Abstract
An initiative of the NIH Blueprint for neuroscience research, the Neuroscience Information Framework (NIF) project advances neuroscience by enabling discovery and access to public research data and tools worldwide through an open source, semantically enhanced search portal. One of the critical components for the overall NIF system, the NIF Standardized Ontologies (NIFSTD), provides an extensive collection of standard neuroscience concepts along with their synonyms and relationships. The knowledge models defined in the NIFSTD ontologies enable an effective concept-based search over heterogeneous types of web-accessible information entities in NIF’s production system. NIFSTD covers major domains in neuroscience, including diseases, brain anatomy, cell types, sub-cellular anatomy, small molecules, techniques, and resource descriptors. Since the first production release in 2008, NIF has grown significantly in content and functionality, particularly with respect to the ontologies and ontology-based services that drive the NIF system. We present here on the structure, design principles, community engagement, and the current state of NIFSTD ontologies.
Keywords: ontologies, ontology reuse, neuroscience ontology, semantic search
Introduction
The Neuroscience Information Framework Project (NIF)1 facilitates the utilization of the growing number of neuroscience-relevant data available through the web. NIF, supported by the National Institutes of Health Blueprint, was initiated in recognition of the current difficulties of locating and searching across the diverse array of web-based resources and databases (Gardner et al., 2008). The NIF was also charged with developing tools and strategies for creating resources that can be integrated across neuroscience domains. The end product is a semantic search engine and a knowledge discovery portal that consists of a framework for describing neuroscience resources and provides simultaneous access to multiple types of information organized by relevant categories. Through its extensive resource catalog and data federation, NIF currently represents the largest source of neuroscience information available on the web.
The semantic framework through which these diverse resources are accessed is provided by the NIF Standardized Ontologies (NIFSTD; Bug et al., 2008). NIFSTD represents an extensive collection of terms and concepts from the major domains of neuroscience. The overall ontology has been assembled in a form that promotes reuse of multiple existing biomedical ontologies and standard vocabulary sources, while allowing for extension and modification over the course of its evolution. This paper presents the development principles of NIFSTD along with its application within the NIF system.
NIFSTD Design Principles
As originally proposed in Bug et al. (2008), NIFSTD was envisioned as an extensive set of ontologies, specific to the domain of neuroscience. NIFSTD started its journey with a carefully designed set of principles which enabled its ontologies to be maximally reusable, extendable, and practically applicable within information systems. Over the course of its evolution, NIFSTD augmented its principles in order to conform to the current, up-to-date trends, and practices recommended by the semantic web communities as well as by the community of standard biomedical ontologies. NIFSTD closely follows the OBO Foundry (Smith et al., 2007) best practices; however, the constraints of the NIF project required that we take a practical approach, designed to easily extend the NIFSTD ontologies, while at the same time mitigating against any disruptions to the production NIF system. Our approach is outlined following the discussion of the NeuroLex Semantic Wiki framework in Section “The NeuroLex Semantic Wiki Framework.”
NIFSTD modular structure
The NIFSTD ontologies are built in a modular fashion, where each module covers a distinct, orthogonal domain of neuroscience (Bug et al., 2008). Modules covered in NIFSTD include anatomy, cell types, experimental techniques, nervous system function, small molecules, and so forth. The upper-level classes in NIFSTD modules are carefully normalized under the classes of Basic Formal Ontology (BFO)2. These normalizations closely follow the guidelines specified in BFO manual (BFO manual)3. Based on the principles described in Rector (2003), NIFSTD utilizes a powerful ontology modularization technique that allows its ontologies to be reusable and easily extendable. Each domain specified in Table 1 has their corresponding module in NIFSTD. The individual module in turn may cover multiple sub-domains. The ingestion strategy for each source in Table 1 is shown in the “Import/Adapt” column, where “import” refers to the BFO compliant sources which were already represented in OWL; “adapt” refers to the sources that required refactoring of the source vocabularies into OWL, and/or required normalization under BFO entities.
Table 1.
The NIFSTD OWL modules and corresponding community sources from which they were built.
| NIFSTD modules | External source | Import/adapt |
|---|---|---|
| Organismal taxonomy | NCBI Taxonomy, GBIF, ITIS, IMSR, Jackson Labs mouse catalog; the model organisms in common use by neuroscientists are extracted from NCBI taxonomy and kept in a separate module with mappings | Adapt |
| Molecules, chemicals | IUPHAR ion channels and receptors, sequence ontology (SO); NIDA drug lists from ChEBI, and imported protein ontology (PRO) | Adapt/import |
| Sub-cellular anatomy | Sub-cellular anatomy ontology (SAO). Extracted cell parts and sub-cellular structures from SAO-CORE. Imported GO cellular component with mapping | Adapt/import |
| Cell | CCDB, NeuronDB, NeuroMorpho.org. Terminologies; OBO cell ontology was not considered as it did not contain region specific cell types | Adapt |
| Gross anatomy | NeuroNames extended by including terms from BIRNLex, SumsDB, BrainMap.org, etc.; multi-scale representation of nervous system, macroscopic anatomy | Adapt |
| Nervous system function | Sensory, behavior, cognition terms from NIF, BIRN, BrainMap.org, MeSH, and UMLS | Adapt |
| Nervous system dysfunction | Nervous system disease from MeSH, NINDS terminology; Imported Disease Ontology (DO) with mapping | Adapt/import |
| Phenotypic qualities | Phenotypic quality ontology (PATO); imported as part of the OBO foundry core | Import |
| Investigation: reagents | Overlaps with molecules above from ChEBI, SO, and PRO | Adapt/import |
| Investigation: instruments, protocols, plans | Based on the ontology for biomedical investigation (OBI) to include entities for biomaterial transformations, assays, data collection, data transformations. OBI-Proxi class still remains. See discussion below | Adapt |
| Investigation: resource type | NIF, OBI, NITRC, biomedical resource ontology (BRO) | Adapt |
| Investigation: cognitive paradigm | Cognitive paradigm ontology (CogPO) was extended from NIF-investigation module | Import |
| Biological process | Gene ontology (GO) biological process | Import |
This table reports the updates of the external sources that were previously used in Bug et al. (2008) paper.
NIFSTD representation formalism
NIFSTD modules are expressed in W3C standard Web Ontology Language (OWL)4; Description Logic (OWL-DL) formalism. Using OWL-DL, NIFSTD provides a balance between its expressivity and computational decidability. OWL-DL also allows the NIFSTD ontologies to be supported by a range of open source DIG compliant reasoners (DIG Group)5 such as Pellet and Fact++. NIFSTD utilizes these reasoners to maintain its inferred classification hierarchies as well as to keep its ontologies in a logically consistent state.
NIFSTD currently supports OWL 2 (OWL 2 Primer)6, the latest ontology language advocated by the W3C consortium. OWL 2 provides improved ontological features such as defining property chain rules to enable transitivity across object properties, specifying reflexivity, asymmetry, and disjointness between object properties, richer data-types, qualified cardinality restrictions, and enhanced annotation capabilities.
Accessing NIFSTD ontologies
NIFSTD is available in OWL format7 for loading in Protégé (Protégé Ontology Editor)8 or other ontology editing tools that use the OWL API. Protégé has been the main editing tool for building the NIFSTD modules. Currently, NIFSTD supports Protégé 4.X versions with OWL 2. On the web, NIFSTD is available through the NCBO BioPortal (NIFSTD in NCBO BioPortal)9, which also provides annotation and various mapping services. NIFSTD is also available in RDF and has its SPARQL endpoint (NIFSTD SPARQL endpoint)10.
Within NIF, NIFSTD is served through an ontology management system called OntoQuest (Gupta et al., 2008, 2010). Originally reported in Chen et al. (2006), OntoQuest generates an OWL-compliant relational schema for NIFSTD ontologies and implements various graph search algorithms for navigating, path finding, hierarchy exploration, and term searching in ontological graphs. OntoQuest provides a collection of web services to extract specific ontological content11. Ontoquest also provides the NIF search portal with automated query expansion (Gupta et al., 2010) for matching NIFSTD terms, including those that are defined through logical restrictions.
Reuse of external sources
One of the founding principles of NIFSTD is to avoid duplication of efforts by conforming to existing standard biomedical ontologies and vocabulary sources. It should also be noted that NIF is not charged with developing new ontological modules but relies on community sources for new contents. Whenever possible, NIFSTD reuses those existing sources as the initial building blocks for its core modules. Essentially, these external sources were selected based on their relevance to neuroscience knowledge models. Table 1 illustrates the modules in NIFSTD that are either adapted, or imported, or extracted from external community sources. NIFSTD reuses a diverse collection of sources for its ontologies. These sources range from fully structured ontologies to loosely structured controlled vocabularies, lexicons, or nomenclatures that exist within the biomedical community. Each module in NIFSTD (Table 1) integrates the relevant terms or concepts from those external sources into a single, internally consistent ontology with a matching standard nomenclature. The process and nature of reusing an external source in NIFSTD varied upon its state. The following rules summarize the basic reuse principles:
If the source is already represented in OWL, normalized under BFO, and is orthogonal to existing NIFSTD modules, the source is simply imported as a new module.
If the source is represented in OWL and orthogonal to NIFSTD modules, but is not normalized under BFO, then an ontology-bridging module (explained later) is constructed before importing the new source. These kinds of bridging modules declare the necessary relational properties to normalize the target ontology source under BFO.
If the source is orthogonal to NIFSTD modules, but is not represented in OWL, or does not use BFO as its foundational layer, then the source should be converted into OWL, and should be normalized under BFO following the Second rule above.
If the source is satisfiable by the above three principles but observed to be too large for NIF’s scope, then a relevant subset is extracted as suggested by NIF domain experts.
For the ontologies that are of type 4 above, NIFSTD currently follows MIREOT principles (Courtot et al., 2009) that allow extracting a required subset of classes from a large ontology, e.g., ChEBI, NCBI Organismal Taxonomy, etc.
Neuroscience Information Framework Project readily accepts contributions from groups working on ontologies in the neuroscience domain. For example, the Cognitive Paradigm Ontology (CogPO; Turner and Laird, 2012), has been imported under the NIF-Investigation module. As we worked through the process of adopting CogPO, we needed to make sure that the upper-level classes in CogPO were BFO compliant and derivable under the same foundational layers of NIFSTD, and the properties were extended from OBO-RO. As part of NIFSTD, CogPO can be used to annotate datasets for specific querying and comparisons and the contents are exposed via NeuroLex for community involvement (see The NeuroLex Semantic Wiki Framework).
At the beginning of the NIF project, the size, format, or immaturity of some community ontologies necessitated that NIF add significant custom content in order to provide coverage in certain modules. Over the last couple of years, the tools for extracting relevant portions of ontologies and for converting ontologies from OBO to OWL format have been improved. Thus, since the last publication (Bug et al., 2008), several of these custom ontologies were swapped for community ontologies. However, it should be noted that the NIF-Investigation module still contains “OBI-proxy” classes that were originally meant to be replaced by the matured version of OBI under BFO 1.0. However, the matured version of OBI entailed many of the original OBI-proxy classes to be retired, changed their identifiers, and sometimes did not replace them by any new classes. As NIF-Investigation continued to add many new concepts under the original obi-proxy classes, directly importing the current OBI to replace the proxy classes was not a reasonable solution. However, we have proposed the NIF-Investigation terms to be added, aligned, and maintained within OBI. We plan to incorporate portions of OBI to be extracted under NIF-Investigation, for the future release of NIFSTD.
Single inheritance for named classes
An asserted named class in NIFSTD can have only one named class as its parent. However, the same named class can be asserted under multiple anonymous classes. This principle promotes the named classes to be univocal to avoid ambiguities. In NIFSTD, classes with multiple parents are derivable via automated classification on defined classes. This approach saves a great deal of manual labor and minimizes human errors inherent in maintaining multiple hierarchies. Also, this approach provides logical and intuitive reasons as to how a class may exist under multiple, different hierarchies. A useful example can be seen in Neuronal type classification in section “Example Knowledge Model: NIFSTD Neuronal Cell Types” where a particular neuron type can be a subclass of multiple different “anonymous” classes, e.g., Neuron X is a Neuron that has GABA as a neurotransmitter. The details about the motivation behind this approach can be found in Alan Rector’s Normalization pattern discussion (Ontology Design Pattern: Normalization)12.
Unique identifiers and annotation properties
NIFSTD entities are named by unique identifiers and are accompanied by a variety of annotation properties. These annotation properties are mostly derived from Dublin Core Metadata (DC) and the Simple Knowledge Organization System (SKOS) model. While several annotation properties still exist from the legacy modules of BIRNLex, from which NIFSTD was built (Bug et al., 2008), currently NIFSTD only requires the following set of annotation properties for a given new class.
rdfs: label – A human-readable name for a class or property. If a class can be named in multiple ways, a label is chosen based on the name most commonly used in literatures as selected by NIF domain experts. Other names for the class can be kept as synonyms.
nifstd: createdDate – The date when the current class or property was created. This property serves as a way to track versioning.
dc: contributor – Name of the curator who has contributed to the definition of a class.
core: definition – A natural language definition of a class. In ideal case, this definition should be written in a standard Aristotelian form.
nifstd: definitionSource – A traceable source for the current definition in a free text form. A source could be a URI, an informal publication reference, a PubMed ID, etc.
owl: versionInfo – A version number associated with NeuroLex category.
The following set of properties is used when necessary:
nifstd: modifiedDate – The date when the current class was last updated.
nifstd: synonym – A lexical variant of the class name.
nifstd: abbreviation – A short name serving as a synonym, consisting of a sequence of letters typically taken from the beginning of words of which either the preferred label or another synonym are composed. Note that this should only be used for standard abbreviation (i.e., those that are commonly used in literatures, e.g., in a PubMed indexed article)13. Many of the abbreviations supplied are actually acronyms, but we no longer distinguish between the two.
rdfs: comment – Anything related to the class or the property that should be noted.
For the current versions of Protégé, the above properties can be set as the default set of properties for NIFSTD. NIFSTD has other annotation properties associated with version control which will be described in Section “Versioning policy.” When extracting external sources using MIREOT principles, NIFSTD keeps the identical source URIs along with the original identifier fragments unaltered. This approach allows NIF to avoid extra mapping efforts with the community sources. Prior to the MIREOT approach, the practice was simply to assign new class ID for any externally sourced classes which led to maintenance difficulty due to too many mapping annotations. We still have some mappings from the BIRNLex vocabularies, as we did not have the MIREOT tool when we started.
NIFSTD object properties
NIFSTD imports the OBO Relations Ontology (OBO-RO) for the standard set of properties as defined by the OBO Biomedical community. Other object properties in NIFSTD are mostly derived from OBO-RO. Based on where the relations are asserted, there are two kinds of relations that exist in NIFSTD: one that are within a same module, i.e., intra-modular relations, and the other that is inter-modular, cross-domain relations that exist as a separate, isolated module between two independent modules.
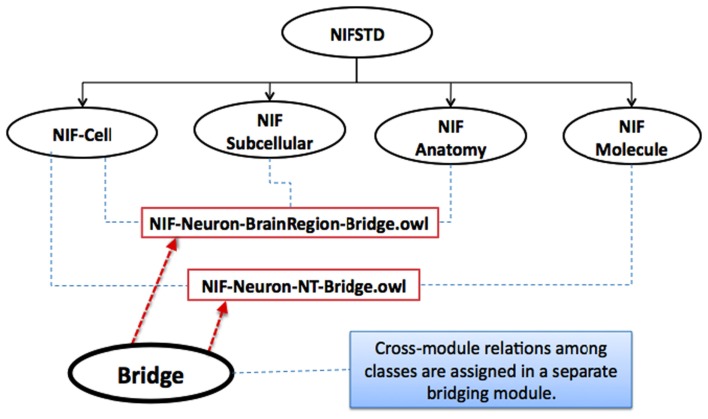
The intra-modular relations are the ones that exist as universally true within the classes of a specific module; these relations are kept integrated together within the same module. The relations between entities that could vary based on a specific application and require domain-dependent viewpoints are kept in a separate bridging module – a module that only contains logical restrictions and definitions on a required set of classes assigned between multiple modules (see Figure 1).
Figure 1.
Two example bridging OWL modules in NIFSTD (rectangular boxes) that contain class property associations between multiple core modules.
The bridging modules allow the core domain modules – e.g., anatomy, cell type, etc., to remain independent of one another. This approach keeps the modularity principles intact, and facilitates broader communities to utilize and extend NIFSTD with reasonable ease. Some of the bridge modules in NIFSTD are constructed in order to include simple semantic equivalencies between ontologies.
New bridging modules can be developed should a user desire a customized ontology of their own application domain based on one or multiple NIFSTD core modules. For example, the Neurodegenerative Disease Phenotype Ontology (NDPO; Maynard et al., submitted) is essentially a bridge module that asserts a number of entity-quality relations (on classes in relevant NIFSTD modules) to specify and define a list of named phenotypes.
As the existing reasoners fail to scale against large ontologies like NIFSTD, modularity in NIFSTD plays an important role. From an ontology development perspective, it is crucial to frequently check the consistency after asserting any new set of classification along with their axioms. Since NIFSTD is divided into smaller independent modules, the task of automated classification and consistency checking becomes much more maintainable while working on a specific module of interest.
Versioning policy
NIFSTD provides various levels of versioning for its content. It allows humans and machine to choose the level of version information required for tracking changes. Various annotation properties are associated with versioning different levels of content, including creation and modified date for each of the classes and files, file level versioning for each of the modules, and annotations for retiring antiquated concept definitions, tracking former ontology graph position, and replacement concepts.
– NIFSTD: has Former Parent Class – the full logical URI of the former parent class of a deprecated class or any other class whose super-class has been changed. This property is typically used for a deprecated/retired class.
– NIFSTD: is Replaced By Class – the full logical URI of the new class that exists as the replacement of the current retired class. This property should only be used if there exist a new replacing class.
The umbrella file nif.owl at http://purl.org/nif/ontology/nif.owl always imports the current versions of the NIFSTD modules. All other versions after the 1.0 release can be accessed from the NIF ontology archive at http://ontology.neuinfo.org/NIF/Archive/.
The NeuroLex Semantic Wiki Framework
One of the largest roadblocks that NIF identified early in the project was the lack of tools for domain experts to view, edit, and contribute their knowledge to the formal ontologies like NIFSTD. When constructing its ontologies, NIF strived to balance the involvement of the neuroscience community for domain expertise and the knowledge engineering community for ontology expertise. By combining several open sourced, semantic media wiki technologies, NIF created NeuroLex, a semantic wiki for the neuroscience community and domain experts. Details about the NeuroLex platform will be included in a separate publication (Larson et al., in preparation). Here we focus on the interplay between the NeuroLex and NIFSTD.
Relation between NIFSTD and NeuroLex
The initial contents of the NeuroLex were derived from NIFSTD which established its neuroscience-centric semantic framework and enabled the semantic relationships among its category pages. NIFSTD OWL classes were automatically transformed into category pages containing simplified, human-readable class descriptions. The category pages are editable and readily available to access, annotate, or enhance by the community or domain experts. Additions of new categories and enhancements to the NeuroLex contents are regularly transformed into NIFTSD in formal OWL-DL expressions. NeuroLex category pages are linked with NIF Search interface where users can quickly view descriptive ontological details about a matching search term.
While the properties in NeuroLex are meant for easier interpretation, the corresponding restrictions in NIFSTD are more rigorous and based on standard OBO-RO relations. For example, the property “soma located in” is translated as “Neuron X” has_part some [“Soma” and (part_of some “Brain region Y”)] in NIFSTD. Sometimes similar kinds of “macro” relations, e.g., “has_neurotransmitter,” are used in NIFSTD, recognizing that these relations can be defined in a more rigorous manner if required. These macro relations can be defined as a composition of multiple transitive properties using OWL 2.0 property chains.
Neuroscience Information Framework Project considers NeuroLex.org as the main entry point for the broader community to access, annotate, edit, and enhance the core NIFSTD content. The peer-reviewed contributions in the media wiki are later implemented in formal OWL modules. As NIF relies on the communities to enhance its ontologies, NeuroLex is an ideal interface for NIF’s current scope. For example, it has proven to be effective in the area of neuronal cell types where NIF is working with a group of neuroscientists to create a extensive list of neurons and their properties.
NIFSTD/NeuroLex curation workflow
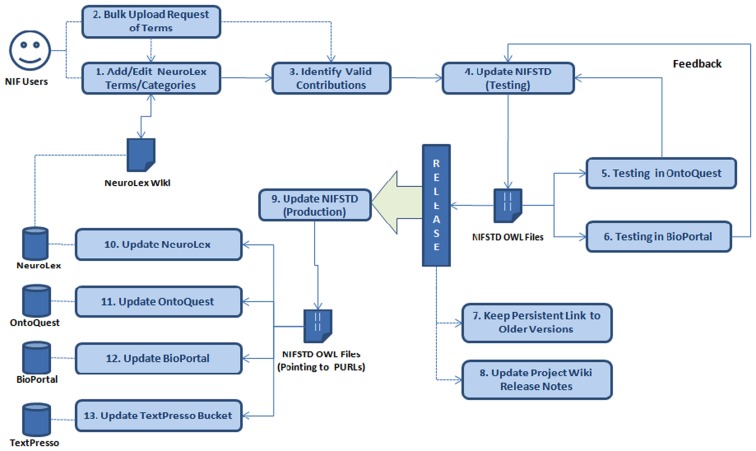
The NIFSTD development/curation workflow includes the tasks mentioned in each of the boxes followed by a number as in Figure 2. Each of the steps along with the associated tasks in the workflow is summarized in the following table, Table 2.
Figure 2.
Transition of contributions between the NeuroLex and the NIFSTD.
Table 2.
The steps and tasks involved in NIFSTD/NeuroLex curation workflow.
| Step | Tasks |
|---|---|
| Add/edit to NeuroLex | This step involves various NIF users/group who are interested in adding, updating, enhancing, or annotating the vocabularies through the NeuroLex wiki |
| Bulk upload | Depending on the number and nature of terms (i.e., adding new large sub-tree of an existing class, or new classes with known parents for a specific NIF module, etc.), we can support bulk upload of terms |
| Identify valid contributions | This step involves identifying the contributions in the previous steps that are valid according to a NIF domain expert. This step should make sure that a term contributed is actually new and not a synonym or duplicate of any existing term. Invalid contributions should be rolled back in NeuroLex during this step |
| Update NIFSTD (testing) | This step involves updating the NIFSTD OWL files or creating new OWL files in testing environment based on the update of contents from previous steps |
| Testing in OntoQuest | After significant updates in NIFSTD (every 1–1.5 months), the OWL implementations should be loaded in OntoQuest testing server for feedbacks |
| Testing in BioPortal | After significant updates in NIFSTD (every 1–1.5 months), the OWL implementations should be tested in BioPortal staging environment for feedbacks |
| Persist links to older versions | After positive feedbacks from Step 5 and 6, we archive the links to the old OWL files and post the links to the project wiki |
| Release notes | Before releasing the production version of NIFSTD, a new release note should be added for the forthcoming version. The release note should include a version number, version specific major changes, major hierarchical changes, newly added module(s), and other technical changes |
| Update NIFSTD (production) | This step involves updating the NIFSTD OWL modules in the production server that are pointing to the Persistent URLs (PURL; http://purl.org) |
| Update NeuroLex Wiki | A new release of NIFSTD should be followed by the updates in NeuroLex wiki which will reflect the implemented additions/changes of the NIFSTD contents merged with the previous iteration |
| Update OntoQuest | A major release of NIFSTD should be followed by an update in OntoQuest production version |
| Update BioPortal | A major release of NIFSTD should be followed by an update in BioPortal production version |
The scope of NeuroLex
NeuroLex can be viewed as a full-fledged information management system that provides a bottom-up ontology development approach where multiple participants can edit the ontology instantly. The semantics of NeuroLex are limited to what is convenient for the domain experts. Essentially, the NeuroLex approach is not a replacement for top-down ontology construction, but critical to increase accessibility for non-ontologist domain experts. NeuroLex provides various simple forms for structured knowledge where communities can contribute and verify their knowledge with ease. It also allows the simple query mechanisms to generate specific class hierarchies, or extraction of a specific portion of the ontology contents based on certain properties in a spreadsheet, without having to learn any complicated ontology tools.
Although NeuroLex does not support many of the standard first-order logic features that are available in standard OWL-DL formalism to support reasoning, we feel that NeuroLex has its place within the process of standard ontology development. NeuroLex can be seen as an interface to initiate the process of conceptualization where the main target is to associate the categories/concepts with the existing set of concepts/categories using simple properties. Users contributing to the NeuroLex are not formal knowledge engineers, but domain scientists tasked with ensuring that the appropriate concepts and relationships are available to the NIF for effective search and description of NIF resources.
Essentially, NeuroLex is a place to accommodate the concepts and entities that are found in literatures and other legitimate sources that are not yet been realized within a formal ontology relevant to Neuroscience. NeuroLex allows a neuroscientist to add a new concept without having to worry about its deep semantic consequence due to incompleteness or partial truth about an asserted. Fundamentally, OWL-DL can only represent a conceptual domain in a rigorous, logical fashion where it can only reason over a set of statements that are asserted to be true. Unlike OWL-DL version in NIFSTD, incomplete, non-rigorous knowledge is fine within the context of NeuroLex. Over time a concept/category in Neurolex can become ideally matured in a collaborative and completely transparent manner. As the conceptual model becomes more mature in NeuroLex, the category pages become more interconnected. While transitioning these NeuroLex contents into NIFSTD, the fundamental idea is to identify and append all the necessary logical constraints on top those “interconnection” properties. The transition of knowledge from NeuroLex to NIFSTD is essentially a context-aware, “structured” transition of knowledge between a group of domain experts and formal oncologists. This, in fact, is a practical approach of developing life science ontologies in a collaborative manner.
NeuroLex vs. Wikipedia
Although both NeuroLex Wiki and Wikipedia projects share some common goals of providing a platform for collaborative knowledge development, they differ significantly in terms of their available functionalities, features, and scopes. In order to expose structured knowledge, WikiPedia utilizes MediaWiki templates through its “info-boxes.” These info-boxes are transformed into RDF graphs by the DBPedia project in order to mine the knowledge structures. Building on top of Semantic MediaWiki (an extension of Mediawiki platform), NeuroLex does not require the two step process of producing the RDF knowledge models. Unlike Wikipedia, where a user must learn the wiki-text syntax to contribute her knowledge, NeuroLex provides “Semantic Forms” option for easy editing. NeuroLex contributors therefore can choose not to be confronted with wiki-text syntax for editing.
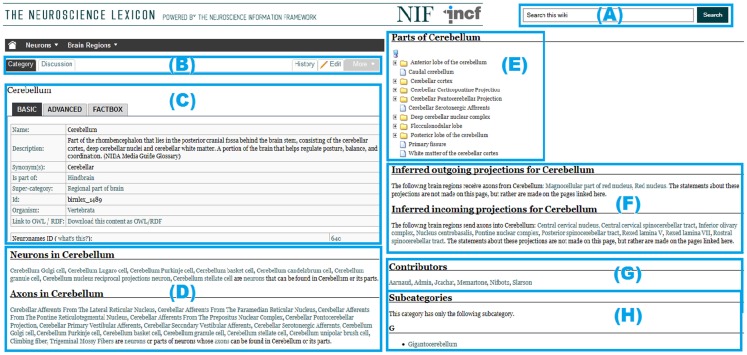
Figure 3 illustrates some of the unique features of the NeuroLex wiki platform. A standard Wikipedia page requires all the knowledge about the page to be entered manually within a single text box. In contrast, as NeuroLex has a semantic backend to structure its overall knowledge, a page in NeuroLex can dynamically call relevant information from other pages. For example, NeuroLex has the ability to automatically assemble related knowledge about Cerebellum as shown in the boxes corresponding to Figures 3D–F. Note that the information contained in Figures 3D–F are not entered as part of the “Cerebellum” page itself, but are automatically assembled from the edits made to other pages, e.g., if a user enters a soma location for a neuron that is a part of cerebellum, the neuron automatically shows up on this page under the “Neurons in cerebellum” in Figure 3D. Analogously, the “Axons in Cerebellum” in Figure 3D is also populated from the edits made in other pages. Finally, NeuroLex is meant to house all concepts of relevance to neuroscience, regardless of whether or not they are particularly noteworthy.
Figure 3.
Structure of contents in a typical NeuroLex category page. (A) The standard input text field for searching the entire NeuroLex wiki contents. (B) Different tabs to display and edit the contents of a particular category page. (C) The structured contents of a category page (e.g., Cerebellum). Boxes corresponding to (D–F) demonstrate the ability of the NeuroLex to automatically assemble related knowledge about a particular category from the edits made in other NeuroLex pages. (G) The list of contributors who made edits to the page. (H) The list of subcategories of a particular category page.
Example Knowledge Model: NIFSTD Neuronal Cell Types
Following the basic NIFSTD principle, NIF neuron types are listed in a simple, flat hierarchy of named classes under the common super-class called “Neuron” within the NIF-Cell module. These cell types were largely contributed by the NIF team, as the Cell Ontology (CL) did not contain many region specific cell types (Bard et al., 2005) at the time NIF-cell was developed. The neurons in NIFSTD are asserted with logical necessary conditions based on a set of properties that characterize mature neurons and provide a reasonable basis on which to classify them. The relational properties relate neuron types in NIF-Cell module with classes in other modules such as NIF-Subcell, NIF-Anatomy, NIF-Quality, and NIF-Molecule. As mentioned earlier in section “NIFSTD Design Principles,” these cross-module relations are kept in separate bridging modules. These modules contain necessary restrictions along with a set of defined classes to infer useful classification of neurons. The following list illustrates some of the key neuron types along with their classification schemes:
Neurons by their soma location in different brain regions – e.g., Hippocampal neuron, Cerebellum neuron, Retinal neuron
Neurons by their neurotransmitter – e.g., GABAergic neuron, Glutamatergic neuron, Cholinergic neuron
Neurons by their circuit roles – e.g., Intrinsic neuron, Principal neuron
Neurons by their morphology – e.g., Spiny neuron
Neurons by their molecular constituents – e.g., Parvalbumin neuron, Calretinin neuron.
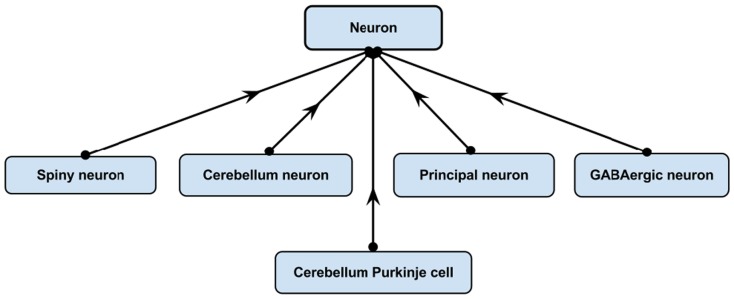
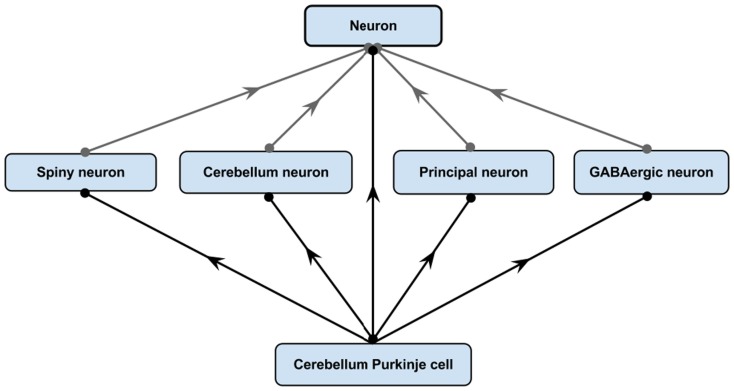
One of the most powerful features of having an ontology is that it allows explicit knowledge of a domain to be asserted from which implicit logical consequences can be inferred using logical reasoners. The following example illustrates the strength and usefulness of this feature. NIFSTD includes various neuron types with an asserted simple hierarchy under the common super-class, “Neuron.” Figure 4 illustrates an example with five neuron types.
Figure 4.
Asserted simple hierarchy of “Cerebellum Purkinje cell.”
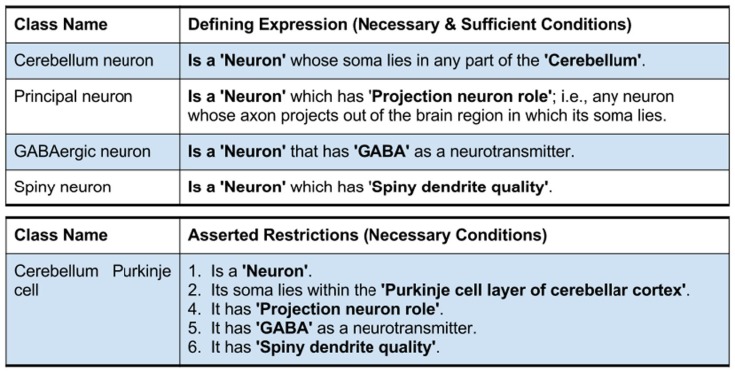
However, as illustrated in Figure 5, logical restrictions about these neurons are asserted in a bridging module along with a set of defined neuron types with necessary and sufficient conditions. The first table in Figure 5 defines three neuron types with logical necessary and sufficient conditions: the Cerebellum neuron, Principal neuron, and GABAergic neuron. The second table in Figure 5 lists a set of necessary restrictions for Cerebellum Purkinje cell. All these restrictions written in a readable format here are expressed in OWL-DL in actual NIFSTD. When the NIF-Cell module along with the bridging modules are passed to a reasoner, the reasoner automatically computes for the asserted neuron types and produces a hierarchy where the neurons are inferred under multiple superclasses. In this example, although the Cerebellum Purkinje cell was not asserted under any specific named neuron types, after invoking the automated reasoner, the neuron becomes an inferred subclass of four different defined neurons – namely, the GABAergic neuron, Cerebellum neuron, Spiny neuron, and Principal neuron as illustrated in Figure 6.
Figure 5.
Typical NIFSTD restrictions asserted for various neuron types.
Figure 6.
After invoking a reasoner NIFSTD Cerebellum Purkinje cell becomes a subclass of four different defined neuron types based on the restrictions specified in Figure 5.
Note that NIF does not currently perform deep logical modeling of neuron types, such that a reasoner would be able to deduce the necessary and sufficient conditions for a neuron to be considered a Purkinje cell. It is currently very difficult to provide universal identifying criteria for identification of particular cell types (Hamilton et al., 2012). Rather, NIF uses the logical restrictions placed on properties to generate useful classifications of neurons based on general properties that can be used to enhance search within the NIF portal, and which allows neurons to be grouped based on common features. As the ontologies are also available in RDF graphs, SPARQL queries can be written to extract a list of data elements that are linked through these simple properties.
Evolution of NIFSTD
Since the first release in 2008, the NIFSTD ontologies have undergone extensive revision and refinements. These updates include simplified structural changes to its import hierarchies, retirement of duplicate classes due to multiple imports from the first release, enforced modularization principles by adopting bridging modules between the core modules, enhancement into the partonomy restrictions in NIF Gross Anatomy, refactoring the modules under more appropriate BFO classes, simplifying the NIFSTD backend module that comprises the common entities shared by all of the NIFSTD modules. As biomedical ontologies from different communities matured, NIFSTD included various new modules such as the Gene Ontology (GO), Protein Ontology (PRO), part of ChEBI, and Human Disease Ontology (DOID). NIFSTD also imported a simplified, slim version of NCBI Taxonomy removing taxon ranks not commonly used by neuroscientists (Gardner et al., 2008). Various equivalency bridge modules have been constructed in order to ensure logical mappings on the overlapping classes between the existing NIFSTD modules and newly added modules. NIFSTD core contents have also been rapidly enhanced from NeuroLex contributions. The vision that was proposed in 2008 (Bug et al., 2008) of building detailed representations of multi-scale brain structure using common and interconnected building blocks has been realized in NIFSTD v1.8 and subsequent versions, as illustrated above with NIFSTD’s representation of neuronal cell types.
An example of how the NIFSTD continues to evolve is shown by the NIFSTD gross anatomy module. While constructing the original gross anatomy module, NIF avoided importing Foundational Model of Anatomy (FMA) or Mouse Anatomy as we wanted the core module to represent generic, species independent parts. NIFSTD extensively adopted and transformed portions of NeuroNames (Bowden et al., 2012) structures into an OWL ontology to represent NIF’s brain anatomy without any species-specific restrictions. Initially, NIFSTD divided up the brain parts into several categorical superclasses. These different categorical classes were established to make it easier to keep different types of brain parts straight, without having to worry too much about assigning other relations. These super categories included the following parts:
– Regional part: A division of a structure that can be recognized by gross anatomical features, cytoarchitecture or chemoarchitecture, e.g., cerebral cortex is a regional part of brain.
– Cytoarchitectural part: A division of a brain structure that is based on the organization of cell bodies, usually revealed by a Nissl stain, e.g., CA1 is a cytoarchitectural part of the hippocampus.
– Chemoarchitectural part: A division of a brain structure based on the distribution of some chemical marker, e.g., the patch/matrix division of the caudate nucleus
– Aggregate part: A brain structure that is composed of many different parts that are distributed in location, e.g., basal ganglia.
– Composite part spanning many brain regions: A brain part whose subdivisions are found throughout the neuraxis, e.g., the corticospinal tract.
For the current version of NIFSTD, these categorical classes are removed from the primary hierarchy of the brain structures, as they have been largely replaced through the assignment of “part of” relationships. NIF currently considers all parts of brain as a “regional part of brain” at the highest level to represent a general reference structure across species. Through the partonomy restrictions, parts comprising groupings of brain structures such as white matter structures, basal ganglia, and circumventricular organs can be generated, so that they can be used in the NIF search system. A more detailed report on the representation of brain parts within NIFSTD, in conjunction with the program on ontologies of the International Neuroinformatics Coordinating Facility14 is in preparation.
Use of NIFSTD within the NIF System
As outlined in the introduction, the NIFSTD provides the semantic framework for searching across the diverse data sources available through the NIF. As such, it was designed to represent high level neuroscience knowledge that is useful for searching data sources. The NIF portal provides simultaneous search across three major sources of information: (1) The NIF Registry; a catalog of >4500 resources (databases, tools, materials, services) categorized according to the NIF Resource module and annotated with keywords derived from other NIFSTD modules; (2) The NIF Data Federation: Deep access to the contents of >150 databases; and (3) NIF Literature: Abstracts of Pub Med and full text of open access articles.
Neuroscience Information Framework Project adopted a very aggressive population strategy to ensure that the system was well populated as rapidly as possible in order to serve its primary mission of providing deep access to neuroscience-relevant data and tools. As is well known, resources are developed with little thought to how they would interoperate within a global information system, leading to a fragmented system of custom resources, each with their own data models and terminologies. Just as with the NIFSTD itself, we designed the system to be able to work with resources in their current state, while building in capacity for us to evolve the system over time, as new tools and technologies became available. The NIFSTD is not meant to represent the information within these sources; rather, it serves as a semantic index for searching across those diverse resources. In other words, the semantic search mechanism in NIF is enhanced through the utilization of NIFSTD; as the ontology becomes richer, search is improved. Through OntoQuest, NIF enhances the search by providing an ontology-based query formulation, source selection, term expansion, and finally better ranking on the search results based on the NIFSTD contents.
Using OntoQuest services, search through the NIF interface auto-completes to terms within the NIFSTD. OntoQuest provides automatic expansion of these terms to their synonyms, abbreviations, and lexical variants as defined in NIFSTD. The NIF system uses a query language inspired by current search engines like Google. In this language, the simplest option is to ask a keyword query, but one can optionally add predicates on metadata and data attributes, specify return structures, and make references to ontologies. An advanced search box allows users to expand terms into their ontologically related terms, e.g., part of, subclasses that can be included within the search. NIF employs Boolean operators to connect these terms in an intelligent fashion, i.e., all synonyms are joined through an “OR” operator as are any related classes selected via the ontology tree. Additional concepts entered into the search box are joined through an “AND.” Thus, if a user enters “Neurodegenerative disease” “drug,” and selects Parkinson’s disease and Alzheimer’s disease as children of neurodegenerative disease, NIF will join them as follows (synonyms are omitted: “Neurodegenerative disease OR Parkinson’s disease OR Alzheimer’s disease” AND “drug”). Typical query expansion constructs are presented in Table 3 illustrating how the contents from ontologies are utilized.
Table 3.
Examples of ontological query expansions in NIF through OntoQuest.
| Example query type | Ontological expansion |
|---|---|
| A single term query for hippocampus and its synonyms | synonyms(Hippocampus);expands to Hippocampus OR “Cornu ammonis” OR “Ammon’s horn” OR “hippocampus proper” |
| A conjunctive query with three terms | transcription AND gene AND pathway |
| A sixth term and/or query with one term expanded into synonyms | (gene) AND (pathway) AND (regulation OR “biological regulation”) AND (transcription) AND (recombinant) |
| A conjunctive query with two terms, where a user chooses to select the subclasses of the second term | synonyms(zebrafish AND descendants (promoter, subclassOf))), zebrafish gets expanded by synonym search and the second term transitively expands to all subclasses of promoter as well as their synonyms |
| A single term query for an anatomical structure where a user chooses to select all of the anatomical parts of the term along with synonyms | synonyms(descendants(Hippocampus, partOf)), expands to all parts of hippocampus and all their synonyms through the ontology. All parts are joined as an “OR” operation |
| A conjunctive query with two terms, where a user chooses to select all the equivalent terms for the second term | synonyms(Hippocampus) AND equivalent(synonyms(memory)), the second term uses the ontology to find all terms that are equivalent to the term memory by ontological assertion, along with synonyms |
| A conjunctive query with two terms, where a user is interested in a specific subclasses for both of the terms | synonyms(x:descendants(neuron, subclassOf) where x.neurotransmitter=“GABA”) AND synonyms(gene where gene. name=“IGF”), x is an internal variable |
| A query to seek all subclasses of neuron whose soma location is in any transitive part of the hippocampus | synonyms(x:descendants(neuron, subclassOf) where x.soma.location = descendants (Hippocampus, partOf)) |
| A query to seek a conceptual term that is semantically equivalent to a collection of terms rather than a single term | “GABAergic neuron” AND equivalent (“GABAergic neuron”), The term gets recognized as ontologically equivalent to any neuron that has GABA as a neurotransmitter and therefore expands to a list of inferred neuron types |
One of the key features of the current NIFSTD is the inclusion and enrichment of various cross-domain bridging modules which include a number of useful defined classes. As illustrated in the neuronal examples in section “Evolution of NIFSTD,” we have been working with domain experts to define relationships between entities within different NIFSTD core modules, e.g., brain region to neuron; neuron to molecule that weave together the different modules in a coherent manner. These defined classes are then used by the NIF system to formulate its useful concept-based queries through OntoQuest. For example, while searching for “GABAergic neuron,” the NIF query expansion through OntoQuest recognizes the term as “defined” from the ontology, and looks for any neuron that has GABA as a neurotransmitter (instead of the lexical match of the search term) and enhances the query over those inferred list of neurons. Searching this defined concept in a Google search would essentially exclude all the GABAergic neurons unless they are explicitly listed within the search box. Other analogous example include query formulation for the defined concepts like Tracer, Anterograde tracer, Retrograde tracer, Neurotransmitter, Neurotransmitter receptor, Non-human primate, Drug of abuse, etc.
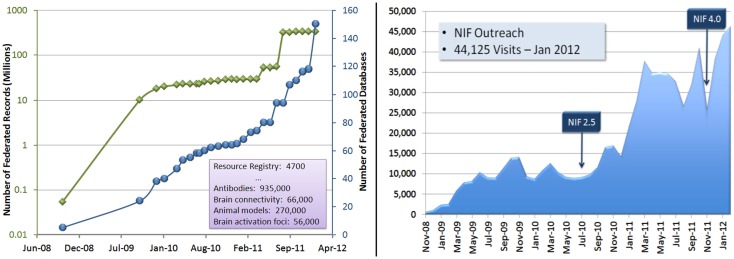
Since the first release in 2008, NIF has grown significantly in contents and community building. The chart on the left in Figure 7 illustrates the growth of federated records and database resources in NIF since June, 2008. The chart on the right illustrates the utilization growth in visits per month across NIF holdings, including NIF search portal, NeuroLex, and NIF services. Currently, NIF search portal has ∼6000 visits per month, and NeuroLex has over 15,000 visits per month. Also, it is worth mentioning that a significant number of current NIF users are successfully finding their desired terms and concepts from the NIFSTD vocabularies. For example, based on the recent Google analytics report (from April 1st to 30th, 2012) on NIF’s user interaction patterns, out of total 7108 search events, 3317 committed auto-complete search (i.e., 46.66% of the desired search terms existed in NIFSTD vocabularies), and 256 of them required advanced ontological query expansion search.
Figure 7.
On the left, the increase of NIF contents in terms of the number of federated records (green) and databases (blue). On the right, the increase of community outreach in terms of the number of visitors to the NIF portal.
Conclusion
The NIF project provides an example of practical ontology development and how it can be used to enhance search and data integration across diverse resources. NIF uses the NIFSTD to provide a semantic index to heterogeneous data sources and the basis of the concept-based query system. Using the upper-level BFO ontologies allowed us to promote a broad semantic interoperability between a large numbers of biomedical ontologies. The modularity principles along with the bridging modules allowed us to limit the complexity of the base ontologies. Users of NIFSTD can exclude the NIF specific bridging modules, which promotes easy extendibility and keeps the modularity principles intact. All of the practices adopted by NIF were designed to allow ontologies to be utilized within an evolving production system with minimum disruption as the ontologies and ontology design principles evolved.
We have defined a process to form complex semantics to various neuroscience concepts through NIFSTD and through NeuroLex collaborative environment. NIF encourages the use of community ontologies for resource providers, and as the project moves forward, we are using NIFSTD to build an increasingly rich knowledge base for neuroscience that integrates the data sources with the larger life science community. Essentially, the key aspects of these knowledge-bases are the integration of necessary semantic layer on top of the data elements found in databases, and literature corpus by linking those data elements with ontological concepts. NIF is closely following the movements such as Open Data, Linked Data, and Web of Data, to provide effective new ways that could semantically integrate data regardless of their sources.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Supported for NIF is provided by a contract from the NIH Neuroscience Blueprint HHSN271200800035C via the National Institute on Drug Abuse.
Footnotes
1NIF, http://neuinfo.org
3BFO manual, http://www.ifomis.org/bfo/manual
5DIG Group, http://dl.kr.org/dig/
6OWL 2 Primer, http://www.w3.org/TR/owl2-primer/
7OWL format, http://purl.org/nif/ontology/nif.owl
8Protégé Ontology Editor, http://protege.stanford.edu/
9NIFSTD in NCBO BioPortal, http://bioportal.bioontology.org/ontologies/40510
10NIFSTD SPARQL endpoint, http://ontology.neuinfo.org/sparql-endpoint.html
11OntoQuest, http://ontology.neuinfo.org/ontoquest-service.html
12Ontology Design Pattern: Normalization, http://ontologydesignpatterns.org/wiki/Submissions:Normalization
14International Neuroinformatics Coordinating Facility, http://incf.org
References
- Bard J., Rhee S. Y., Ashburner M. (2005). An ontology for cell types. Genome Biol. 6, R21. 10.1186/gb-2005-6-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden D. M., Song E., Kosheleva J., Dubach M. F. (2012). NeuroNames: an ontology for the BrainInfo portal to neuroscience on the web. Neuroinformatics 10, 97–114 10.1007/s12021-011-9128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug W. J., Ascoli G. A., Grethe J. S., Gupta A., Fennema-Notestine C., Laird A. R., Larson S. D., Rubin D., Shepherd G. M., Turner J. A., Martone M. E. (2008). The NIFSTD and BIRNLex vocabularies: building extensive ontologies for neuroscience. Neuroinformatics 6, 175–194 10.1007/s12021-008-9032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Martone M. E., Gupta A., Fong L., Wong-Barnum M. (2006). “Ontoquest: exploring ontological data made easy,” in Proceedings 31st International Conference on Very Large Database (VLDB), Seoul, 1183–1186 [Google Scholar]
- Courtot M., Gibson F., Lister A., Malone J., Schober D., Brinkman R., Ruttenberg A. (2009). MIREOT: The Minimum Information to Reference an External Ontology Term. Available at: http://dx.doi.org/10.1038/npre.2009.3576.1
- Gardner D., Goldberg D. H., Grafstein B., Robert A., Gardner E. P. (2008). Terminology for neuroscience data discovery: multi-tree syntax and investigator-derived semantics. Neuroinformatics 6, 161–174 10.1007/s12021-008-9024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Bug W. J., Marenco L., Condit C., Rangarajan A., Müller H. M., Miller P. L., Sanders B., Grethe J. S., Astakhov V., Shepherd G., Sternberg P. W., Martone M. E. (2008). Federated access to heterogeneous information resources in the neuroscience information framework (NIF). Neuroinformatics 6, 205–217 10.1007/s12021-008-9033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Condit C., Qian X. (2010). BioDB, an ontology-enhanced information system for heterogeneous biological information. Data Knowl. Eng. 69, 1084–1102 10.1016/j.datak.2010.07.003 [DOI] [Google Scholar]
- Hamilton D. J., Shepherd G. M., Martone M. E., Ascoli G. A. (2012). An ontological approach to describing neurons and their relationships. Front. Neuroinformatics 6:15. 10.3389/fninf.2012.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A. (2003). “Modularisation of domain ontologies implemented in description logics and related formalisms including OWL,” in Proceedings of K-CAP 2003, Sanibel Island [Google Scholar]
- Smith B., Ashburner M., Rosse C., Bard J., Bug W., Ceusters W., Goldberg L. J., Eilbeck K., Ireland A., Mungall C. J., OBI Consortium. Leontis N., Rocca-Serra P., Ruttenberg A., Sansone S. A., Scheuermann R. H., Shah N., Whetzel P. L., Lewis S. (2007). The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 25, 1251–1255 10.1038/nbt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. A., Laird A. R. (2012). The cognitive paradigm ontology: design and application. Neuroinformatics 10, 57–66 10.1007/s12021-011-9126-x [DOI] [PMC free article] [PubMed] [Google Scholar]