a magnified view of forelimb at E12.5 (D) and E15.5 (E).
Sox9 Binds to Upstream Region of miR-140 and Enhances Its Expression in Concert with L-Sox5 and Sox6
Previous studies have clarified that L-Sox5 and Sox6 are critical regulators of chondrogenesis and function as Sox9 co-operators in chondrocytes (4, 14). It has also been reported that expression of chondrogenic genes regulated by Sox9, such as Col2a1 and Agc1, is enhanced by L-Sox5 and Sox6 (8, 14). Therefore, we explored whether L-Sox5 and Sox6 were involved with Sox9 in regulation of miR-140 expression.
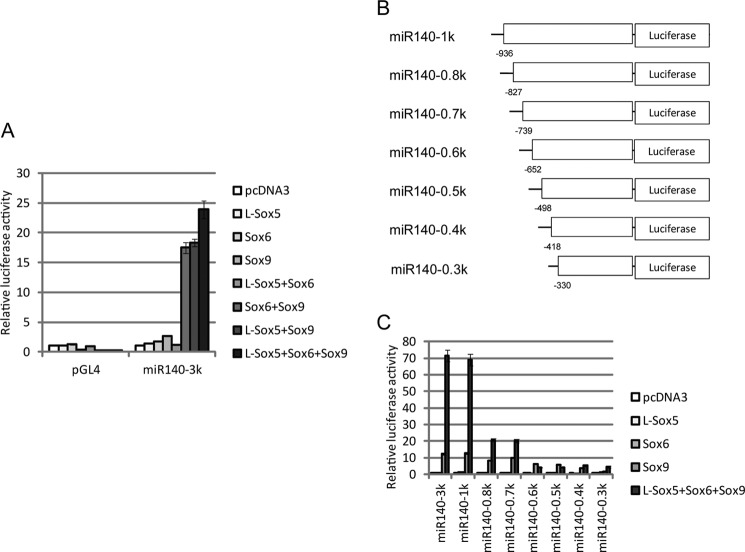
We first examined promoter activity from the miR140–3k reporter plasmid coupled with overexpression of L-Sox5, Sox6, and/or Sox9 in 293T cells. Luciferase activity was greater with overexpression of the Sox trio than with Sox9 alone, and L-Sox5 and/or Sox6 did not enhance activity without Sox9 overexpression (Fig. 7A). L-Sox5 and Sox6 can thus promote miR-140 expression through activating Sox9.
FIGURE 7.
L-Sox5 and Sox6 enhance Sox9-dependent proximal promoter activation of miR-140. A, 293T cells were co-transfected with pGL4 as a negative control or with the miR140–3k reporter plasmid and the indicated expression plasmids. Luciferase activity is presented as the mean ± S.D.; n = 3. B, luciferase reporter constructs containing different lengths of the miR-140 upstream region are shown. Numbers indicate position from the pri-miR-140 transcription start site. C, luciferase activity using the reporter plasmids displayed in B and indicated expressing plasmids is presented as the mean ± S.D.; n = 3.
A series of deletion constructs in the miR-140 upstream region was then created to identify the Sox trio response element (Fig. 7B). Luciferase activity up-regulated by the Sox trio decreased with a deletion from −936 to −827 bp upstream of the miR140 TSS and was almost abolished with a deletion from −739 to −652 bp (Fig. 7C). Remarkably, an additional increase in activity by L-Sox5 and Sox6 co-transfected with Sox9 was not shown with deletion of the −739 to −652 bp upstream region (Fig. 7C).
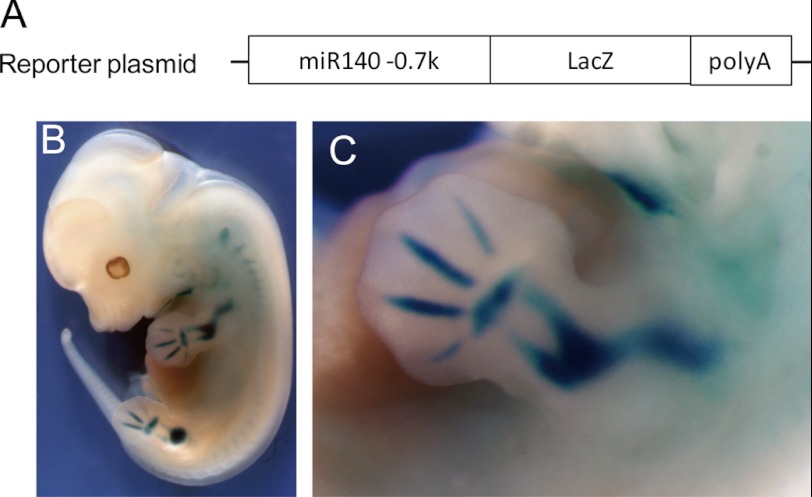
Our luciferase assay analysis indicates that the −739 bp upstream region from miR-140 TSS is minimally required for its regulation by L-Sox5, Sox6, and Sox9 (Sox trio). We then constructed a LacZ reporter plasmid containing −739 bp upstream region from miR-140 TSS (Fig. 8A) and generated Tg mice (miR140–0.7k-LacZ) to examine whether the minimal region is essential for its chondrogenic expression. We observed that the four other miR140–0.7k-LacZ mice expressed LacZ in cartilaginous tissues that expression was similar to miR140–3k-LacZ mice (Fig. 8, B and C). These results indicate that the −739-bp proximal upstream region of miR-140, which is minimally required for its regulation by Sox trio, is essential for its chondrogenic expression.
FIGURE 8.
The 0.7-kb upstream region of miR-140 has chondrogenic promoter activity. A, shown is the LacZ reporter construct containing −739 bp upstream region of pri-miR-140 transgene. B, shown are LacZ expression patterns in miR140–0.7k-LacZ Tg mouse of E12.5 embryos. C, shown is a magnified view of forelimb.
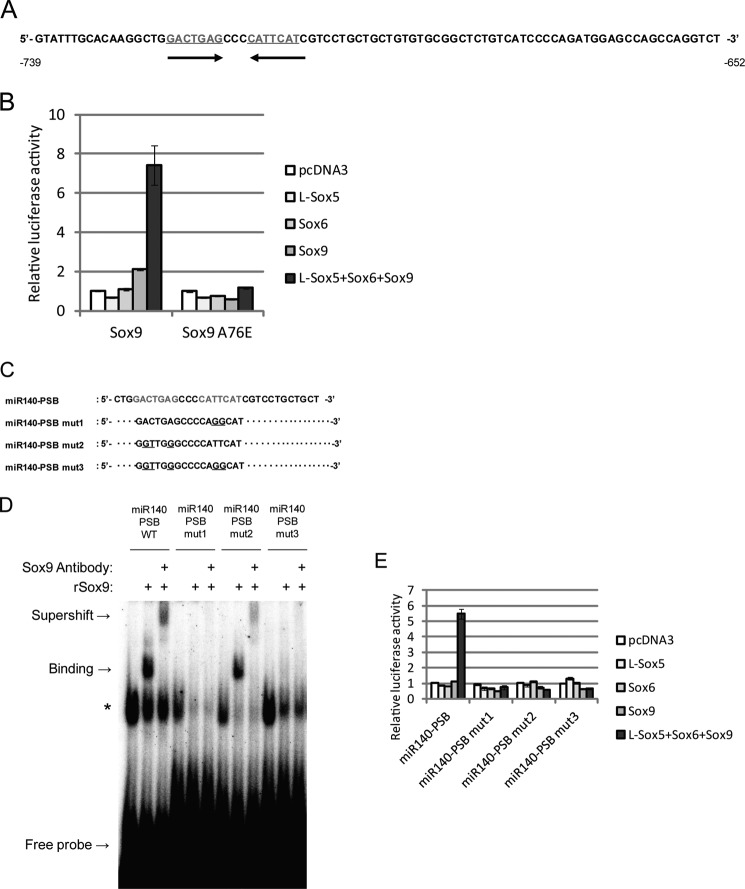
The −739 to −652-bp upstream region was then explored in detail because up-regulation by the Sox trio was abolished with its deletion. We looked for consensus Sox9 binding sequences because Sox trio up-regulation was still Sox9-dependent and found a putative imperfect palindromic sequence in the binding site (Fig. 9A). Previous reports indicated that Sox9 homodimers bind to enhancer regions that contain inverted Sox9 binding sites separated by 3–4 bp and represent a palindromic motif for chondrocyte-specific gene regulation (30–32). To assess whether Sox9 homodimer binding was critical for miR-140 regulation, a miR140–0.7k reporter construct and Sox9 A76E expression plasmid that does not form homodimers were used (30–32). There was no obvious up-regulation of luciferase activity by Sox9 A76E alone or with co-expression in the Sox trio, in contrast to the Sox9 wild type plasmid (Fig. 9B). To confirm that Sox9 binds to the putative palindromic sequence, we performed EMSA using oligonucleotide probes for the putative Sox binding (PSB) sequences (Fig. 9C). miR140-PSB probes detected a band when incubated with recombinant Sox9 protein, and additional incubation with anti-Sox9 antibody reduced the band with an upward supershift (Fig. 9D). Utilizing probes with point mutations in one or both Sox9 binding sequences (Fig. 9C) resulted in band reduction, where miR140-PSB mut1 almost abolished the band, and mutations in both motif sites completely abolished the band (Fig. 9D). The same results occurred with the supershift band produced by incubation with anti-Sox9 antibody (Fig. 9D). To further confirm Sox9 binding and promoter activity in the −739 ∼ −652-bp region upstream of the miR-140 TSS, another luciferase reporter assay was performed with a vector containing point mutations in one or both Sox9 motif sites. Dual mutations and a single Sox9 motif mutation diminished up-regulation of promoter activity by Sox trio expression (Fig. 9E).
FIGURE 9.
The L-Sox5 and Sox6 cooperative effect for miR-140 promoter activation is repressed by blockage of Sox9 homodimer formation. A, shown is a sequence of the region −739 and −652 bp upstream from the pri-miR-140 transcription start site. A putative Sox9 binding site is indicated in red; arrows indicate site orientation. B, 293T cells were co-transfected with the miR140–0.7k reporter plasmid displayed in Fig. 7B and the indicated expressing plasmids. Wild type Sox9 (left) or Sox9 with a dimerization missense mutation (A76E) (right) were used as expression plasmids. Luciferase activity is presented as the mean ± S.D.; n = 3. C, shown are wild type and mutant oligonucleotide probe sequences corresponding to Sox9 binding sites in the miR-140 promoter. D, binding of oligonucleotide probes with wild type and mutated PSB consensus sequences complexed to recombinant Sox9 protein (rSox9) was detected by electrophoretic mobility shift assay. Mutations in one (mut1, mut2) or both (mut3) Sox9 binding motifs were compared with the wild type (WT) in the presence or absence of anti-Sox9 antibody. E, 293T cells were co-transfected with reporter plasmids containing the −739- and −652-bp pri-miR-140 wild type upstream region (miR140-PSB) or the mutations displayed in C and the indicated expressing plasmids. Luciferase activity is presented as the mean ± S.D.; n = 3.
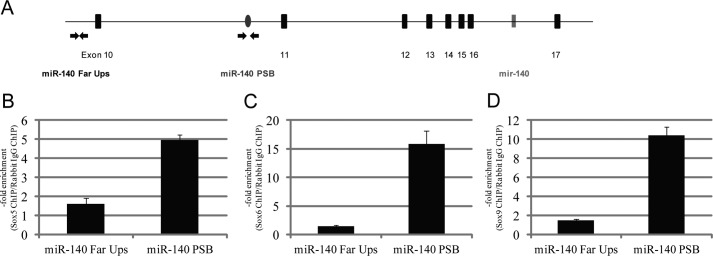
Finally, the ability of Sox9 to bind to the endogenous miR-140 promoter was examined in E13.5 mouse limbs by a ChIP assay using far upstream (approximately −5 kb) (miR140 Far Ups) and putative Sox9 binding site (miR140 PSB) primer sets as shown in Fig. 10A. The enrichment value of the putative Sox9 binding site was confirmed in Sox9 chromatin-immunoprecipitated DNA, although the far upstream site did not show enrichment (Fig. 10B). Additionally, the use of antibodies showed that L-Sox5 and Sox6 also bound to the putative Sox9 binding site but not the far upstream site (Fig. 10, C and D). Taken together, these results suggest that Sox9 enhances miR-140 expression by cooperating with L-Sox5 and Sox6 to bind −739 ∼ −652 bp upstream from the miR-140 TSS.
FIGURE 10.
The Sox trio binds to the miR-140 promoter in E13.5 mouse limbs. A, location of site-specific primer sets for PSB sites and the −5 kb upstream (Far Ups) region of the pri-miR-140 TSS as a negative control are displayed by arrows. B, chromatin-immunoprecipitated DNA and whole cell extract DNA in E13.5 mouse limbs were quantitatively analyzed by real-time PCR. The enrichment value of L-Sox5 (B), Sox6 (C), and Sox9 (D) chromatin-immunoprecipitated DNA was calculated by dividing rabbit IgG chromatin-immunoprecipitated DNA data as a negative control. Data are presented as the mean ± S.D.; n = 3.
DISCUSSION
Most miRNAs, which frequently are located within the intron region of genes, originate from the spliced intronic RNA of a host gene transcript (33). The cartilage-specific intronic miRNA miR-140 is located in intron 16 of Wwp2, a host gene that functions as a critical regulator of craniofacial development and is also expressed in cartilaginous tissues (34, 35). Wwp2 expression is directly regulated by the binding of Sox9 to intron 3 (35), which suggests that miR-140 expression can be similarly generated and regulated from the Wwp2 transcript. In this study we demonstrated that the proximal upstream region of pri-miR-140 located in intron 10 of Wwp2 has in vivo promoter activity. The results suggest that miR-140 may be derived from its own specific transcript during chondrogenesis.
Previous studies found miR-140 to be specifically expressed in chondrocytes and a critical regulator of cartilage development and homeostasis (21–23). Our in situ hybridization analysis confirmed that pri-miR-140 expression was chondrocyte-specific in mouse embryos and mirrored Sox9 expression patterns as shown in limb buds. Also, miR-140 expression was abolished in Sox9-deficient limb buds and chondrocytes and enhanced in cultured chondrocytes with overexpressed Sox9. These findings show that Sox9 is a direct regulator of miR-140 as well as Wwp2. A recent study indicated that Sox9 binds to intron 10 of Wwp2 and up-regulates miR-140 expression (25). By further exploring this mechanism, we found that Sox9 activates a promoter region upstream of miR-140 on intron 10 with in vivo chondrogenic activity. This strongly suggests that miR-140 in vivo chondrogenic expression is directly regulated in part through activation of its promoter region by Sox9 instead of splicing itself out of the host transcript.
Most previously known Sox9 targets were extracellular matrix genes, and only a few, such as Bapx1, S100A1, and S100B, were shown to be direct regulators of it except for extracellular matrix genes (29, 36). Our in vivo and in vitro analyses demonstrated that miR-140 is also a downstream target of Sox9. Previous studies reported that miR-140 repressed Hdac4 to block chondrocyte maturation, Adamts-5 to promote cartilage development and homeostasis, and Sp1 to maintain chondrocyte proliferation (21, 23, 25). These diverse roles are similar to Sox9, which also promotes early chondrogenesis but represses chondrocyte maturation (2). Furthermore, Sox9 expression was significantly lower in osteoarthritis cartilage compared with normal cartilage (37), whereas miR-140 knock-out mice exhibited age-related, osteoarthritis-like phenotypes (23). These studies strongly support miR-140 as both a downstream and a critical target of Sox9 due to its diverse roles in chondrogenesis. In fact, miR-140-deficient mice have a mild skeletal phenotype with a short stature (23).
In this study we showed that L-Sox5 and Sox6 were also involved in regulation of miR-140 expression with Sox9. Results showed that expression levels did not change in the presence of L-Sox5 and/or Sox6 without Sox9 co-expression, making control of miR-140 expression by L-Sox5 and Sox6 completely Sox9-dependent. L-Sox5 and Sox6 participate in the regulation of cartilage-specific genes such as Col2a1 and Agc1, although they have no known transactivation or transrepression domains (8, 14). A recent study suggested that L-Sox5 and Sox6 may facilitate Sox9 DNA binding by an unknown mechanism (8). A comparable regulatory event by the Sox trio may also occur with the miR-140 promoter.
Our detailed luciferase, EMSA, and ChIP analyses identified the precise Sox trio response element and Sox9 binding site. Sox9 was previously reported as bound to intron 10 of Wwp2 to regulate miR-140 expression (25). However, the Sox9 binding site exhibited in this study is different and suggests the possibility that miR-140 expression is controlled by multiple Sox9 and/or the Sox trio binding in its promoter region, as shown with Col11a2 expression that is directed by Sox9 at least three cartilage-specific enhancer elements (38).
Sox9 is critical to chondrogenesis because it regulates cartilage-specific genes such as Col2a1, Col11a2, and Agc1 (7, 10, 11). In addition, Sox9 plays a critical role in testicular development by regulating targets such as Amh and Ptgds in Sertoli cells (39–41). However, the regulatory mechanism of tissue-specific target distribution is less understood. Previous reports indicated a human SOX9 missense mutation (A76E) in an XY patient exhibiting skeletal abnormalities without sex reversal; this mutation perturbed Sox9 dimerization, resulting in dysregulation of chondrocyte-specific genes (30–32). Sox9 homodimers are believed to bind to enhancer regions containing inverted Sox9 binding sites separated by 3 or 4 bp (30–32). The Sox9 binding site identified in this study is also a Sox9 palindromic binding motif and again indicates that miR-140 is a critical chondrocyte-specific target. In fact, LacZ expression was not detected in the male gonads of any miR140–3k-LacZ Tg embryonic mice (data not shown). We further showed that Sox9 A76E did not up-regulate miR-140 promoter activity even when L-Sox5 and Sox6 were co-expressed. Only one site mutation of the Sox9 palindromic motif completely abolished the ability of the Sox trio to up-regulate promoter activity despite a Sox9 motif mutation that decreased but retained Sox9 binding status. These findings suggest that DNA-dependent Sox9 homodimer formation is needed for L-Sox5 and Sox6 assembly to enhance its DNA binding and/or transactivation ability, but a more detailed mechanism remains undetermined. As previously mentioned, binding of the Sox9 homodimer to its palindromic motif is critical for cartilage-specific gene regulation, and the regulatory mechanism suggested in this study could apply to other cartilage targets. In fact, we observed that Sox9 A76E did not up-regulate luciferase activity from a Col2a1 enhancer reporter vector with L-Sox5 and Sox6 co-transfection (data not shown).
In conclusion, we report that a proximal region upstream of miR-140 has chondrogenic promoter activity in vivo, and its cartilage-specific expression is generated from its transcript within the host gene Wwp2. miR-140 promoter activity is up-regulated by the critical transcription factors L-Sox5, Sox6, and Sox9, and their response elements and detailed binding sites were identified. Our findings suggest that the DNA binding and/or transactivation ability of Sox9 in its homodimer form is dependently boosted by L-Sox5 and Sox6.
Acknowledgments
We thank Dr. Makoto Taketo of Kyoto University for the Ck19-Cre mice. We thank Akane Nakamura and Hideki Tsumura for help with generation of miR140–3k-LacZ transgenic mice and Arisa Igarashi and Moe Tamano for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AR050631 and AR056120. This work was also supported by Health and Labor Sciences research grants, grants-in-aid for Scientific Research (Ministry of Education, Culture, Sports, Science, and Technology of Japan), and Japan Science and Technology Agency, Core Research for Evolutional Science and Technology. This work was also supported by a grant from the Deutsche Forschungsgemeinschaft (to G. S.).
- miRNA
- microRNA
- RACE
- rapid amplification of cDNA ends
- Tg
- Transgenic
- TSS
- transcriptional start site
- PSB
- putative Sox binding.
REFERENCES
- 1. Provot S., Schipani E. (2005) Molecular mechanisms of endochondral bone development. Biochem. Biophys. Res. Commun. 328, 658–665 [DOI] [PubMed] [Google Scholar]
- 2. Lefebvre V., Smits P. (2005) Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res. C Embryo Today 75, 200–212 [DOI] [PubMed] [Google Scholar]
- 3. Goldring M. B., Tsuchimochi K., Ijiri K. (2006) The control of chondrogenesis. J. Cell. Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 4. Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akiyama H., Lyons J. P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J. M., Taketo M. M., Nakamura T., Behringer R. R., McCrea P. D., de Crombrugghe B. (2004) Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes Dev. 18, 1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. (1999) Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89 [DOI] [PubMed] [Google Scholar]
- 7. Bridgewater L. C., Lefebvre V., de Crombrugghe B. (1998) Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 273, 14998–15006 [DOI] [PubMed] [Google Scholar]
- 8. Han Y., Lefebvre V. (2008) L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol. Cell. Biol. 28, 4999–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kou I., Ikegawa S. (2004) SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J. Biol. Chem. 279, 50942–50948 [DOI] [PubMed] [Google Scholar]
- 10. Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro-α1(II) collagen gene. Mol. Cell. Biol. 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sekiya I., Tsuji K., Koopman P., Watanabe H., Yamada Y., Shinomiya K., Nifuji A., Noda M. (2000) SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275, 10738–10744 [DOI] [PubMed] [Google Scholar]
- 12. Xie W. F., Zhang X., Sakano S., Lefebvre V., Sandell L. J. (1999) Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J. Bone Miner. Res. 14, 757–763 [DOI] [PubMed] [Google Scholar]
- 13. Smits P., Li P., Mandel J., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B., Lefebvre V. (2001) The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1, 277–290 [DOI] [PubMed] [Google Scholar]
- 14. Lefebvre V., Li P., de Crombrugghe B. (1998) A new long form of Sox5 (L-Sox5), Sox6, and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17, 5718–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai X., Hagedorn C. H., Cullen B. R. (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu N., Williams A. H., Kim Y., McAnally J., Bezprozvannaya S., Sutherland L. B., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. U.S.A. 104, 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S., Lodish H. F. (2006) Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 103, 8721–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Y., Samal E., Srivastava D. (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 [DOI] [PubMed] [Google Scholar]
- 21. Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M. K., Clark I., Dalmay T. (2006) The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 580, 4214–4217 [DOI] [PubMed] [Google Scholar]
- 22. Miyaki S., Nakasa T., Otsuki S., Grogan S. P., Higashiyama R., Inoue A., Kato Y., Sato T., Lotz M. K., Asahara H. (2009) MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 60, 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S., Takada S., Lotz M. K., Ueno-Kudo H., Asahara H. (2010) MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24, 1173–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakamura Y., He X., Kato H., Wakitani S., Kobayashi T., Watanabe S., Iida A., Tahara H., Warman M. L., Watanapokasin R., Postlethwait J. H. (2012) Sox9 is upstream of microRNA-140 in cartilage. Appl. Biochem. Biotechnol. 166, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Qin S., Yi C., Ma G., Zhu H., Zhou W., Xiong Y., Zhu X., Wang Y., He L., Guo X. (2011) miR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 585, 2992–2997 [DOI] [PubMed] [Google Scholar]
- 26. Yokoyama S., Hashimoto M., Shimizu H., Ueno-Kudoh H., Uchibe K., Kimura I., Asahara H. (2008) Dynamic gene expression of Lin-28 during embryonic development in mouse and chicken. Gene Expr. Patterns 8, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawakami Y., Tsuda M., Takahashi S., Taniguchi N., Esteban C. R., Zemmyo M., Furumatsu T., Lotz M., Belmonte J. C., Asahara H. (2005) Transcriptional coactivator PGC-1α regulates chondrogenesis via association with Sox9. Proc. Natl. Acad. Sci. U.S.A. 102, 2414–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrionuevo F., Taketo M. M., Scherer G., Kispert A. (2006) Sox9 is required for notochord maintenance in mice. Dev. Biol. 295, 128–140 [DOI] [PubMed] [Google Scholar]
- 29. Yamashita S., Andoh M., Ueno-Kudoh H., Sato T., Miyaki S., Asahara H. (2009) Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp. Cell Res. 315, 2231–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernard P., Tang P., Liu S., Dewing P., Harley V. R., Vilain E. (2003) Dimerization of SOX9 is required for chondrogenesis but not for sex determination. Hum. Mol. Genet. 12, 1755–1765 [DOI] [PubMed] [Google Scholar]
- 31. Coustry F., Oh C. D., Hattori T., Maity S. N., de Crombrugghe B., Yasuda H. (2010) The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin DNA template. Nucleic Acids Res. 38, 6018–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sock E., Pagon R. A., Keymolen K., Lissens W., Wegner M., Scherer G. (2003) Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 12, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 33. Baskerville S., Bartel D. P. (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura Y., He X., Kobayashi T., Yan Y. L., Postlethwait J. H., Warman M. L. (2008) Unique roles of microRNA140 and its host gene WWP2 in cartilage biology. J. Musculoskelet. Neuronal Interact. 8, 321–322 [PMC free article] [PubMed] [Google Scholar]
- 35. Zou W., Chen X., Shim J. H., Huang Z., Brady N., Hu D., Drapp R., Sigrist K., Glimcher L. H., Jones D. (2011) The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 13, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saito T., Ikeda T., Nakamura K., Chung U. I., Kawaguchi H. (2007) S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO Rep. 8, 504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haag J., Gebhard P. M., Aigner T. (2008) SOX gene expression in human osteoarthritic cartilage. Pathobiology 75, 195–199 [DOI] [PubMed] [Google Scholar]
- 38. Bridgewater L. C., Walker M. D., Miller G. C., Ellison T. A., Holsinger L. D., Potter J. L., Jackson T. L., Chen R. K., Winkel V. L., Zhang Z., McKinney S., de Crombrugghe B. (2003) Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 31, 1541–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arango N. A., Lovell-Badge R., Behringer R. R. (1999) Targeted mutagenesis of the endogenous mouse Mis gene promoter. In vivo definition of genetic pathways of vertebrate sexual development. Cell 99, 409–419 [DOI] [PubMed] [Google Scholar]
- 40. De Santa Barbara P., Bonneaud N., Boizet B., Desclozeaux M., Moniot B., Sudbeck P., Scherer G., Poulat F., Berta P. (1998) Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell. Biol. 18, 6653–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilhelm D., Hiramatsu R., Mizusaki H., Widjaja L., Combes A. N., Kanai Y., Koopman P. (2007) SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 282, 10553–10560 [DOI] [PubMed] [Google Scholar]