Abstract
Mutational events as well as the selection of the optimal variant are essential steps in the evolution of living organisms. The same principle is used in laboratory to extend the natural biodiversity to obtain better catalysts for applications in biomanufacturing or for improved biopharmaceuticals. Furthermore, single mutation in genes of drug-metabolizing enzymes can also result in dramatic changes in pharmacokinetics. These changes are a major cause of patient-specific drug responses and are, therefore, the molecular basis for personalized medicine. MuteinDB systematically links laboratory-generated enzyme variants (muteins) and natural isoforms with their biochemical properties including kinetic data of catalyzed reactions. Detailed information about kinetic characteristics of muteins is available in a systematic way and searchable for known mutations and catalyzed reactions as well as their substrates and known products. MuteinDB is broadly applicable to any known protein and their variants and makes mutagenesis and biochemical data searchable and comparable in a simple and easy-to-use manner. For the import of new mutein data, a simple, standardized, spreadsheet-based data format has been defined. To demonstrate the broad applicability of the MuteinDB, first data sets have been incorporated for selected cytochrome P450 enzymes as well as for nitrilases and peroxidases.
Database URL: http://www.MuteinDB.org
Introduction
One of nature’s fundamental mechanisms to create genetic diversity in living organisms is the creation of mutants, which, in turn, leads to evolution. Mutational events and selection of the optimal variant are essential to obtain a better catalyst. In human medicine, enzyme polymorphisms arising from evolutionary events have been identified since the 1960s (1). Physicians recognized that patients with the same disease responded differently to drugs, according to which allelic variant their genomes were carrying. This opened the road to what is nowadays called ‘personalized medicine’ (2). Additionally, industry desires to artificially improve enzymes through mutation and selection. To this end, efficient protein engineering tools to create tailor-made enzyme variants, named ‘muteins’, have been developed over the past decades (3). Muteins generated either by rational design or by directed or designed evolution were adapted to the needs of industrial processes or for completely new applications.
Increasing interest in personalized medicine and in tailor-made enzymes in the fast-growing biocatalysis industry has led to an exponential increase of literature about muteins and their influence on enzymes’ kinetic properties. In a plethora of examples, the artificial substitution of one or more amino acids in a polypeptide resulted in a significant increase or decrease of stability, turnover rate or substrate specificity for the enzyme (4). Even new reactions or activities on molecules that were not substrates for the natural parental enzyme can be caused by just a few or every single mutation. For example, esterases could be changed to hydroxynitrile lyases and epoxide hydrolases (5,6). Papain, a protease, was modified to an enzyme with efficient nitrile hydratase activity (7). Furthermore, the fatty acid hydroxylase CYP102A1 and the camphor hydroxylase CYP101 were redesigned to efficient alkane hydroxylases (8–10). By a single mutation, the broadly applied lipase CalB was modified to perform aldol additions and epoxidations (11). More recently, a transaminase showing almost no activity for a commercially interesting substrate was mutated to a highly active and selective catalyst enabling a new efficient industrial process for sitagliptin production (12).
Information about specific proteins and their muteins are widely spread in the literature. Many studies only describe single mutation and its effects without comparison to already known muteins. Possible additive effects of single amino acid changes are scarcely described or used. Even after a thorough and time-consuming literature search, researchers face the problem of assembling and presenting the data in an easy understandable and comprehensive way. Essential information may be lost such as details about potentially cooperative mutations or reactions one would not expect in certain protein families. Therefore, a web-accessible database combining available knowledge about a specific enzyme and its muteins in a single place are highly desirable. Such a database would allow researchers to access relevant information about their protein of interest in a fast and easy way and accelerate the engineering of new and improved variants.
Existing, comprehensive enzyme engineering databases such as CYPED are mainly focused on enzyme sequences and their structures (13). Only a few databases go beyond that and contain, to some extent, information about muteins and their properties. The most recently published database introducing mutein information is SuperCYP (14), which exclusively addresses human cytochrome P450s. Another example is SPROUTS (15), which provides details on the influence of point mutations on protein stability. The Protherm database (16) contains experimental thermodynamic data, and BRENDA (17), a well-known enzyme databases, includes only a small section about muteins. Finally, the Protein Mutant Database (18) includes references to mutant proteins from the literature. However, none of these databases provides kinetic characteristics of muteins and allows a fast, systematic and user-friendly way to search for known mutations and catalyzed reactions of interest. All these databases focus on enzymes and provide information about their variants from the view of the protein. Additionally, none of the existing databases is searchable by substrate or product molecule structures allowing comparison of muteins with respect to their catalytic properties.
In this article, we present the novel database MuteinDB (http://www.MuteinDB.org). It is a user-friendly graphically appealing database devoted to provide easy access to detailed information on naturally occurring and laboratory-evolved muteins as well as on the influence of mutations on kinetics of catalyzed reactions, including inhibition. It allows to search for the best biocatalyst for a given substrate, reaction or product simply by substrate name, Chemical Abstracts Service (CAS) number or molecule structure. In addition, a structure search tool offers the possibility to predict muteins which most likely accept a new substrate, if no enzyme/substrate properties were described so far.
MuteinDB overview
The MuteinDB is a platform to collect, catalog, and store experimentally derived data about muteins from publicly available sources as well as data directly submitted by the scientists. Additionally, it allows flexible searches by reaction type, molecular (sub) structures, substrate, product or mutein name. MuteinDB provides details on catalyzed reactions, kinetic data (activity, kinetic resolution) and experimental conditions used for data generation as well as for possible substrates or products consumed or produced by a reaction of choice including relevant scientific publication or patent information. Furthermore, it is possible to screen all enzyme variants for known interactions with specific inhibitors. Substrate, product and inhibitor data are linked to CAS and/or CID number (PubChem) as a unique identifier for unambiguous reference. The use of these distinct identification numbers allows even to extract information about (comparative) stereo- or enantioselectivity of individual muteins. Furthermore, once the user has identified a mutein of interest, information about its sequence, the employed expression hosts, cofactors, cosubstrates, and coproteins can be directly shown. In the sequence view, all mutations of a specific mutein are highlighted and liked to other muteins with known mutations of the same position. Additionally, the wild type sequence including all known amino acid exchanges which are again linked to muteins containing the modified position is illustrated for all muteins.
For first-time users, a comprehensive frequently asked question (FAQ) section and in-depth tutorial movies are provided. The key features are described in more detail below.
Search options
In contrast to other databases, MuteinDB allows users not only to query for muteins and mutations but also for substrates, products, or inhibitors, using the corresponding name or CAS number as well as catalyzed reaction types. The database also provides means to easily and efficiently search the data (e.g. by allowing to enter wildcards in the values) and display it in a clear, tabular form.
Structural search
Another important difference to other existing databases is the fully integrated (sub) structure search tool. It allows searching for substances with a similar structure by drawing an arbitrary chemical structure in the JME Molecule Editor (19). The database will provide all possible hits related to the drawn structure, and the user can navigate amongst them to refine the search. Based on knowledge about mutein/substrate combinations and their specific products, this for the first time also allows predictions of other possible substrates and products for known muteins which were not experimentally evaluated so far.
Individual features of MuteinDB
MuteinDB uses a mutein-based classification. A unique ID is assigned to each mutein and is linked to the reference source, the catalyzed reaction and the corresponding wild type protein. This mutein-centric approach allows more flexible and specific searches compared to the publication-based classification of the Protein Mutant Database (18) or the reaction based classification of BRENDA (17). Each reaction and publication reference can be independently surveyed, which is especially important when amino acid changes result in new functionalities. An example for such a case is the lipase CALB that was modified to a C–C bond forming enzyme for aldol additions (11).
Basic information about underlying wild type protein sequences, structures and source organism as well as compound structures, their respective references and reactions are retrieved from the public databases GenBank, PDB, UniProt, PubChem, PubMed, CrossRef, and KEGG (20–24). Wherever third party data is presented, it is linked to the corresponding database entry.
Standardized format for data collection and import
The recent introduction of experimental high-throughput techniques required the development of standardized formats for data from biological experiments. They facilitate exchange of data, their storage in publicly accessible repositories, increase experimental transparency and allow reproduction of bioinformatic analyses from publications. For example, such formats are available for DNA-microarray experiments (25), proteomics studies (26), and data deriving from qPCR experiments (27). Most of them are XML based, which can be difficult to create and manipulate. Therefore, simpler, spreadsheet-based formats have been introduced which are more accessible for the individual researcher. A prominent representative is the MAGE-TAB format for 45 DNA-microarray experiments (28).
Here, we propose a standardized spreadsheet-based data exchange format for muteins and related experimental kinetic data. The MuteinDB import spreadsheet comprises seven sections for each entry: (i) basic data; (ii) signal sequences; (iii) pH conditions; (iv) temperature conditions; (v) storage stability; (vi) reaction data and (vii) activity data. The basic data section includes the enzyme’s name, the GenBank protein ID and the PDB ID (if available). Additionally, the corresponding wild-type name and the sequence mutations are illustrated for muteins. The reaction section contains the substrate and the product of the reaction (both with CAS number and name), the enzyme classification (EC) number of the reaction and the reaction type. The activity section can cover one of following types: conversion activity, enatiomeric excess or inhibition. All three types are followed by the corresponding kinetic values and the experimental conditions. The provided standards for kinetic data necessitate a minimum quality of biochemical protein data (e.g enzyme activity provided in µmol product made by µmol enzyme per minute).
A detailed description of the fields along with guidelines for data collection and a template spreadsheet are available on the MuteinDB homepage. Standardized entry of data into the spreadsheet is ensured by drop-down lists for fields with a defined value set. Drop-down lists can be extended if new values for a field are required.
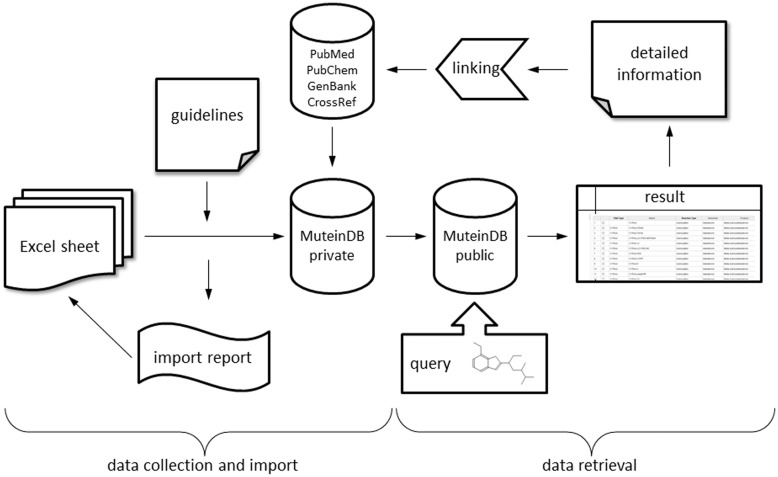
For data import, the files are checked for data consistency according to the guidelines and compared with the already existing mutein data to prevent duplicate entries (Figure 1). A detailed report on the import is provided, allowing focused modification of the data to adjust it conforming to the guidelines. Upon successful import, the data is reviewed by an expert team at Graz University of Technology and feedback is provided to the submitter. After all inconsistencies are resolved, the new content is publicly released. New data can be submitted any time and is made available immediately after the review.
Figure 1.
Schematic diagram of database structure. MuteinDB structure can be divided into two major parts. Firstly, the data collection and import structure within MuteinDB, illustrated on the left. Detailed guidelines structure and specify the correct and unified data collection as well as the data import. The standardized excel data import template guarantees data quality and consistency. During the automated data import from the data import excel sheet, metadata from third party databases such as PubMed, PubChem, GenBank and CrossRef are retrieved and added. The data import procedure ends either with a summary including imported muteins, molecules, reactions, activities or with a detailed error report. Secondly, stored public mutein data can be easily retrieved via various search mechanisms. For example, chemical structures can be used for identifying molecules of interest and their catalyzed reactions. Results are presented in tabular listings with links to third party databases or to detailed information contained in MuteinDB.
MuteinDB structure and implementation
The MuteinDB is implemented using Java, an object-oriented and platform-independent programming language. The application is based on a 3-tier architecture with an Oracle database as the persistence tier, an application server (JBoss) as the middle tier and a WEB interface as the client tier. Business logic is implemented using Enterprise JavaBeans 3. The web interface depends on JavaServer Faces 2, Asynchronous JavaScript and XML and JBoss Seam. The relational database schema has been designed to accommodate controlled vocabularies in form of a data dictionary. Attributes with a defined value set are linked to data dictionary entries to facilitate standardized content in the database.
For substructure search (5), the JME Molecule Editor (19) and the Chemistry Development Kid (CDK)—an open-source Java library—are used.
Use of MuteinDB
The MuteinDB was developed as a user-friendly and intuitive resource of mutein-related properties for scientists in the fields of biology, biotechnology, organic chemistry and pharmaceutical sciences. The top information bar offers ‘FAQs’ where users will find helpful information. Furthermore, first-time users will find tutorial movies explaining the database usage and the different MuteinDB sections.
The simplest search option ‘Search by Substrate’ is directly accessible via the home screen. The left side navigation bar gives access to further querying options.
Search options
Substrate: enables the user to search for muteins that convert a certain substrate of interest.
Reaction: enables the user to search for specific reactions by entering a molecule name or a CAS number for the substrate and/or the product (including single enantiomers)
Structure: enables the user to draw chemical structures to search for similar or exact (sub) structure matches in either one or all of the molecule categories (substrate, product and/or inhibitor).
Inhibitor: enables the user to search for inhibitors of muteins and wild-type enzymes by entering a molecule name or a CAS number.
Mutation: enables the user to search for muteins containing mutations at a certain position.
Wild type: enables the user to browse all muteins and their reactions for a defined wild-type enzyme.
Mutein: enables the user to search for all relevant reactions for a defined mutein name.
To keep the additional querying options simple and flexible, further refinements of the query can, but do not have to, be specified or selected. For example, the search can be restricted amongst others to the reaction type, the underlying wild type protein, or to a specific organism.
All text fields are equipped with ‘suggest input’. While typing a box will appear and provide suggestions one can choose from. Furthermore, selected fields allow ‘wildcard search’ with ‘*’ as a placeholder.
Example workflow
The ability to search for exact or similar structures is one of the unique main features of the MuteinDB. Therefore, we will describe this search type in more detail and use it as example to demonstrate the ability of MuteinDB for valuable data retrieval (Figure 2).
Figure 2.
MuteinDB structure search, its results and the capabilities of the MuteinDB webinterface. (A) The MuteinDB (sub) structure search uses the JME editor, which allows users to draw arbitrary molecular structures. (B) The user-drawn structure is used as seed for the following database search and shown on top of the structure search result table. In this table all molecules, substrates, products or inhibitors which contain the query structure are presented. A selection of these molecules can be used for a subsequent ‘Search by Reaction’. (C) All wild type enzymes and muteins which catalyze the selected molecules are shown. (D) For each row of the tabular result, further information can be obtained via the mutein or wild type name. The detailed information is organized in four main categories: (i) basic data; (ii) properties; (iii) substrate and (iv) sequence. (E) The ‘Sequence’ tab of the selected mutein allows to explore the sequence of the mutein as well as the wild type sequence. Known mutations are highlighted and linked to the corresponding entries of MuteinDB. (F) Information in the ‘Substrate’ tab is linked to third party databases. For example, (F) molecules are linked to PubChem, (H) EC-Numbers to Brenda and (G) literature to PubMed or to its DOI location. For muteins, experimental settings and wild type activity values are available from the ‘Substrate’ tab.
Selecting ‘Search by Structure’ will open the JME Molecule Editor Applet (19) and allows the user to draw an arbitrary chemical structure (Figure 2A). After submitting the search, results will be presented as a table listing all molecules containing the drawn structure (Figure 2B). The ‘structure result’ page proposes several related substrates, products and inhibitors with similar structures to the drawn molecule structure. In all result views, moving the cursor over a molecule name will show its chemical structure. Additionally, each molecule name is linked to PubChem (22). This also facilitates the search if the CAS number or the exact molecule name is unknown or if different trivial names of the molecule are commonly used.
One or several molecules can be selected via checkboxes and can be used for a subsequent search by substrate/product or inhibitor. The results are shown again in tabular form listing all muteins that convert the selected substrates or produce the selected products or are inhibited by the chosen inhibitors.
Selecting ‘testosterone’ from the list for a subsequent search reveals several muteins that are able to convert this steroid (Figure 2C). This supports predictions about possible transformations of testosterone derivatives where no experimental data is available so far. Hits from such searches are preferred muteins for experimental evaluation.
Information about the catalyzed reactions such as substrate, product and reaction type are presented in the ‘substrate view’ (Figure 2D). A link to KEGG reaction (29) is provided when a corresponding entry exists. As the kinetic data are one of the most important pieces of information stored in the database, kinetic parameters such as Km and kcat are given. Furthermore, enantiomeric access and E-values are provided if available. The view can be customized using ‘edit display settings’.
As multiple publications may have reported the same reaction for a given mutein, the one stating the highest activity is shown in the main result screen. By using the expand button the data from the other reports are also shown. Clicking on the mutein name will bring up the ‘mutein view’ where detailed information about the mutein and the reaction are provided.
In the ‘substrate section’, several mouse-over buttons (Figure 2D) give further information about the catalyzed reaction. ‘C’ shows comments on the reaction, ‘W’ gives activity data of the underlying wild-type reference, ‘R’ shows information about reaction conditions and analysis and ‘L’ provides detailed information about the corresponding literature. The PubMed ID or the digital object identifier (DOI, http://crossref.org) of the publications are given and directly linked to PubMed or to the webpage associated with the digital object identifier, respectively. Additionally, the EC number is provided and linked to the comprehensive enzyme database BRENDA (Figure 2F–H).
The ‘sequence section’ shows the mutein sequence aligned with its corresponding wild-type sequence (Figure 2E). In the mutein sequence, the mutations are highlighted in violet. The sequence can be downloaded as FASTA format. The amino acids of the wild-type sequence highlighted in blue mark the positions of known mutations. These positions are linked to the ‘enzyme mutation view’. In this view, all muteins that contain a mutation at this position are listed. Via the mutein name it is possible to navigate to the mutation view of the corresponding mutein.
Another highlight of the MuteinDB is the ability to select two or more muteins, which convert the substrate of interest or form the product of interest, for comparison in side by side view. In the ‘compare view’ the kinetic data of the catalyzed reaction as well as information about the mutations, expression system and involved cofactors and coproteins are displayed.
Inhibitors have a special status and may have been reported in the ‘structure result’ page for the inhibitor search. The results are shown in tabular form (Figure 3) listing muteins that are inhibited by the chemical compound. Instead of kinetic data, the inhibitor constant Ki or the IC50 value are provided. Additionally, the underlying reaction used to determine the inhibitor constant is shown.
Figure 3.
Result display of the MuteinDB web-interface for testosterone as a substrate. Information within the result listing for each mutein is by default grouped into catalyzed reaction and kinetic data. Reaction information comprises the reaction type as well as the catalyzed substrate and product. Molecules are directly linked to their corresponding PubChem entry. Additionally, the molecule structure can be displayed by moving over the compound’s name. Important kinetic parameters such as K value, activity value including its unit as well as the relative activity in (%) are directly available in the result view. All presented information and further links for each mutein or wild type is directly linked by its name.
As the same inhibitor measurements can be found in different publications, only the one with the highest inhibition constant is shown as the main result. Via the expand button, the data of the other literature sources is shown. The mutein name is again linked to the ‘mutein view’, where detailed information on the mutein and the inhibition reaction are provided.
Results and conclusions
MuteinDB is a comprehensive and carefully curated database for specific muteins and their kinetic data of catalyzed reactions including inhibition. It provides in-depth information on mutein properties combined with flexible search capabilities. The MuteinDB has been designed to be broadly applicable to proteins and their muteins from any enzyme class including those with no known catalytic function. We demonstrated this by entering data sets of several enzymes and their variants of different enzyme classes.
Presently, the understanding of the structure–function relationship of proteins is still limited. Scientists are trying to tackle the problem from different perspectives (from medicine and pharmacokinetics, to structural biology or applied biocatalysis) and are, therefore, interested in how mutations can influence catalytic properties.
By means of MuteinDB a user can find enzymes that catalyze a particular reaction not only in expected enzyme classes but also in others [e.g. a C–C bond forming mutein derived from a hydrolase (30)]. This feature helps to identify potential starting points for further enzyme engineering. Moreover, medical scientists can get information about the influence of mutations on the drug metabolism and the in vivo activation. This helps to predict a patient’s personal response to certain administered drugs. In addition, the implemented structure search for substrates, products and inhibitors allows the prediction of structure scaffolds that could be accepted by muteins. This might provide helpful information for the development of new biocatalysts and, most probably, will facilitate drug metabolite prediction in pharmaceutical research and development.
At present MuteinDB contains several thousand reactions (Table 1) for muteins of different enzyme classes. It is the largest collection of kinetic data of muteins compiled in a single database. To demonstrate the general applicability of the database, different types of enzymes from different origins have been searched in literature and imported into MuteinDB. Data were collected by searching SciFinder (www.cas.org) and PubMed (21) abstracts for specific keywords. Detailed data from texts, tables and figures were manually extracted from the matching full-text publications and were curated by a team of scientists, who enriched the published information with first-hand kinetic data wherever possible.
Table 1.
MuteinDB data overview
| Wild-type Name | Muteins | Reactions | Activities | Publications |
|---|---|---|---|---|
| CYP102A1 | 168 | 909 | 995 | 42 |
| CYP102A2 | 0 | 4 | 4 | 1 |
| CYP2D6 | 98 | 648 | 1259 | 213 |
| CYP3A4 | 124 | 825 | 1908 | 220 |
| HAPMO | 6 | 106 | 114 | 5 |
| HRP C1 | 17 | 32 | 45 | 8 |
| Nitrilases | 8 | 26 | 26 | 3 |
| NITAf | 11 | 42 | 42 | 2 |
| P3H | 0 | 1 | 12 | 1 |
| P3H type1 | 0 | 21 | 31 | 3 |
| P3H type2 | 0 | 16 | 23 | 2 |
| P4H | 0 | 21 | 53 | 4 |
| PAMO | 31 | 309 | 385 | 10 |
| Total: 11 | 444 | 2892 | 4829 | 422 |
CYP2D6 and CYP3A4 are human liver enzymes and known to be involved in drug metabolism. Both enzymes have been chosen as primary data sets due to their pronounced polymorphism and high importance for human drug and xenobiotic metabolism. We selected CYP102A1 (BM-3) from Bacillus megaterium as a prokaryotic representative. This protein is one of the most mutated and investigated proteins known.
To import the data, we used the standardized spreadsheet-based import file format described previously. It contains all attributes necessary to describe a mutein and its properties.
In order to augment the database content, data collection is on-going. To make the database as comprehensive and up-to-date as possible, we are addressing the research community with a request to aid us in the collection of kinetic data sets for enzymes of different type and origin. We appreciate any contribution to the database both updates to existing data and new kinetic data sets.
Future directions
In the course of integrating new data sets, the MuteinDB will be adapted, and the guidelines for data collection will be adjusted. Feedback from end users and data collectors will ensure a continued focus on a user-friendly development.
The collection of data sets was carried out as part of the OXYGREEN (www.oxygreen.org) project, a research collaboration funded by the European Commission Seventh Framework Programme (EU FP7), and will be continued to do so. MuteinDB will be used and extended in the context of BIONEXGEN, a recently funded EU project. To ensure continuation of data collection and curation of the database, MuteinDB will be integrated into future projects.
A downloadable version of the MuteinDB is in preparation. It will be provided for companies or universities that would like to store their own data in-house. The data can be integrated into the public online database on request. The download will be available in exchange for new mutein data sets or for a fee for database curation and data collection.
Funding
EU-FP7 project OXYGREEN (EC grant 212281); the Austrian Ministry of Science and Research GEN-AU project BIN (FFG grant 820962). The Austrian Centre of Industrial Biotechnology (ACIB) contribution was supported by FFG, bmvit, mvwfi, ZIT, Zukunftsstiftung Tirol and Land Steiermark within the Austrian COMET programme (FFG grant 824186). Funding for open access charge: EU-FP7 project OXYGREEN (EC grant 212281).
Conflict of interest. None declared.
Acknowledgements
We would like to thank Andrea Camattari, Peter Remler, Norbert Klempier, Kurt Faber, Peter Macheroux, Helmut Schwab and the whole OXYGREEN team for assistance and fruitful discussions. Furthermore, we want to thank Peter Ertl for providing the JME molecular editor applet.
References
- 1.Ford EB. Genetic polymorphism. Proc. R. Soc. Lond. B Biol. Sci. 1966;164:350–361. doi: 10.1098/rspb.1966.0037. [DOI] [PubMed] [Google Scholar]
- 2.Shastry BS. Pharmacogenetics and the concept of individualized medicine. Pharmacogenomics J. 2006;6:16–21. doi: 10.1038/sj.tpj.6500338. [DOI] [PubMed] [Google Scholar]
- 3.Lutz S, Bornscheuer TU. Protein Engineering Handbook. Weinheim, Germany: Wiley-VCH GmbH & Co.KGaA; 2008. [Google Scholar]
- 4.Brannigan JA, Wilkinson AJ. Protein engineering 20 years on. Nat. Rev. Mol. Cell Biol. 2002;3:964–970. doi: 10.1038/nrm975. [DOI] [PubMed] [Google Scholar]
- 5.Pan K, Zhang R, Sun H, et al. Proceedings of the Third International Multi-symposiums on Computer and Computational Sciences (IMSCCS'08) IEEE Computer Society Press; 2008. An implementation of substructure search in chemical database management system; pp. 203–206. [Google Scholar]
- 6.Padhi SK, Fujii R, Legatt GA, et al. Switching from an esterase to a hydroxynitrile lyase mechanism requires only two amino acid substitutions. Chem. Biol. 2010;17:863–871. doi: 10.1016/j.chembiol.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Reddy SY, Kahn K, Zheng YJ, et al. Protein engineering of nitrile hydratase activity of papain: molecular dynamics study of a mutant and wild-type enzyme. J. Am. Chem. Soc. 2002;124:12979–12990. doi: 10.1021/ja020918l. [DOI] [PubMed] [Google Scholar]
- 8.Urlacher V, Schmid RD. Biotransformations using prokaryotic P450 monooxygenases. Curr. Opin. Biotechnol. 2002;13:557–564. doi: 10.1016/s0958-1669(02)00357-9. [DOI] [PubMed] [Google Scholar]
- 9.Peters MW, Meinhold P, Glieder A, et al. Regio- and enantioselective alkane hydroxylation with engineered cytochromes P450 BM-3. J. Am. Chem. Soc. 2003;125:13442–13450. doi: 10.1021/ja0303790. [DOI] [PubMed] [Google Scholar]
- 10.Glieder A, Farinas ET, Arnold FH. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 2002;20:1135–1139. doi: 10.1038/nbt744. [DOI] [PubMed] [Google Scholar]
- 11.Branneby C, Carlqvist P, Magnusson A, et al. Carbon-carbon bonds by hydrolytic enzymes. J. Am. Chem. Soc. 2003;125:874–875. doi: 10.1021/ja028056b. [DOI] [PubMed] [Google Scholar]
- 12.Savile CK, Janey JM, Mundorff EC, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 13.Fischer M, Knoll M, Sirim D, et al. The Cytochrome P450 Engineering Database: a navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 2007;23:2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- 14.Preissner S, Kroll K, Dunkel M, et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010;38:D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonquety M, Lacroix Z, Papandreou N, et al. SPROUTS: a database for the evaluation of protein stability upon point mutation. Nucleic Acids Res. 2009;37:D374–D379. doi: 10.1093/nar/gkn704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar MD, Bava KA, Gromiha MM, et al. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34:D204–D206. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schomburg I, Chang A, Ebeling C, et al. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ertl P. Molecular structure input on the web. J. Cheminform. 2010;2:1. doi: 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Clark K, et al. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler DL, Church DM, Edgar R, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Xiao J, Suzek TO, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Uniprot-Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman H, Henrick K, Nakamura H, et al. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 26.Taylor CF, Paton NW, Lilley KS, et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 27.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 28.Rayner TF, Rocca-Serra P, Spellman PT, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotera M, Hirakawa M, Tokimatsu T, et al. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol. Biol. 2012;802:19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Hassler M, Bugg TD. Catalytic promiscuity in the alpha/beta-hydrolase superfamily: hydroxamic acid formation, C–C bond formation, ester and thioester hydrolysis in the C–C hydrolase family. Chembiochem. 2008;9:71–76. doi: 10.1002/cbic.200700428. [DOI] [PubMed] [Google Scholar]