Background: iLOV is a fluorescent flavoprotein engineered from the plant blue light receptor phototropin.
Results: Structures reveal altered protein-chromophore interactions within the flavin-binding cavity of iLOV when compared with its progenitors. Directed evolution further anchored the chromophore to increase iLOV photostability by an order of magnitude.
Conclusion: Improving iLOV photostability by constraining its fluorophore establishes a framework for fine-tuning fluorescence.
Significance: Enhanced photostability increases iLOV utility as an oxygen-independent fluorescent reporter.
Keywords: Flavin, Fluorescence, Molecular Evolution, Photobiology, Photoreceptors
Abstract
Fluorescent proteins derived from light, oxygen, or voltage (LOV) domains offer advantages over green fluorescent protein (GFP) from their small size and efficacy under anaerobic conditions. The flavoprotein improved LOV (iLOV) was engineered from the blue light receptor phototropin as a reporter of viral infection. To inform the molecular basis for the improved, photoreversible, fluorescent properties of iLOV, we employed directed evolution and determined five LOV crystallographic structures. Comparative structural analyses between iLOV and its progenitors reveal mutation-induced constraints in the environment of the flavin mononucleotide (FMN) chromophore; in iLOV, the methyl group of Thr-394 “crowds” the FMN isoalloxazine ring, Leu-470 triggers side chain “flipping” of Leu-472, and the terminal FMN phosphate shows increased anchoring. We further engineered iLOV variants that are readily detectable in bacterial and mammalian cells due to order-of-magnitude photostability increases. Structure determination of a resulting representative photostable iLOV (phiLOV) variant reveals additional constraints on the chromophore. Aromatic residues Tyr-401 and Phe-485 in phiLOV sandwich the FMN isoalloxazine ring from both sides, whereas Ser-390 anchors the side chain of FMN-interacting Gln-489 Our combined structural and mutational results reveal that constraining the FMN fluorophore yields improved photochemical properties for iLOV and its new photostable derivative. These findings provide a framework for structural fine-tuning of LOV scaffold proteins to maximize their potential as oxygen-independent fluorescent reporters.
Introduction
Fluorescent proteins (FPs)4 have revolutionized the imaging of dynamic processes within living cells (1). Despite their wide range of applications, however, green fluorescent protein (GFP) and its derivatives have three key limitations. First, their use is restricted to aerobic systems as chromophore formation is oxygen-dependent (2). Second, for certain applications, the relatively large size (∼25 kDa) of GFP is limiting (3). Generation of smaller GFP derivatives is unlikely as the 11-stranded β-barrel structure of the protein is intrinsic to its function (2). Third, GFP fluorescence is unstable at pH < 5, prompting the development of pH-resistant variants (1). As a result of these factors, there is considerable interest in developing alternative protein-based fluorescent probes.
LOV (light, oxygen, or voltage) domains derived from plant and bacterial blue light receptors exhibit considerable potential as green fluorescent probes with advantages over the current GFP technology (3–5). LOV domains form a subset of the versatile Per-ARNT-Sim (PAS) superfamily (6, 7) characterized by the prototypic photoactive yellow protein (PYP) structure, which has yielded key information on photocycle intermediates (8) and conformational changes (9). LOV domains typically bind FMN noncovalently as a UV-A/blue light-absorbing chromophore (10). Upon photoexcitation, LOV domains undergo a reversible photocycle involving the formation of a covalent bond between the FMN chromophore and a conserved cysteine residue within the protein (11) reminiscent of the cysteine connection in PYP (12). This light-induced covalent flavin adduct thermally decays to the dark state within tens to thousands of seconds depending on the specific LOV domain environment (13).
The FMN chromophore provides the LOV domain with a weak intrinsic green fluorescence when excited with UV-A/blue light (14, 15). Mutagenesis of the active-site cysteine residue abolishes LOV domain photochemistry and increases its fluorescence (11, 16), making photochemically inactivated LOV domains useful as in vivo reporters. For example, because of their small size (∼11 kDa), LOV-based FPs outperform GFP as reporters for monitoring local and systemic infections of plant RNA viruses and confer improved functionality when fused to viral proteins essential for movement (3). Furthermore, the dependence on a cellular cofactor for fluorescence does not limit the use of the LOV domain in subcellular targeting in plant or mammalian cells (3, 5).
LOV-based FPs offer other favorable attributes besides their smaller size. LOV-mediated fluorescence is stable over a wide pH range in vitro (16) and may circumvent the pH sensitivity commonly associated with GFP-related FPs (2). Similarly, LOV fluorescence, unlike that of GFP, is independent of molecular oxygen (4, 5). This property is particularly advantageous for studies of biological processes that operate under anaerobic conditions. Consequently, LOV-based FPs hold promise over GFP and related protein scaffolds as real-time reporters for probing cell biomass and product formation in micro and macro bioreactors (17, 18). More recently, a LOV-based FP that efficiently generates singlet oxygen upon blue light illumination has been reported that can facilitate correlative light and electron microscopy of intact tissues (5).
Photochemically inactivated LOV domains are inherently fluorescent, but additional mutagenesis was necessary to generate variants with improved fluorescence and photostability (3, 5). One such variant, designated iLOV, contains multiple amino acid changes as a consequence of molecular evolution (3). Structural analysis of this variant is therefore important to understand the impact of these substitutions on protein-chromophore interactions that fine-tune fluorescence.
Here, we uncover a molecular framework underlying the enhanced fluorescent properties of the iLOV reporter by comparative structural analysis. Moreover, we demonstrate that further fine-tuning of the iLOV scaffold can be achieved through DNA shuffling to yield variants with increased photostability. Together, these results enhance our understanding of protein-chromophore interactions within the LOV scaffold and how these can be altered to control its fluorescence in predetermined ways.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
LOV domain constructs based on the sequence of the LOV2 domain of Arabidopsis thaliana phototropin 2 (National Center for Biotechnology Information (NCBI) Accession NP_851211.1; amino acids 387–496) were expressed in Escherichia coli and purified as fusions to glutathione S-transferase (GST) using pGEX-6P-1 or pGEX-4T-1 (GE Healthcare). Amino acid substitutions were introduced using the QuikChange site-directed mutagenesis kit (Agilent) and verified by DNA sequencing. Transformed E. coli were grown at 37 °C; protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to 0.4 mm at A600 0.6–0.8. After overnight incubation at 24 °C, cells were harvested by centrifugation for 15 min at 5,000 × g, and the pellet was resuspended in solution containing 50 mm Tris-HCl, pH 8.0. Cells were sonicated on ice five times for 1 min with regular resting intervals. The clarified lysate obtained after centrifugation at 12,000 × g for 1 h was applied to a GST resin column and washed with 5 bed volumes of solution containing 50 mm Tris-HCl, pH 8.0. Bound protein was eluted with reduced glutathione in 50 mm Tris-HCl (pH 8.0) and 250 mm NaCl. Selected fractions were pooled and treated with PreScission (GE Healthcare) or thrombin (Sigma) protease overnight at 4 °C. After cleavage, the protein was dialyzed against 50 mm Tris-HCl, pH 8.0, and 50 mm NaCl, and the cleaved GST tag was removed using the GST purification resin column. The protein was further purified by gel filtration on Sephacryl S-100 (GE Healthcare), eluted with a solution of 50 mm Tris-HCl, pH 8.0, and 50 mm NaCl, and concentrated to 20 mg/ml.
Protein Crystallography
Crystals were obtained by hanging drop vapor diffusion against a reservoir solution at 20 °C. Drops contained 1 μl each of the concentrated protein and the reservoir solution: 37.5% methoxypolyethylene glycol 2000, 0.2 m imidazole malate, pH 4.8, for wild-type LOV2, C426A, and iLOV (form I), 25% PEG 3350, 5% PEG 400, 0.1 m sodium acetate, pH 5.5, for iLOV (form II), and 25% methoxypolyethylene glycol 2000, 0.2 m imidazole malate, pH 8.6, for phiLOV2.1. Crystals were flash-cooled in liquid nitrogen before data collection. X-ray diffraction data were collected at the SIBYLS beamline 12.3.1 at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, Berkeley, CA or at the Stanford Synchrotron Research Laboratory (SSRL) beamline 11-1 and processed with HKL2000 (19). The structure of iLOV crystal form I was determined by molecular replacement, using Protein Data Bank (PDB) 2V1A as a search model. Phaser (20) gave a clear solution with a log likelihood gain of 2,385, whereas the runner-up was 959. This structure was used to determine phases for the remaining structures. Models were fitted manually with COOT (21) and refined with Phenix (22) for phiLOV and Refmac (23) for the others. Diffraction data collection and refinement statistics are summarized in supplemental Table S1.
Mutagenesis and Screening
Mutagenesis and DNA shuffling were performed as described previously (3). Ligation reactions were transformed into electrocompetent BL21(DE3) E. coli (Novagen), plated on LB-agar containing 100 μg/ml ampicillin (Sigma) and 100 μm isopropyl β-d-1-thiogalactopyranoside (Roche Applied Science), and incubated in darkness at 30 °C for 24 h, as described (24). LOV-associated fluorescence was visualized with a Blak-Ray lamp (UVP, LLC). Colonies were photobleached using a bluephase 16i dental curing light (Ivoclar Vivadent) for 5–10 s (λmax 443 nm, half-bandwidth 10 nm; 0.16 W/cm2).
Spectroscopic Analysis
Fluorescence emission spectra were recorded using a FluoroMax-3 spectrofluorometer (Horiba Scientific). For quantitative in vivo fluorescence measurements, overnight cultures of E. coli were diluted 10-fold in 50 mm Tris-HCl, pH 7.0, and the A600 was recorded. Fluorescence measurements were determined and normalized based on the A600. Photobleaching was analyzed by irradiating with a bluephase 16i dental curing light (Ivoclar Vivadent) for 5 s (λmax 443 nm, half-bandwidth 10 nm; 0.06 W/cm2) or 10 s (0.16 W/cm2).
Cell Culture, Transfection, and Fluorescence Microscopy
iLOV and phiLOV2.1 codon usage was optimized for mammalian cell expression by de novo gene synthesis (GenScript) and cloned into pEGFP-N1 (Clontech) via SalI and NotI to replace the GFP encoding sequence. BSC1 monkey kidney cells were grown under 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium supplemented with 20 mm HEPES, pH 7.0, 10 mg/ml streptomycin, 66 μg/ml penicillin, and 10% (v/v) fetal calf serum (FCS, HyClone). Cells were transfected with iLOV or phiLOV2.1 DNA using Lipofectamine 2000 (Invitrogen), according to manufacturer's instructions. Cells were trypsinized 24 h after transfection and seeded onto cover slides at a density of 8,500 cells/cm2. The next day, cells were imaged at room temperature in a home-made imaging chamber consisting of a coverslip mounted on a slide with two strips of double-sided tape as spacer before sealing the chamber with VALAP (1:1:1 of Vaseline, lanolin, and paraffin). Samples were imaged on an inverted fluorescence microscope (Olympus model IX71) using a Plan Apo N 60X (NA 1.42) objective, equipped with a Hamamatsu Photonics ORCA-ER camera (model C4742-80-12AG) and motorized excitation and emission filter wheels (Sutter Instrument Co., Chroma filters: excitation 403 ± 12 nm, emission 528 ± 38 nm). The open source microscopy software Micro-Manager (25) was used to control image acquisition. Fixed exposure times (500 ms) were used, and images were acquired at a frame rate of 1 s. For quantification, cells edges were drawn manually in ImageJ, and mean fluorescent intensities of the regions of interest were determined. After background subtraction, intensity values were normalized to initial values in frame 1.
RESULTS
Overall Structural Comparisons of iLOV with Its Progenitors
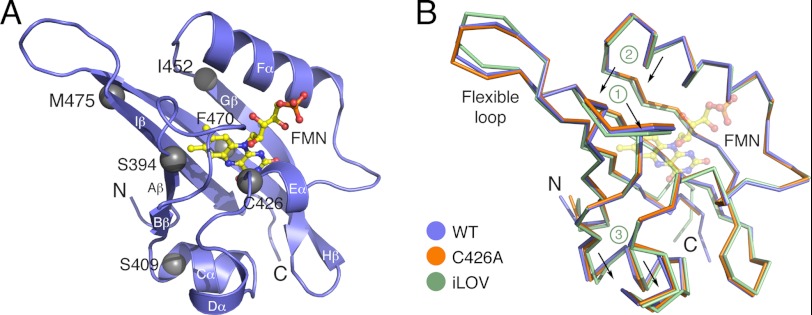
iLOV was engineered from the LOV2 domain of A. thaliana phototropin 2, for which structural information is lacking. To better understand the structural basis for iLOV fluorescence, we determined and compared x-ray crystallographic structures of iLOV with its wild-type progenitor (residues 387–496 of Arabidopsis phototropin 2 (WT)) and its photochemically inactivated C426A variant, from which iLOV was derived (3). To aid comparisons, we obtained isomorphous crystals belonging to space group P43212 for all three proteins under identical conditions, with one molecule per asymmetric unit (supplemental Table S1). Structures were determined and refined to 1.8 Å resolution for iLOV and 1.7 Å resolution for WT and C426A. All exhibit the LOV/PAS fold comprising five antiparallel β-strands (Aβ, Bβ, Gβ, Hβ, and Iβ) framed by four α-helices (Cα, Dα, Eα, and Fα) (Fig. 1A). The edge of the FMN isoalloxazine ring rests on the Hβ- and Iβ-strands, whereas the ring itself is sandwiched between the Eα- and Fα-helices (Fig. 1A).
FIGURE 1.
Overall structures for wild-type Arabidopsis phototropin 2 LOV2 (WT), its C426A derivative, and iLOV. A, WT structure presented as ribbons (β-strands and α-helices are labeled). Residues mutated in iLOV are shown as gray spheres. FMN is presented as a yellow ball-and-stick model with red oxygen and blue nitrogen atoms. B, superposition of WT (blue), C426A (orange), and iLOV (green) structures. The carbon α trace reveals that the loop region between Aβ and Bβ (1), the β-hairpin between Hβ and Iβ (2), as well as the Cα and Dα-helices (3) are shifted in iLOV.
The overall structure of iLOV exhibits noticeable differences from those of WT and C426A, particularly within the flexible loop extension (residues 476–483) between Hβ and Iβ (Fig. 1B). iLOV contains the following substitutions, relative to WT: C426A; S409G; F470L; S394T; I452T; and M475V (3). The M475V substitution in iLOV is located at the edge of the flexible Hβ/Iβ loop extension (Fig. 1B) and likely contributes to the observed structural difference in this region. The S409G and I452T mutations in iLOV also appear to introduce subtle structural changes associated respectively with Cα and Dα and with Gβ and preceding Fα of the LOV scaffold (Fig. 1B).
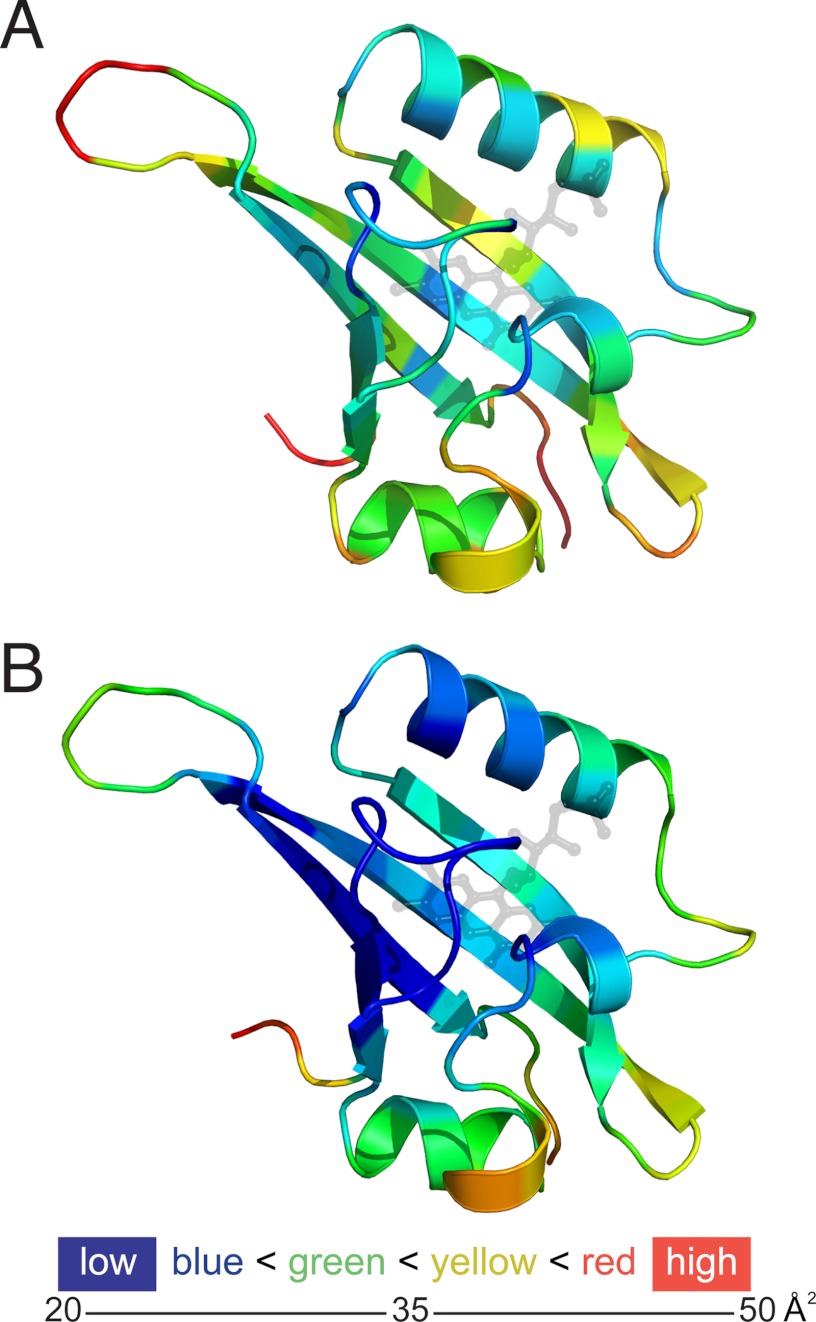
Mapping crystallographic temperature factors (B values) onto protein structures has proven useful for observing differences in mobility with functional implications (26). When B values were mapped onto the WT structure (Fig. 2A), we found that core flexible regions (Cα, Dα, and the adjacent β-hairpin) matched regions of the archetypal LOV/PAS domain protein PYP that showed increased flexibility during light-induced signaling, as measured by enhanced hydrogen/deuterium exchange mass spectrometry (9). B value mapping on the iLOV structure (Fig. 2B) showed a similar pattern, but suggested more order in the vicinity of the chromophore and less overall flexibility.
FIGURE 2.
Crystallographic temperature factors (B values) mapped onto the structures of Arabidopsis phototropin 2 LOV2 (WT) (A) and iLOV (B) suggest differences in relative flexibility, both within and between these structures. Many factors can influence B values, including crystal size, x-ray exposure time, radiation dose, and crystal mosaicity, which can vary with cryo-protection protocols, yet the overall pattern of B values appears consistent with the increased packing interactions in the flavin-binding cavity, supporting decreased flexibility in iLOV relative to WT (also see supplemental Fig. S6).
The iLOV structure is more compact than its progenitors (supplemental Table S2), consistent with enhanced rigidity. The reduced molecular volume of iLOV does not appear to result from crystal packing as the same property was observed for different nonisomorphous crystals (form II; 1.2 Å) (supplemental Tables S1 and S2). Thus, both iLOV crystal forms provide evidence for compaction relative to its progenitors. The presence of two molecules per asymmetric unit in form II crystals (supplemental Table S1) suggests that iLOV has the propensity to dimerize. However, iLOV was found to be monomeric by size exclusion chromatography (supplemental Fig. S1).
iLOV Exhibits Altered Protein-Chromophore Interactions
Given the overall changes evidenced in both iLOV structures, we paid particular interest to structural differences that impacted protein backbone and/or side chain interactions with the FMN chromophore. Introduction of the C426A mutation does not appear to alter the backbone position of Eα (Fig. 1B). iLOV contains two additional changes within the vicinity of the FMN isoalloxazine ring: S394T and F470L. Ser-394 faces away from the isoalloxazine ring (Fig. 3, A and B) and hydrogen-bonds with Asn-401 in the structures of WT and C426A. In iLOV, the methyl group of Thr-394 “crowds” this region adjacent to the isoalloxazine ring, thereby restricting the mobility of the FMN (Fig. 3C). The F470L mutation, located on the opposite side of the isoalloxazine ring, imparts further steric restrictions on the FMN (Fig. 3C). Notably, Leu-472 (which is not mutated in iLOV) becomes reoriented or “flipped” toward the isoalloxazine ring to accommodate contacts with the FMN and Leu-470 (Fig. 3C).
FIGURE 3.
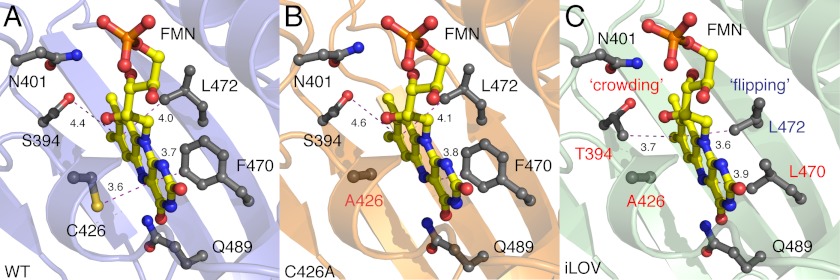
FMN-binding cavities of wild-type Arabidopsis phototropin 2 LOV2 (WT) (A) its C426A derivative (B) and iLOV (C). Main chains are shown as ribbons. FMN is presented as a yellow ball-and-stick model with red oxygen and blue nitrogen atoms. Relevant side chains are indicated in gray as ball-and-stick models. Residues mutated in C426A and iLOV are labeled red. Critical distances between the FMN chromophore and specific residues are highlighted with dashed magenta lines. The S394T mutation in iLOV results in “crowding” of the chromophore environment at this region, whereas the side chain of Leu-472 undergoes “flipping” as a consequence of the F470L mutation.
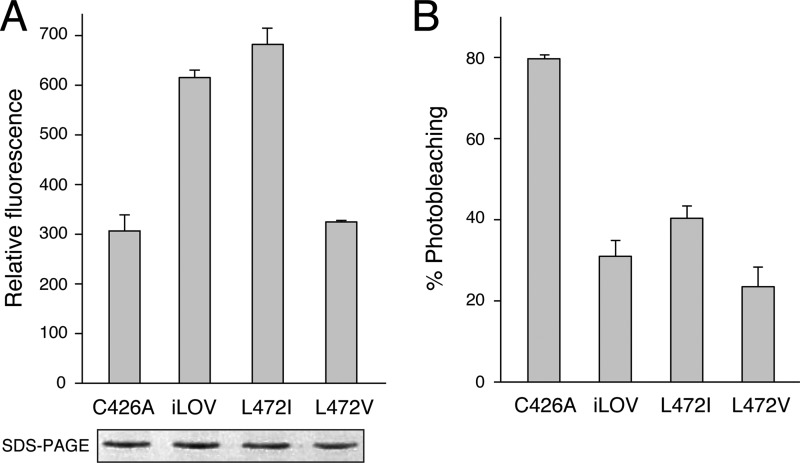
Mutations in Leu-472 were introduced to further probe its contribution to iLOV fluorescence. Substitution to smaller valine reduced iLOV fluorescence in liquid cultures of E. coli to a level comparable with C426A, whereas mutation to size-equivalent isoleucine was without effect (Fig. 4A). Neither replacement enhanced the photostability of iLOV over C426A when irradiated with medium intensity blue light (0.06 W/cm2; Fig. 4B), indicating that side chain rotation of Leu-472 contributes to the improved fluorescence of iLOV over C426A, but has little impact on photostability.
FIGURE 4.
Impact of Leu-472 on iLOV fluorescence. A, in vivo fluorescence of E. coli liquid cultures expressing C426A, iLOV, or iLOV harboring L472V or L472L mutations. B, fluorescence photobleaching of iLOV and respective mutants expressed in E. coli liquid culture. Fluorescence emission intensities were recorded at 495 nm upon excitation with blue light (450 nm). Values represent the mean ± S.D. (n = 3). Protein expression levels are shown in A.
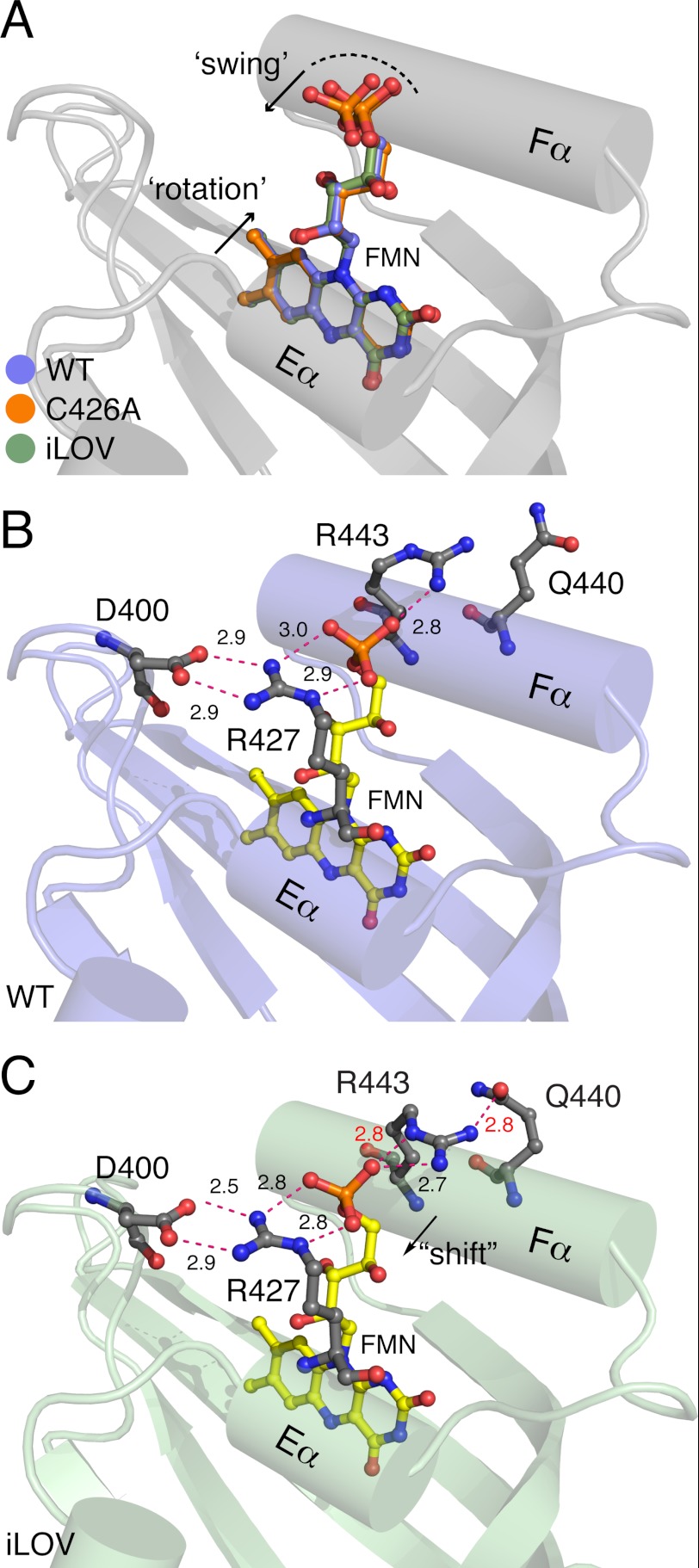
Superposition of WT, C426A, and iLOV structures revealed that the terminal phosphate and ribityl side chain of the FMN chromophore are shifted in iLOV away from the flanking Fα-helix (Fig. 5A). The terminal phosphate of the FMN is typically anchored in LOV domains by conserved salt bridges. In WT, Arg-427 at the Eα-helix links Asp-400 in the Aβ/Bβ loop to the FMN phosphate, which is tethered at the other side by Arg-443 of the Fα-helix (Fig. 5B). Shifting of the FMN phosphate and ribityl tail in iLOV is coupled to side chain movements of Arg-443 and Glu-440 in the Fα-helix that restrict the phosphoribityl tail of the FMN (Fig. 5C). Together, these changes tend to rigidify or reduce the dynamics of the chromophore.
FIGURE 5.
FMN movement in iLOV when compared with wild-type Arabidopsis phototropin 2 LOV2 (WT) and its C426A derivative. Relevant side chains are indicated in gray as ball-and-stick models. Critical distances between residues and the FMN chromophore are highlighted with dashed red lines. A, superposition of the FMN of iLOV (green), WT (pale blue), and C426A (orange) reveals that the isoalloxazine ring is tilted and undergoes some “rotation” when compared with that of WT and C426A. In addition, the phosphoribityl tail of iLOV appears to “swing” and is shifted within the LOV scaffold. B and C, when compared with WT (B), the terminal phosphate of the FMN in iLOV (C) is tightly packed by two salt bridges that comprise two highly conserved arginine residues, Arg-427 and Arg-443, respectively.
Enhancements in iLOV Photostability
Besides being 10-fold brighter than WT, iLOV also exhibits greater photostability (3). With this in mind, we wished to test directed evolution strategies to further fine-tune iLOV for photostability enhancements.
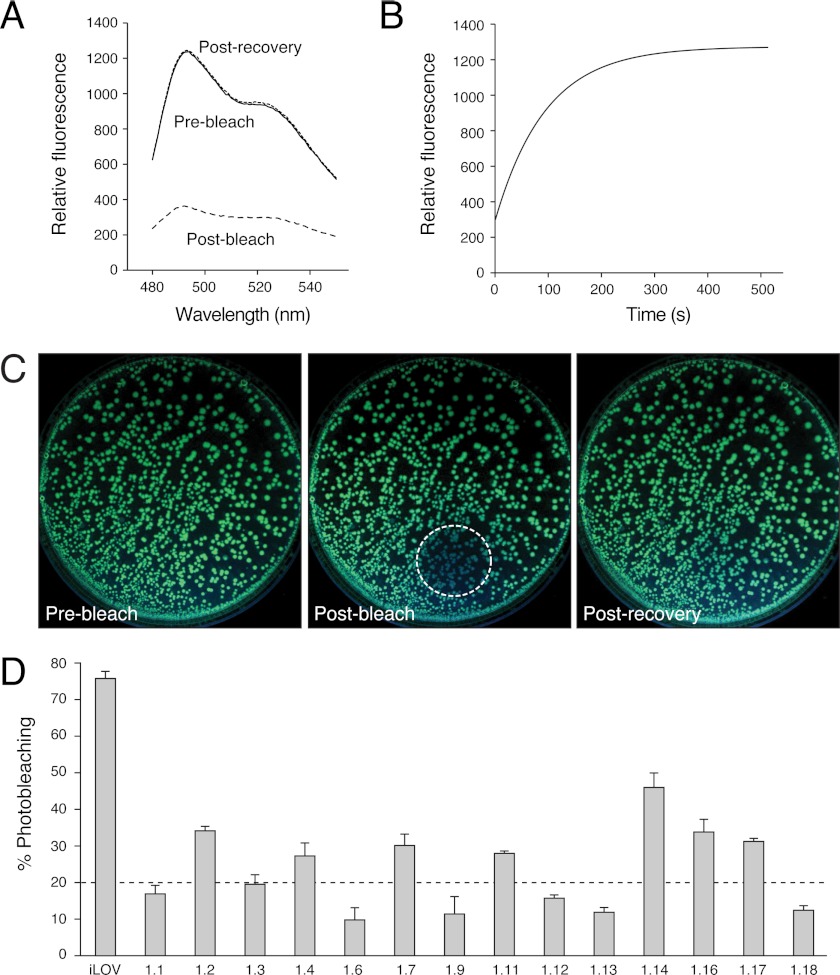
An in vivo screening method was developed in E. coli to assay for improvements in iLOV photostability. When iLOV is expressed in liquid culture, its fluorescence can be photobleached (Fig. 5A) by using a high intensity blue light source (0.16 W/cm2). Consistent with our previous findings in tobacco epidermal cells (3), iLOV fluorescence in E. coli is fully recoverable following photobleaching (Fig. 6A), with a half-maximal recovery time of 75 s (Fig. 6B). Reversible photobleaching of iLOV was also detected in E. coli colonies grown on agar medium (Fig. 6C), providing a medium throughput means to screen for variants with resistance to photobleaching.
FIGURE 6.
Photobleaching of iLOV and screening for improved photostability. A, fluorescence emission spectra of iLOV in liquid cultures of E. coli before photobleaching, after bleaching, and after recovery after 5 min. Spectra were recorded by using an excitation wavelength of 450 nm. B, recovery kinetics for iLOV fluorescence in E. coli after photobleaching. Recovery fits to a first exponential and indicates a half-maximal recovery time of 75 s. C, iLOV fluorescence in E. coli colonies before photobleaching, after bleaching (dashed area), and after recovery after 5 min. D, fluorescence photobleaching of iLOV mutants isolated after one round of mutagenesis and screening for improved photostability. The dashed line represents the cut-off whereby mutant sequences exhibiting greater than 4-fold improvements in photostability were used for a second round of mutagenesis. Fluorescence emission intensities were recorded at 495 nm upon excitation with blue light (450 nm). Values represent the mean ± S.D. (n = 3).
Random mutagenesis was employed to generate a library of iLOV-encoding sequences that were screened for photostability improvements. Fourteen variants from the first round of screening were selected and subjected to further analysis. Comparative in vivo photobleaching measurements in liquid cultures confirmed that all variants selected showed improved photostability when compared with iLOV (Fig. 6D). These variants were designated phiLOV1 (for photostable iLOV, round 1).
phiLOV1.3 and phiLOV1.6 displayed notable blue shifts of ∼10 nm in their fluorescence emission when compared with iLOV (supplemental Fig. S2). Sequence analysis of phiLOV1 variants (supplemental Fig. S3) revealed several amino acid changes that occurred at high frequency (supplemental Table S3). These included mutations in Asn-425 and Glu-430, which hydrogen-bond with the ribityl side of the FMN chromophore. Point mutations of these residues were introduced into iLOV to determine their individual impacts on photostability. Both N425S and Q430R mutations improved iLOV photostability in vivo (supplemental Fig. S4A). However, when these mutations were combined, a severe reduction in fluorescence was observed (supplemental Fig. S4B) as a consequence of reduced flavin binding (supplemental Fig. S4C). PhiLOV1 variants showing >4-fold photostability enhancements (Fig. 6D) were therefore selected as templates for a second round of mutagenesis and DNA shuffling in an attempt to further enhance photostability.
Isolation of iLOV Variants with 10-Fold Enhancements in Photostability
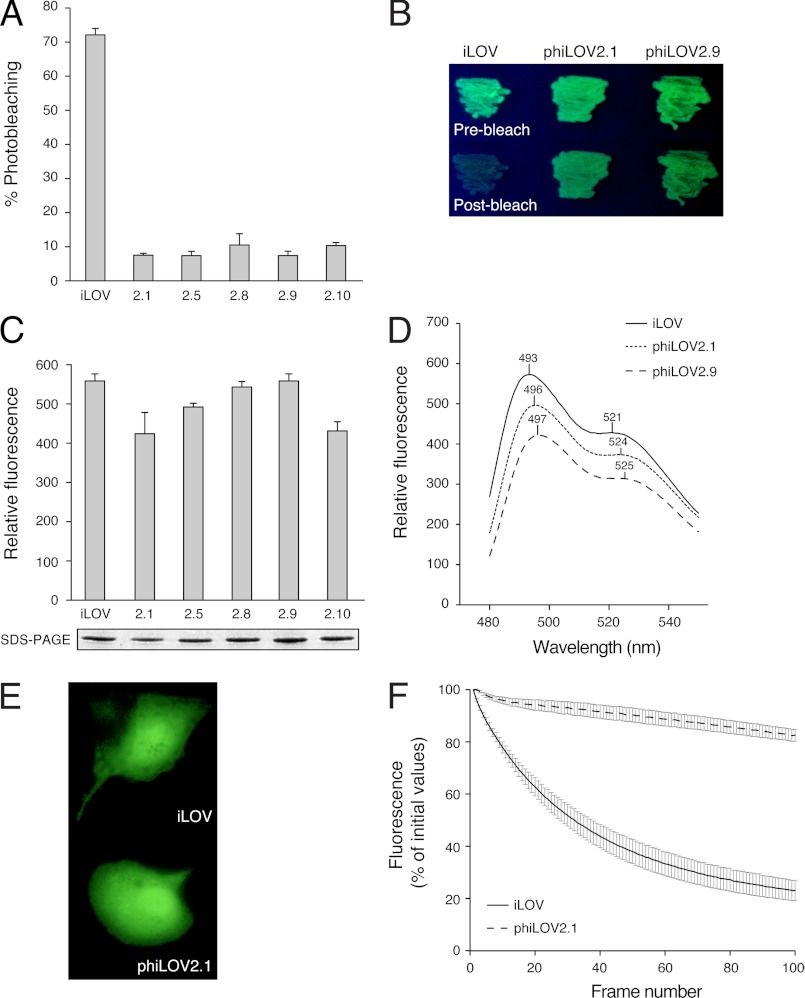
Screening after a second round of mutagenesis on pooled phiLOV1 variants 1.1, 1.3, 1.6, 1.9, 1.12, 1.13, and 1.18 (Fig. 6D) produced further photostability enhancements. These variants, designated phiLOV2, showed ∼10-fold enhancements in photostability when compared with iLOV in liquid culture (Fig. 7A). The improved photostable properties were also clearly visible in E. coli grown on solid agar medium (Fig. 7B).
FIGURE 7.
Characterization of phiLOV2 variants. A, fluorescence photobleaching of variants isolated after a second round of mutagenesis and screening for improved photostability. Fluorescence emission intensities were recorded at 495 nm upon excitation with blue light (450 nm). Values represent the mean ± S.D. (n = 3). B, fluorescence of phiLOV2 variants in E. coli grown on agar medium before photobleaching and after bleaching. C, fluorescence of E. coli liquid cultures expressing phiLOV2 variants. Fluorescence emission intensities were recorded as in A. Protein expression levels are shown. D, fluorescence emission spectra of phiLOV2 variants in liquid cultures of E. coli. Spectra were recorded by using an excitation wavelength of 450 nm. E, fluorescence imaging of free iLOV and phiLOV2.1 expressed in BSC1 monkey kidney cells. F, photobleaching kinetics of iLOV and phiLOV2.1 expressed in BSC1 monkey kidney cells. Fluorescence loss in response to repeated scanning was monitored as described under “Experimental Procedures.” Values represent the mean ± S.E. (n = 2 for iLOV, n = 3 for phiLOV2.1).
Sequence analysis of phiLOV2 variants revealed multiple substitutions throughout the domain (supplemental Fig. S5). These mutations, however, had little impact on the level of in vivo fluorescence of phiLOV2 variants when compared with iLOV (Fig. 7C). Subtle differences were seen in the fluorescence emission spectra for particular variants (phiLOV2.1; 2.9), suggesting alterations within their chromophore-binding environment (Fig. 7D).
phiLOV2.1 was selected for codon optimization to determine whether its improved photostability could be detected in mammalian cells. Although the transfection efficiency for phiLOV2.1 was slightly reduced when compared with that for iLOV, both proteins exhibited robust fluorescence in BSC1 monkey kidney cells (Fig. 7E). However, iLOV fluorescence decreased rapidly after repeated exposures, reaching a steady-state level close to background. In contrast, phiLOV2.1 showed a greater resistance to photobleaching (Fig. 7F), demonstrating that its photostable properties were extendable to mammalian cells.
Structural Characterization of phiLOV2.1
Confirmation of its enhanced photostability in mammalian cells prompted us to determine the structure of phiLOV2.1. Consistent with providing optimal structural comparisons between iLOV and its progenitors, we prepared crystals of phiLOV2.1 belonging to space group P43212 and then solved and refined its structure to 1.4 Å resolution (supplemental Table S1).
phiLOV2.1 contains six mutations when compared with iLOV (supplemental Fig. S5; N390S, N401Y L422M, Q468R, E471G, and D491V). Nonetheless, the overall structure of the LOV/PAS fold is maintained in phiLOV2.1, as are FMN binding features found in iLOV, including side chain rotation of Leu-472 (Fig. 8A).
FIGURE 8.
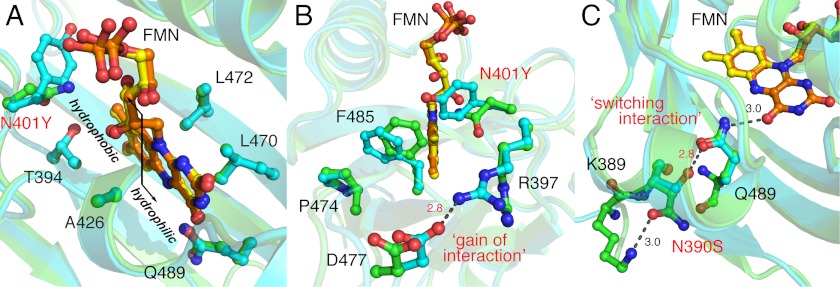
Structural evolution of phiLOV2.1. Superposition of the FMN-binding cavity of iLOV (green) and phiLOV2.1 (pale blue) is shown. FMN is presented as a yellow ball-and-stick model for iLOV and orange for phiLOV2.1. Relevant side chains are indicated as ball-and-stick models. A, phiLOV2.1 maintains iLOV-like FMN binding properties including the flipping of Leu-472 and crowding of the immediate FMN environment by Thr-394. B, the N401Y substitution in phiLOV2.1 (labeled red) influences the chromophore binding environment at the hydrophobic side of the FMN isoalloxazine ring. Arg-397 in phiLOV2.1 is oriented to bridge the edge of the FMN-binding cleft by “gaining interaction” and hydrogen-bonding with Asp-477. C, the N390S mutation in phiLOV2.1 (labeled red) indirectly stabilizes the FMN chromophore by gaining interaction with the side chain of the FMN-interacting residue Gln-489. Critical distances between residues are highlighted with dashed lines.
The crowding effect in iLOV as a consequence of the S394T mutation (Fig. 3C) is similarly preserved in phiLOV2.1 despite the N401Y mutation (Fig. 8A). Introduction of a tyrosine ring at position 401 (Fig. 8A) causes the side chain of Phe-485 to move closer and more parallel to the ring of Pro-474 (Fig. 8B). As a result, the aromatic side chains of Tyr-401 and Phe-485 in phiLOV2.1 cap the hydrophobic end of the FMN isoalloxazine ring and further sequester it from solvent. In addition Val-392, Ile-446, Leu-472, and the methyl group of Thr-394 provide an enriched hydrophobic environment surrounding the isoalloxazine ring. The fluorophore is more constrained in phiLOV2.1 and iLOV than in their progenitors, consistent with decreased B values suggesting enhanced rigidity (supplemental Fig. S6). In contrast to iLOV, positions of Arg-397 and Asp-477 are altered to gain a hydrogen bond that bridges the edge of the FMN-binding cleft (Fig. 8B).
All phiLOV2 variants contain the N390S mutation near the N terminus of the domain (supplemental Fig. S5). In phiLOV2.1, the side chain of S390 introduces a hydrogen bond with neighboring Gln-489 (Fig. 8C), which anchors the FMN at the O4 position in iLOV and its predecessors (Fig. 3). Consequently, interactions between Gln-489 and the FMN chromophore are altered in phiLOV2.1 by impacting the chemical environment surrounding the hydrophilic side of FMN isoalloxazine ring.
DISCUSSION
FPs based on flavin-binding LOV domains represent attractive candidates as fluorescent reporters because of their small size and innate ability to acquire their ubiquitous organic cofactor from the cellular environment. Indeed, their utility as fluorescent biomarkers has already been demonstrated (3–5). Hence, characterization of these flavin-based FPs at the biophysical and structural level to better understand their fluorescent properties is invaluable for extending their utility. The comparative structural and mutational analyses presented here provide a molecular basis for the improved photochemical properties of iLOV and its new photostable derivative phiLOV.
Thr-394 and Leu-470 directly pack against the chromophore (Fig. 3C). Leu-472 contributes to iLOV fluorescence (Fig. 4A) and flips its side chain to pack more closely with the FMN chromophore and Leu-470 (Fig. 3C). Repositioning of a conserved salt bridge between Arg-443 and the FMN phosphate also increases interactions between the protein scaffold and the FMN ribityl side chain (Fig. 5). We therefore propose that these added restrictions limit the dynamics of the FMN chromophore in iLOV and phiLOV and thus its ability to dissipate energy arising from light excitation.
Ser-394 (Thr-394 in iLOV) is conserved in the LOV1 and LOV2 domains of plant phototropins (27). In contrast to the predominant light-sensing role of LOV2 (28), LOV1 shows minimal structural alterations upon photoexcitation (29, 30) and is proposed to mediate receptor dimerization (31, 32). In our structures, Ser-394 forms a hydrogen bond with Asn-401, directing the Ser hydroxyl away from the FMN ring. In the crystal structure of Arabidopsis phototropin 2 LOV1 (31), the corresponding serine (Ser-138) cannot form this hydrogen bond because the asparagine partner is replaced by cysteine (Cys-145) (supplemental Fig. S7). Instead, the serine hydroxyl is oriented toward the isoalloxazine ring, potentially forming tighter contacts with the FMN chromophore (supplemental Fig. S7). Within this region, Thr-394 in iLOV and phiLOV appears to incorporate the structural properties of both LOV1 and LOV2 by forming contacts with the isoalloxazine ring while maintaining a hydrogen bond with Asn-401 (Fig. 3C).
The protein environment on the opposite side of the isoalloxazine ring in iLOV (Fig. 3C) also resembles that in LOV1 (supplemental Fig. S7) because of the F470L mutation. Acquisition of LOV1-like residues in iLOV is not unexpected as both LOV1 and LOV2 sequences were included in the DNA shuffling procedure (3). As a consequence of the F470L mutation, light-induced structural changes in LOV2 are markedly reduced and become minimal like those associated with LOV1 (33). In this regard, directed evolution appears to have targeted regions of the LOV core that are important for LOV2 signal transmission (30, 34), but are dispensable for fluorescent reporter function.
Although iLOV exhibited reduced photobleaching in vivo when compared with its predecessors (3), we showed that it was still amenable to improvements by our photostability selection method (Fig. 6). Consistent with previous studies, photobleaching in vivo is reversible and likely correlates with the formation of a nonfluorescent neutral semiquinone species (3). High light intensities induce the formation of a neutral flavin semiquinone in LOV domains in which the photoactive cysteine has been mutated (35, 36). Flavin semiquinone formation has been proposed to involve intraprotein electron transfer between a conserved tryptophan residue within the LOV domain and the FMN chromophore (35). However, mutation of the corresponding tryptophan residue in iLOV (Trp-467) to arginine, a mutation identified in phiLOV1.6 (supplemental Fig. S3), had no impact on fluorescence photobleaching in liquid cultures of E. coli (supplemental Fig. S4A), indicating the presence of an alternative electron donor pathway.
Under the conditions used in our analysis (∼0.1–0.2 W/cm2), enhancements in FP photostability usually qualitatively predict improved performance under conditions typical for laser scanning microscopy (37). Spectral shifts in fluorescence emission of up to 10 nm were detected during the selection method for phiLOV1.3 and phiLOV1.6 (supplemental Fig. S2). Both these variants contain the Q489L substitution (supplemental Fig. S3) shown previously to account for this spectral property (30, 34). However, spectral shifting beyond this range is unlikely as fluorescence emission of the protein is dictated by the FMN chromophore.
Structural characterization of phiLOV2.1 revealed the potential for further modifications within the LOV/PAS fold that can positively impact the fluorescent properties of this domain (Fig. 8). The structures presented here are relevant to the newly engineered singlet oxygen generator known as miniSOG, which was also derived from the LOV2 domain of Arabidopsis phototropin 2 (5). Four of the five mutation sites in miniSOG are virtually identical to the mutations present in iLOV (supplemental Fig. S8: S394T, S409G, C426G, and F470L in miniSOG when compared with S394T, S409G, C426A, and F470L in iLOV). The fifth mutation in miniSOG, N390S, is present in all phiLOV2 variants (supplemental Fig. S5). The N390S mutation, as well as the N401Y mutation found in phiLOV2.1, had a significant impact on iLOV photostability in vivo (supplemental Fig. S4A). Ser-390 tethers the neighboring FMN-interacting side chain of Gln-489, which in WT rotates 180° following photoexcitation to propagate structural changes at the β-sheet surface important for signaling (30, 34, 38). In phiLOV2.1 and miniSOG (5), Ser-390 restricts Gln-489, thus providing additional, indirect anchoring and rigidifying of the FMN chromophore.
Further optimization of the selection method used here and for other FPs (37) could yield additional improvements in photostability. In this regard, our work highlights a structural framework and new amino acid targets suitable for saturation mutagenesis. Notably, LOV domains are widespread throughout nature and are found in organisms adapted to a diverse range of habitats (39). These represent as yet untapped sources of sequence diversity that could be used to further enhance the properties of genetically encoded LOV FPs for specific cell and biological applications.
Acknowledgments
We are very grateful to Sandra Schmid for providing cell culture and imaging facilities. The SIBYLS beamline facilities at the Advanced Light Source of Lawrence Berkeley National Laboratory are supported in part by the United States Department of Energy (DOE) program Integrated Diffraction Analysis Technologies under Contract DE-AC02-05CH11231, with John Tainer's effort on anaerobic tags by DOE program ENIGMA. We also thank Hideki Kandori and Tatsuya Iwata for helpful discussions and Chiharu Hitomi for technical assistance.
This work was supported by the award of a Royal Society University Research Fellowship (to J. M. C.), National Institutes of Health Grant GM37684 (to E. D. G.), and a grant from the Skaggs Institute for Chemical Biology (to K. H.).

This article contains supplemental Tables S1–S3 and Figs. S1–S8.
- FP
- fluorescent protein
- LOV
- light, oxygen, or voltage domains
- iLOV
- improved LOV
- phiLOV
- photostable iLOV
- PAS
- Period (Per)-Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT)-Single-minded (Sim)
- PYP
- photoactive yellow protein
- W
- watt
- miniSOG
- mini singlet oxygen generator.
REFERENCES
- 1. Shaner N. C., Patterson G. H., Davidson M. W. (2007) Advances in fluorescent protein technology. J. Cell Sci. 120, 4247–4260 [DOI] [PubMed] [Google Scholar]
- 2. Tsien R. Y. (1998) The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 3. Chapman S., Faulkner C., Kaiserli E., Garcia-Mata C., Savenkov E. I., Roberts A. G., Oparka K. J., Christie J. M. (2008) The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. U.S.A. 105, 20038–20043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drepper T., Eggert T., Circolone F., Heck A., Krauss U., Guterl J. K., Wendorff M., Losi A., Gärtner W., Jaeger K. E. (2007) Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445 [DOI] [PubMed] [Google Scholar]
- 5. Shu X., Lev-Ram V., Deerinck T. J., Qi Y., Ramko E. B., Davidson M. W., Jin Y., Ellisman M. H., Tsien R. Y. (2011) A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS. Biol. 9, e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pellequer J. L., Wager-Smith K. A., Kay S. A., Getzoff E. D. (1998) Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. U.S.A. 95, 5884–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor B. L., Zhulin I. B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Genick U. K., Soltis S. M., Kuhn P., Canestrelli I. L., Getzoff E. D. (1998) Structure at 0.85 Å resolution of an early protein photocycle intermediate. Nature 392, 206–209 [DOI] [PubMed] [Google Scholar]
- 9. Brudler R., Gessner C. R., Li S., Tyndall S., Getzoff E. D., Woods V. L. (2006) PAS domain allostery and light-induced conformational changes in photoactive yellow protein upon I2 intermediate formation, probed with enhanced hydrogen/deuterium exchange mass spectrometry. J. Mol. Biol. 363, 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christie J. M. (2007) Phototropin blue light receptors. Annu. Rev. Plant Biol. 58, 21–45 [DOI] [PubMed] [Google Scholar]
- 11. Salomon M., Christie J. M., Knieb E., Lempert U., Briggs W. R. (2000) Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39, 9401–9410 [DOI] [PubMed] [Google Scholar]
- 12. Baca M., Borgstahl G. E., Boissinot M., Burke P. M., Williams D. R., Slater K. A., Getzoff E. D. (1994) Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry 33, 14369–14377 [DOI] [PubMed] [Google Scholar]
- 13. Möglich A., Yang X., Ayers R. A., Moffat K. (2010) Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 61, 21–47 [DOI] [PubMed] [Google Scholar]
- 14. Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., Briggs W. R. (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701 [DOI] [PubMed] [Google Scholar]
- 15. Christie J. M., Salomon M., Nozue K., Wada M., Briggs W. R. (1999) LOV (light, oxygen, or voltage) domains of the blue light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. U.S.A. 96, 8779–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swartz T. E., Corchnoy S. B., Christie J. M., Lewis J. W., Szundi I., Briggs W. R., Bogomolni R. A. (2001) The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276, 36493–36500 [DOI] [PubMed] [Google Scholar]
- 17. Drepper T., Huber R., Heck A., Circolone F., Hillmer A. K., Büchs J., Jaeger K. E. (2010) Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76, 5990–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tielker D., Eichhof I., Jaeger K. E., Ernst J. F. (2009) Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryot. Cell 8, 913–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christie J. M., Corchnoy S. B., Swartz T. E., Hokenson M., Han I. S., Briggs W. R., Bogomolni R. A. (2007) Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry 46, 9310–9319 [DOI] [PubMed] [Google Scholar]
- 25. Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. (2010) Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit 14.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., Hendrickson W. A. (1984) The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature 312, 127–134 [DOI] [PubMed] [Google Scholar]
- 27. Crosson S., Moffat K. (2001) Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christie J. M., Swartz T. E., Bogomolni R. A., Briggs W. R. (2002) Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32, 205–219 [DOI] [PubMed] [Google Scholar]
- 29. Iwata T., Nozaki D., Tokutomi S., Kandori H. (2005) Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry 44, 7427–7434 [DOI] [PubMed] [Google Scholar]
- 30. Nozaki D., Iwata T., Ishikawa T., Todo T., Tokutomi S., Kandori H. (2004) Role of Gln-1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry 43, 8373–8379 [DOI] [PubMed] [Google Scholar]
- 31. Nakasako M., Zikihara K., Matsuoka D., Katsura H., Tokutomi S. (2008) Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 381, 718–733 [DOI] [PubMed] [Google Scholar]
- 32. Salomon M., Lempert U., Rüdiger W. (2004) Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 572, 8–10 [DOI] [PubMed] [Google Scholar]
- 33. Yamamoto A., Iwata T., Tokutomi S., Kandori H. (2008) Role of Phe-1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry 47, 922–928 [DOI] [PubMed] [Google Scholar]
- 34. Jones M. A., Feeney K. A., Kelly S. M., Christie J. M. (2007) Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J. Biol. Chem. 282, 6405–6414 [DOI] [PubMed] [Google Scholar]
- 35. Eisenreich W., Fischer M., Römisch-Margl W., Joshi M., Richter G., Bacher A., Weber S. (2009) Tryptophan 13C nuclear-spin polarization generated by intraprotein electron transfer in a LOV2 domain of the blue light receptor phototropin. Biochem. Soc. Trans. 37, 382–386 [DOI] [PubMed] [Google Scholar]
- 36. Kay C. W., Schleicher E., Kuppig A., Hofner H., Rüdiger W., Schleicher M., Fischer M., Bacher A., Weber S., Richter G. (2003) Blue light perception in plants: detection and characterization of a light-induced neutral flavin radical in a C450A mutant of phototropin. J. Biol. Chem. 278, 10973–10982 [DOI] [PubMed] [Google Scholar]
- 37. Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W., Tsien R. Y. (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nash A. I., Ko W. H., Harper S. M., Gardner K. H. (2008) A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry 47, 13842–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pathak G. P., Ehrenreich A., Losi A., Streit W. R., Gärtner W. (2009) Novel blue light-sensitive proteins from a metagenomic approach. Environ. Microbiol. 11, 2388–2399 [DOI] [PubMed] [Google Scholar]