Abstract
Micelle-based siRNA carriers (“micelleplexes”) were prepared from the A-B-C triblock copolymer, poly(ethylene glycol)-poly(n-butyl acrylate)-poly(2-(dimethylamino)ethyl methacrylate) (PEG-PnBA-PDMAEMA), and their in vitro performance and in vivo biodistribution properties were compared with the benchmark PEGylated and basic polycation systems, PEG-PDMAEMA and PDMAEMA, respectively. The micelle architecture, incorporating increased PEG shielding and a larger particle size (~50 nm) than polycation-based complexes (polyplexes; ~10 nm), enhances siRNA delivery performance in two important aspects: in vitro gene silencing efficiency, and in vivo tumor accumulation. The in vitro gene silencing efficiency of the micelleplexes (24% in HeLa cells) was significantly better than the statistically-insignificant levels observed for PDMAEMA and PEG-PDMAEMA polyplexes under identical conditions. This enhancement is linked to the different mechanisms by which micelleplexes are internalized (i.e., caveolar, etc.) compared to PDMAEMA and PEG-PDMAEMA polyplexes. Folate-functionalization significantly improved micelleplex uptake but had negligible influence on gene silencing efficiency, suggesting that this parameter is not limited by cellular internalization. In vivo biodistribution analysis revealed that siRNA delivered by micelleplexes was more effectively accumulated and retained in tumor tissues than that delivered by PEGylated polyplexes. Overall, the micelle particle size and architecture appear to improve in vitro and in vivo delivery characteristics without significantly changing other properties, such as cytotoxicity and resistance to enzymes and dissociation. The self-assembled nature of micelleplexes is expected to enable incorporation of imaging modalities inside the hydrophobic micelle core, thus combining therapeutic and diagnostic capabilities. The findings from the present study suggest that the micelleplex-type carrier architecture is a useful platform for potential theranostic and tumor-targeting applications.
Keywords: nano-carrier, siRNA delivery, in vitro performance, in vivo biodistribution, polyplex, micelleplex
Discovery of the RNA interference (RNAi)2 mechanism by Fire, Mello and coworkers in the late 1990s provided new direction and gave renewed promise to the field of gene therapy. RNAi utilizes double-stranded, 21-25 base pair (bp) long, small interfering RNA (siRNA) to mediate sequence-specific gene silencing.2-3 As with all methods of gene delivery, a protective carrier, preferably non-viral, is desired to help deliver intact siRNA to the site of action, while ensuring stability and nuclease protection along the way.
Polymers, particularly polycations, have become very popular components of non-viral gene carriers.4 Being positively charged, polycations can spontaneously associate with the negatively-charged phosphate backbone of nucleic acids to form “polyplexes”. Charge-neutralized polyplexes, however, lose their hydrophilicity and thus water solubility of these complexes becomes an issue. Upon addition of excess amounts of cationic polymer to a nucleic acid, net positively-charged polyplexes are formed which are believed to promote uptake by negatively-charged cell membranes. On the other hand, under physiological salt conditions, where repulsive forces between like-charged particles are screened, aggregation of cationic polyplexes can result.5 Nonspecific interactions between cationic complexes and negatively-charged blood components, such as serum proteins, can also be a cause of undesired aggregation1, 6-7 A common solution to this conundrum is the conjugation of poly(ethylene glycol) (PEG) to a polycation to alter its properties. PEG has been shown to confer stability to polyplexes by sterically shielding excess positive charges and preventing aggregation.5-9 PEG also increases the overall hydrophilicity of the polyplexes, keeping them soluble in aqueous solutions.8 Both polycation (poly(2-(dimethylamino)ethyl methacrylate) or PDMAEMA) and PEGylated polycation (PEG-PDMAEMA) formulations were employed in this study as benchmark polymeric vehicles for in vitro siRNA delivery. PDMAEMA and several of its copolymer variants have been widely studied throughout the literature10-14 as transfection agents for plasmid DNA with varying results. Binding between PDMAEMA and DNA is often too strong to effectively release DNA inside the cell whereas PEGylation of this polycation can weaken binding with DNA10, 13 to the point where the complexes are susceptible to enzymatic degradation and premature dissociation, both of which limit DNA transfection efficiency in vivo. There is a wealth of existing knowledge pertaining to the delivery performance of DNA polyplexes, but only relatively recently have researchers begun to focus on siRNA delivery mediated by PDMAEMA-based polymers.15-18 Hence, there is still a need for further characterization and understanding of how to optimize PDMAEMA-based siRNA delivery systems both in vitro and in vivo.
Uncomplexed siRNA and siRNA nanoparticles smaller than 10-20 nm in size19-20 in systemic circulation are susceptible to premature clearance from the bloodstream.21 Consequently, there may be an advantage to using particulate rather than molecule-based carriers where the same properties (e.g., PEG shielding) can be incorporated into a larger, more stealthy system. In this work, we have explored one architecture modification based on triblock copolymer micelles formed from poly(ethylene glycol)-poly(n-butyl acrylate)-poly(2-(dimethylamino)ethyl methacrylate) (PEG-PnBA-PDMAEMA). The micelle-based design is different from traditional polymer carriers in that the building blocks for complexes are particles, not individual polymer chains, which consequently give them a very distinct architecture. For in vivo delivery of gene silencing complexes, it has been shown that size is a very important factor determining the efficiency of each step in the delivery process.21 For instance, size can influence capillary navigation , blood clearance kinetics,21 intracellular uptake,22 and biodistribution.21, 23 More specifically, in cancer-related applications, size can also affect the degree of nanoparticle accumulation in tumor tissues.21, 24 It is therefore plausible that the architecture of complexes, which can affect size and the types of interactions with other particles, may also influence the uptake, delivery and gene silencing processes. Furthermore, the degree of PEGylation may regulate particle size23, 25 in the bloodstream, where the ionic conditions are favorable for aggregation of charged complexes. Thus, the potential of a nanoparticulate system to be delivered systemically is largely governed by its basic physicochemical properties. The implications of the results in this study are useful in beginning to understand the fundamental issue of how particle architecture is linked to performance and the importance of its role in the design of siRNA delivery vehicles. We also explored a preliminary modification of the micellplexes in the form of the cancer cell-targeting ligand, folate, to determine whether the gene silencing abilities of the micelleplexes would be enhanced due to the receptor-mediated internalization mechanism26 facilitated by folate. This was in attempt to understand the extent to which internalization efficiency limits the ultimate siRNA delivery and gene silencing efficiency of the complexes. While the gene silencing efficiency of our micelleplex system still requires further optimization, our study provides unique insights into the gene silencing and tumor-targeting advantages to be had by exploiting a micelle-based platform versus carriers possessing a similar chemistry but without the micelle architecture.
Results and Discussion
Morphology of Micelles and Micelleplexes
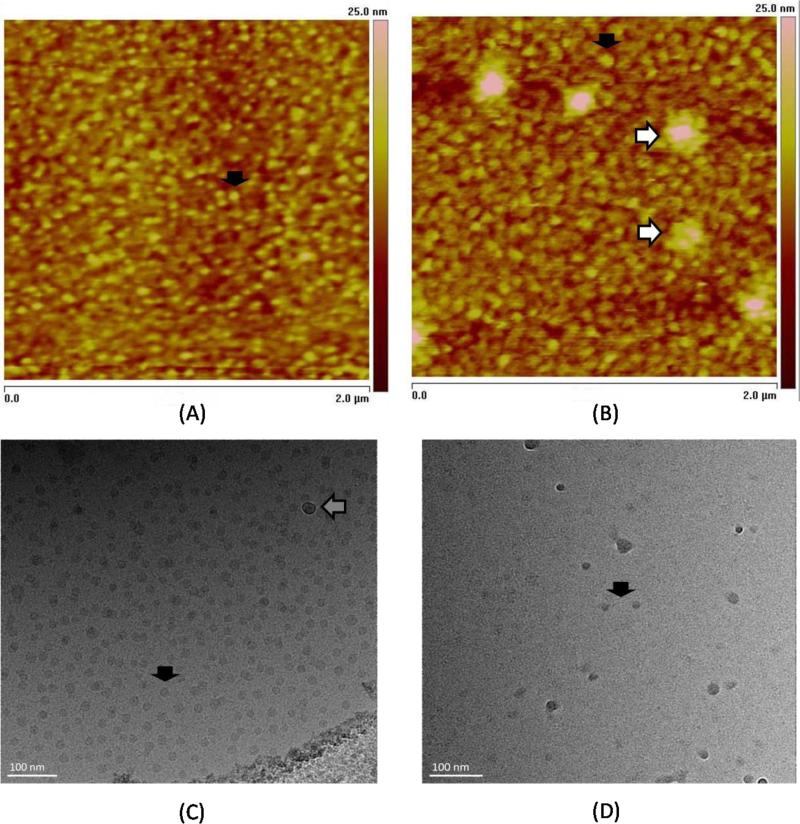
As the PEG-PnBA-PDMAEMA micelles used in this study possess a novel chemistry not yet reported in the literature by researchers outside of our lab, we took some time to characterize these particles and their complexes with siRNA with regard to their approximate size and shape. Fluid AFM imaging allowed us to visualize the micelles and micelleplexes to understand more about their morphology. Panels (A) and (B) of Figure 1 are representative fluid-cell images of the micelles and micelleplexes at N/P 8, respectively, adsorbed onto a negatively-charged substrate. The overall size of particles in the micelleplex solution are visibly larger than those in the micelle solution, indicating that there is a size increase upon siRNA binding. Because they are significantly smaller than micelles (6 nm vs. 50 nm), several siRNA molecules are able to adsorb to the outer surface of a single micelle, accounting for the increase in diameter upon complexation. As indicated in Figure 1(B), there is also a small population of micelleplexes that form aggregates, which are not present in the pure micelle solution shown in Figure 1(A), however, the vast majority exist as individual particles; as indicated in Figure S8.1 of the Supporting Information, the number of aggregates is neglibible relative to the overall population (i.e., less than a few percentage points by volume according to dynamic light scattering analysis). The measured diameters of the micelles and micelleplexes as determined by AFM (47 ± 10 nm and 56 ± 15 nm, respectively) were very consistent with the hydrodynamic diameters measured by DLS (48 ± 6 nm, and 56 ± 3 nm, respectively, as shown in Scheme 1). Figures 1 (C) and (D) are representative Cryo-TEM images of micelles and micelleplexes at N/P 8, respectively. Cryo-TEM, which is unable to capture the diffuse, hydrophilic polymer brush due to insufficient electron density, instead gives us a measure of the electron-dense, hydrophobic core of the micelles and micelleplexes which is approximately 17 ± 3 nm. Please note that full particle size distributions for the AFM and Cryo-TEM images are represented as histograms in Section S2 of the Supporting Information (SI).
Figure 1.
2 μm × 2 μm height-mode AFM images of PEG-PnBA-PDMAEMA micelles (A) and micelleplexes (B) at N/P 8 in 10 mM Tris-HCl buffer, pH 7.5. Cryo-TEM images of micelles (C) and micelleplexes (D). Representative micelle and micelleplex cores are indicated by filled black arrows in their respective figures. A small population of micelleplex aggregates, indicated by the white arrows in (B), were detected by AFM. Ice condensation in Cryo-TEM images, denoted by a gray arrow in (C), can be distinguished from particles of interest by the white rings which appear around the spots. These rings, called “Fresnel fringes”, occur when objects are in a different focal plane than the particles of interest, which is typical of contamination. Visible at the lower right of (C) is the edge of a hole on the TEM grid.
Scheme 1.
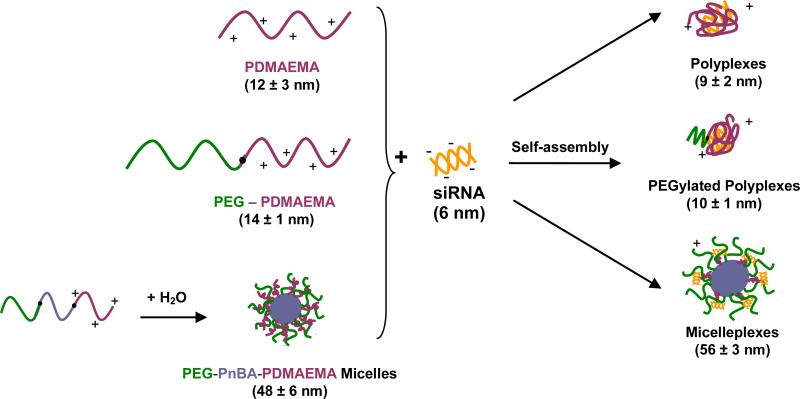
Proposed structure of siRNA/polymer complexes at N/P 8 after self-assembly. In parentheses are the hydrodynamic diameters of the siRNA, polymers and complexes as determined by Dynamic Light Scattering (DLS).
siRNA Condensation Studies
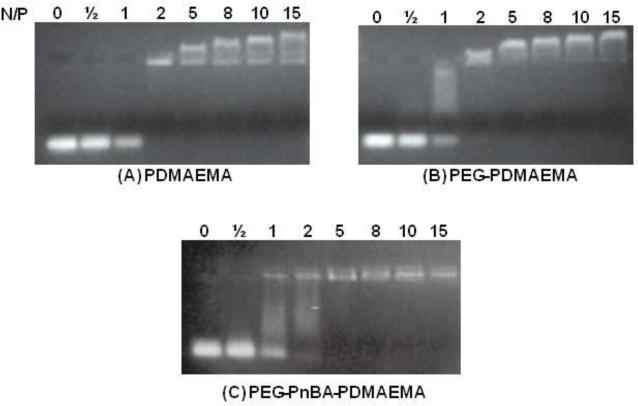
The goal of these experiments was to determine the critical N/P ratio for each type of complex at which the charges on siRNA are completely neutralized by PDMAEMA. Using gel electrophoresis, this was observed as the ratio at which the electrophoretic mobility of the complexes was completely retarded due to the net charge being close to neutral. Complexes were prepared at N/P = 0, 0.5, 1, 2, 5, 8, 10 and 15 and the points of neutrality were observed at N/P = 2 for polyplexes and PEGylated polyplexes and N/P = 3.5 ± 1.5 (i.e. between N/P 2 and 5; DLS analysis (data not shown) of micelleplexes indicated that stable, small-sized complexes are not formed until N/P 4, so this is most likely the point of neutralization.) for the micelleplexes (Figure 2). For the polyplex and PEGylated polyplex cases, neutrality occurring at N/P = 2 instead of 1 may have arisen from the fact that PDMAEMA is a weak polyelectrolyte (effective pKa ~ 7.5 for the polymer under physiological NaCl concentrations (≈ 150 mM)27-28) and is not fully charged at the pH of the medium (10 mM Tris-HCl buffer, pH 7.5). As a result, more polymer (in this case twice as much) was needed to provide enough cationic charge to neutralize the siRNA. For the micelleplex case, this issue is likely to be exacerbated by PDMAEMA chains being even less charged and/or inaccessible in the micelle conformation.
Figure 2.
siRNA Complexation. For all three images, lanes 1-8 correspond to N/P = 0, 0.5, 1, 2, 5, 8, 10 and 15, respectively. In Figure (A) PDMAEMA is shown to fully complex and neutralize siRNA at N/P = 2. The same is true for PEG-PDMAEMA (B). In (C), PEG-PnBA-PDMAEMA complexes and neutralizes siRNA at N/P = 5. Note that all cases (A – C) show some charge inversion after the neutralization point.
Below the neutralization point (e.g., N/P ½ and 1), siRNA appeared as either a vivid band at the marker position or as a smeared band representing varying degrees of partial complexation. At these low N/P ratios, there remains detectable amounts of uncomplexed siRNA molecules due to the insufficient amounts of added polycation molecules. In Figures 2(A) and 2(B), the intensity of the uncomplexed siRNA band gradually decreases as N/P is increased from 0 to 1, and it eventually disappears at N/P = 2. As demonstrated in Figure 2(B), partially complexed siRNA is detected as a smear. At the critical N/P ratio, the polyplexes and micelleplexes are observed in the loading position, indicating neutrality. Notice, also, that in all three cases, above the neutralization point, the complexes underwent a charge inversion in which the net positively-charged polyplexes began migrating towards the anode (Figures 2(A), 2(B) and 2(C), lanes 5-8). In the micelleplex case, the charge inversion is less pronounced due to the relatively larger size of the complexes retarding their migration.
Particle Size of Polymers and Complexes
Dynamic light scattering was used to obtain the particle size (hydrodynamic diameter, DH) of each of the polymers and their corresponding complexes with siRNA at N/P 8. At this N/P stoichiometry, the micelleplexes contain about 33 siRNA molecules per complex whereas for the polyplexes and PEGylated polyplexes, there are approximately 2 polymer chains for every siRNA molecule. For the three different types of polymers and their respective complexes, the DH values were respectively measured to be 12 ± 3 nm and 9 ± 2 nm for the PDMAEMA case, 14 ± 1 nm and 10 ± 1 nm for the PEG-PDMAEMA case, and 48 ± 6 nm and 56 ± 3 nm for the PEG-PnBA-PDMAEMA micelle case; see Scheme 1 for a summary of these results. After complexation with siRNA, the hydrodynamic diameters of the polyplexes and PEGylated polyplexes are found to decrease, which is expected as the cationic PDMAEMA chains collapse upon complexation and charge neutralization with anionic siRNA. In the case of the micelleplexes, the hydrodynamic diameter increases slightly, indicating that due to the coexistence of PEG chains in the micelle corona layer, the hydrodynamic thickness of the micelle corona increases after the binding of the siRNA molecules to the PDMAEMA chains on the outer surface of the micelle (see the AFM-based micelleplex size distribution presented in Figure S2.1 of the Supporting Information). The small population of aggregates observed in the AFM (Figure 1(B)) and cryo-TEM (Figure S2.2) of the micelleplexes is also expected to contribute to the slightly larger particle size of the micelleplexes detected by DLS, compared to micelles (also see the micelleplex size distribution in Figure S2.1). A notable observation is that there is no significant size difference between polyplexes and PEGylated polyplexes but the micelleplexes are much larger than them both. This is indicative of the high degree of chain assembly involved in micellization, which results in larger particles. The data also show that siRNA is smaller than both PDMAEMA and PEG-PDMAEMA and much smaller than PEG-PnBA-PDMAEMA micelles, a potentially important implication for how well-contained siRNA is within the complexes, and the inspiration behind the structures proposed in Scheme 1.
Cytotoxicty of Polymer/siRNA Complexes
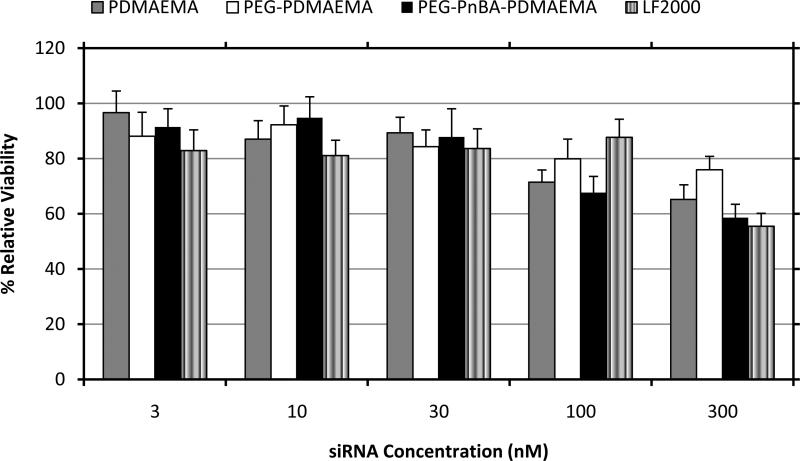
To study the cytotoxic effects of polymer (or micelle)/siRNA complexes on HeLa cells, an MTT assay was performed. Polyplexes and micelleplexes at N/P 8 containing various concentrations of siRNA were exposed to HeLa cells over a period of 24 hours. As shown in Figure 3 within the range of siRNA concentrations tested, the toxicity of the polymer/siRNA complexes was measured to be dose-dependent and at the most experimentally-relevant siRNA concentrations (~ 30 nM), the viability remains > 80% for all PDMAEMA-based complexes, which is comparable to LF2000 complexes at the same conditions. At higher siRNA concentrations, the toxicity of the micelleplexes does increase indicating that their high molecular weight and charge density become more problematic above a certain point. It should be noted that because of the chemistry of the polymers, the concentration by weight of triblock micelles needed to make complexes at a given N/P ratio is always higher than the concentrations required of PDMAEMA or PEG-PDMAEMA. Indeed, there have been several reports showing that high cationic charge density can cause cytotoxicity in a concentration-dependent manner,29-30 although the exact mechanisms of this toxicity are poorly understood. Furthermore, by separate measurements of the cytotoxicities of siRNA and the individual polymers, we have confirmed that siRNA itself does not cause any appreciable toxicity, whereas it is indeed the polymers that are mainly responsible for the cytotoxicity (Figure S5.1). Therefore, we can deduce that the mild toxicity exhibited by the polyplexes and micelleplexes is a concentration-dependent effect of the polymer which can be alleviated by working with lower siRNA doses. For information regarding the corresponding polymer concentrations used in this cytotoxicity study, please see Table S5.1 of the SI.
Figure 3.
MTT cell viability assay in HeLa cells. The absorbance of all samples was quantified relative to that of an untransfected sample, which was set to 100% viability.
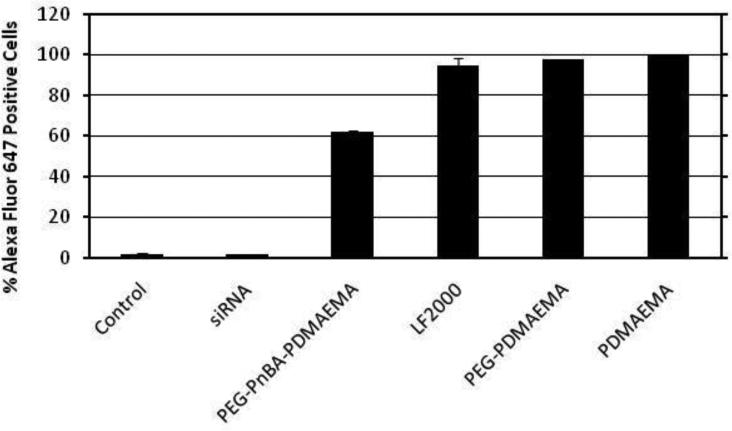
Measuring Uptake via Flow Cytometry
The efficiency of intracellular siRNA delivery by PDMAEMA-based carriers was evaluated by tracking uptake of fluorescently-labeled siRNA in HeLa cells via flow cytometry. The analysis of cellular uptake of uncomplexed siRNA as well as polymer and Lipofectamine 2000 complexes with Alexa Fluor 647-labeled siRNA is summarized in the bar graph in Figure 4, in comparison to untreated HeLa cells; the intensity distribution histograms are also presented in Figure S6.1. Free siRNA, as expected, exhibits very poor uptake efficiency (1.7 ± 0.3%) mostly likely due to electrostatic repulsion from the negatively-charged cell membrane impeding internalization. Complexes with Lipofectamine 2000 have a high uptake efficiency (94.6 ± 2.1%), consistent with its reputation for being a highly efficient transfection reagent, and its lipophilic nature. Of the PDMAEMA-based polymers, the PDMAEMA homopolymer has the highest uptake efficiency (99.5 ± 0%). This is thought to be due to its highly cationic nature which facilitates strong electrostatic interactions with the negatively-charged cell membrane. Similar to the homopolymer, the PEG-PDMAEMA diblock also has a very high uptake efficiency (97.9 ± 0.01%), whereas the PEG-PnBA-PDMAEMA micelleplexes have a more moderate level of uptake (62.1 ± 0.8%). The comparatively lower level of uptake achieved by the micelleplexes appears to indicate better PEG shielding of the structure than what is achievable with the PEG-PDMAEMA polyplexes. Notably, while internalization efficiency may have an impact on gene silencing, the gene silencing efficiency of a carrier was found to not necessarily correlate with its efficiency of uptake (as will be shown in the next subsection), since there are several other factors (e.g., unpackaging of siRNA from the carrier and release from the endosome) required for successful gene delivery.
Figure 4.
Cellular uptake, as determined by flow cytometry, of Alexa Fluor 647-labeled siRNA and complexes formed with polymer or Lipofectamine 2000 as compared to untreated HeLa cells (control). Error bars represent the standard error of the mean.
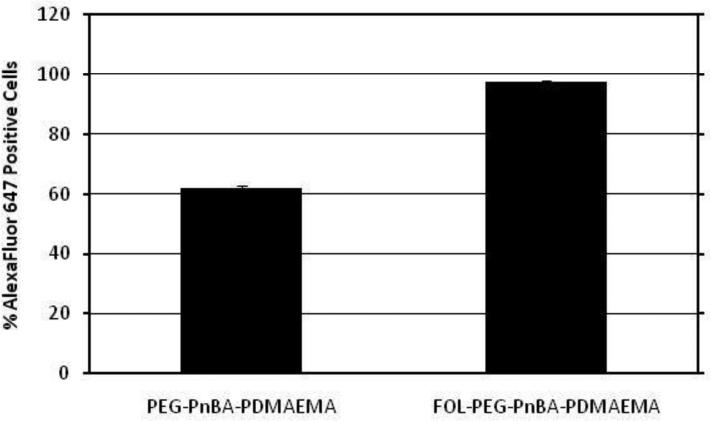
To determine whether the uptake efficiency of the micelleplexes could be improved by conjugation of a cell-targeting ligand, we tested the uptake profile of folic acid-conjugated micelleplexes which can obtain entry to HeLa cells by receptor-mediated endocytosis via the folate receptor, which they are known to over-express at their surfaces.26 As we can see from Figure 5, the internalization efficiency of micelleplexes is indeed enhanced, from 62.1 ± 0.8% to 97.5 ± 0.6% by conjugation of folic acid to the surface of the micelleplexes. Additionally, adding free folic acid to the cell culture media to saturate folate receptors prior to adding the complexes effectively suppresses the uptake of FOL-PEG-PnBA-PDMAEMA micelleplexes (see Figure 7) whereas uptake of unmodified micelleplexes is unaffected by the presence of free folic acid. This indicates that the enhanced uptake afforded by foalte conjugation of the micelleplexes is a direct result of the exploitation of the folate receptor-mediated endocytosis pathway.
Figure 5.
Cellular internalization efficiency comparison between unmodified PEG-PnBA-PDMAEMA micellplexes and folate-conjugated micelleplexes in folate receptor over-expressing HeLa cells.
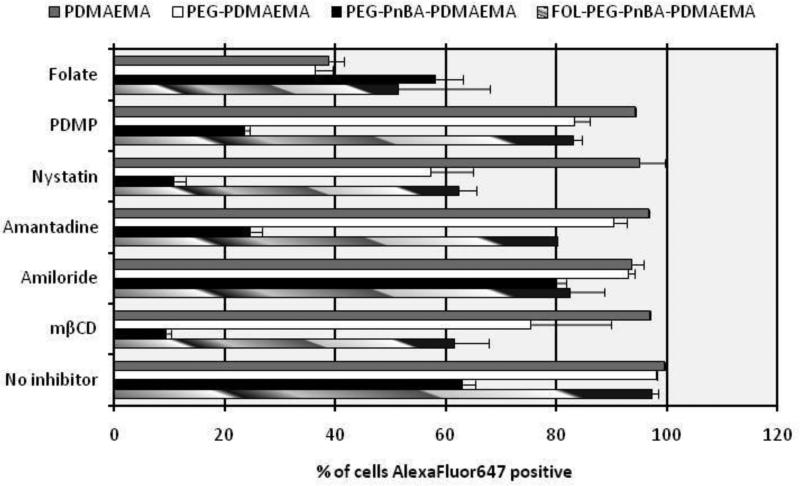
Figure 7.
Flow cytometry analysis of AlexaFluor647 uptake in HeLa cells facilitated by various delivery vehicles in the presence or absence of endocytosis inhibitors. Amiloride, amantadine, nystatin, methyl β-cyclodextrin, folic acid, and PDMP hydrochloride are inhibitors for the macropinocytosis, clathrin-dependent, membrane fusion, caveolar, folate receptor-mediated, and lipid raft endocytosis processes, respectively.
In Vitro Silencing of Endogenous GAPDH
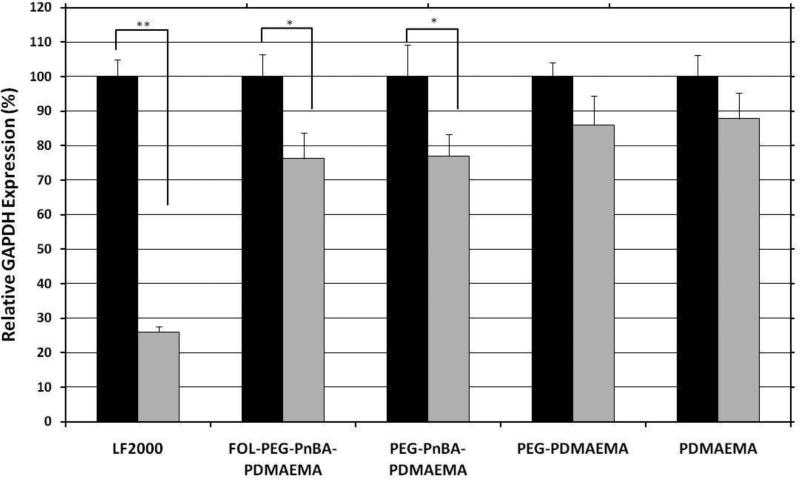
The endogenous gene silencing efficiency of our polymer systems was studied in HeLa cells and quantified by qRT-PCR of GAPDH mRNA levels 24 hours post-transfection (Figure 6). LF2000-transfected samples elicit the highest levels of GAPDH mRNA silencing, 74.1% ± 1.5%, consistent with having a very high level of intracellular uptake. The next most effective carrier is the micelleplex system, which mediates 23.1% ± 6.4% silencing of GAPDH mRNA. Even though conjugation of folate drastically enhanced cellular internalization, no noticeable improvements were observed in the ability of folate-modified micelleplexes to mediate gene silencing, as their efficiency was identical to unmodified micelleplexes. PDMAEMA and PEG-PDMAEMA complexes mediate comparable levels of GAPDH mRNA expression, 14.0% ± 8.4% and 12.2% ± 7.5%, respectively, however the standard error of the measurements suggests that these values are not statistically significant relative to negative control-treated samples. Therefore, we can conclude that the micelleplex architecture enables an advantageous improvement in endogenous gene silencing efficiency compared to its simple polycation and PEGylated polycation counterparts. In the case of PDMAEMA homopolymer complexes, a high level of cellular uptake is achieved, presumably due to strong electrostatic interactions between the cationic polymer and the anionic cell membrane which facilitate internalization. One possible explanation for the lackluster gene silencing mediated by the PDMAEMA homoplymer is its inability to escape the endosomal compartment upon cellular internalization to complete the siRNA delivery process. A recent report31-32 has highlighted the shortcomings of linear PDMAEMA polymers in disrupting the endosomal compartment and observed in their system as well that a high uptake efficiency is not necessarily an indicator of a high transfection efficiency. The authors of Reference 31 also detected an increase in DNA transfection efficiency with star-shaped and highly-branched PDMAEMA compared to linear PDMAEMA, although the cellular uptake efficiencies were mostly independent of architecture. This suggests that the improvement in transfection efficiency was probably due to enhanced endosomal release afforded by the modified PDMAEMA architectures. It also suggests that it is reasonable in the present study to attribute the observed differences in gene silencing efficiency to the architectural/structural dissimilarities of our PDMAEMA-based systems and their uptake mechanisms as well as their relative abilities to escape the endosome and release siRNA, since internalization efficiency does not appear to be the rate-limiting step. While differing internalization mechanisms may play a role as well, the lack of enhanced gene silencing for FOL-PEG-PnBA-PDMAEMA (23.8 ± 7.5%) compared to PEG-PnBA-PDMAEMA (23.1 ± 6.4%) micelleplexes further minimizes the possibility of internalization efficiency being the factor inhibiting efficient gene knockdown. Our results indicate that the micelle architecture facilitates better overall siRNA uptake and release that the basic polycation or PEGylated polycation systems are not able to achieve, thus resulting in a more potent gene silencing effect. This effectively proves the concept of the importance of polymer architecture in siRNA delivery, however further improvements to the micelleplex chemistry to more dramatically enhance their siRNA delivery efficiency remain the subject of ongoing work in our lab.
Figure 6.
qRT-PCR quantification of endogenous GAPDH mRNA silencing in HeLa cells by various siRNA delivery vehicles. Data for anti-GAPDH siRNA-treated samples (gray bars) is normalized against negative-control siRNA data (black bars) for each delivery vehicle. Error bars represent standard error of the mean of at least 3 experiments (** p < 0.0005, * p < 0.1).
Endocytosis Mechanistic Study
We have clearly shown that endogenous silencing of GAPDH at the mRNA level was most efficiently achieved by the PEG-PnBA-PDMAEMA micelle system with about 23% silencing compared to the PDMAEMA homopolymer and PEG-PDMAEMA diblock systems whose silencing was indistinguishable from negative controls. Furthermore, the lack of improvement in silencing efficiency afforded by conjugation of a folate targeting ligand highlights the improbability that the overall potency of gene silencing is controlled by cellular internalization. To elucidate this hypothesis, we employed blockers of various endocytosis pathways to determine wether the mechanism(s) of uptake for the PEG-PnBA-PDMAEMA micelleplexes could be different from those used by the homopolymer and diblock systems, possibly contributing to their higher gene silencing efficiency.
Figure 7 shows the percentage of cells which have taken up AlexaFluor647-tagged siRNA delivered by various vehicles either in the presence or absence of a particular endocytosis inhibitor (see Figure S7.1 for the analysis based on mean AlexaFluor647 intensity per cell). A salient feature of the data is the low level (9.5%) of uptake for the micelleplexes after inhibition of caveolae-mediated endocytosis with methyl β-cyclodextrin (mβCD). In fact, uptake of micelleplexes is inhibited to a greater extent by mβCD than any other inhibitor, indicating that the caveolar pathway is the major pathway contributing to their internalization. On the other hand, mβCD has little to no effect on the uptake of PDMAEMA homopolymer or PEG-PDMAEMA diblock complexes, indicating that these two systems do not use the caveolar pathway (to any great extent) for internalization. Recently, Gabrielson and coworkers have shown33 that polymer complexes internalized via the caveolar pathway have a higher gene delivery efficacy due to the avoidance of lysosomes which would otherwise degrade the genetic material and render it useless. It should also be noted that the micelleplex uptake involves several other endocytosis mechanisms such as membrane fusion, lipid raft and clathrin-dependent mechanisms, whereas all other systems examined appear to be internalized mainly by the receptor-mediated endocytosis process. Thus, we can reasonably conclude that the enhanced gene silencing efficiency of the micelleplexes compared to PDMAEMA homopolymer and PEG-PDMAEMA diblock complexes may be due to their exploitation of the caveolar and other pathways. Furthermore, the difference in internalization mechanisms utilized by the micelleplexes as opposed to the homopolymer and diblock complexes is likely to be a result of their distinct size and architectural properties.
In Vivo Biodistribution
To estimate the likely “in vivo size”, that is the true size of the complexes in the bloodstream, we performed a DLS study of the complexes as a function of time following exposure to a physiologically-relevant concentration (34.3 mg/ml)7 of (Bovine) Serum Albumin (SA), 69 kilo Daltons (kDa),34 the most prevalent serum protein. While the PEGylated polyplexes more than double in size, the aggregates are still relatively small-sized, ~ 25-30 nm, whereas the micelleplexes form aggregates ~ 130 nm. For both types of complexes, stable aggregates form within 5 minutes and do not increase in size during the 90 minute time interval studied (See Figure S8.1).
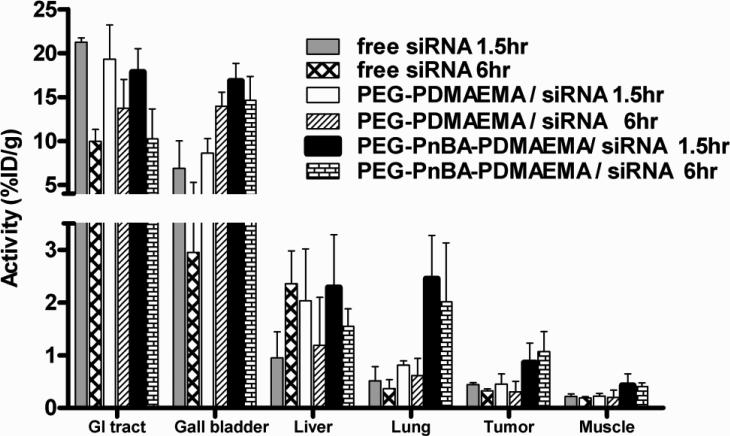
Following the systemic administration of I124-labeled siRNA delivered by either PEG-PDMAEMA PEGylated polyplexes or PEG-PnBA-PDMAEMA micelleplexes at N/P 8, a biodistribution analysis was performed to quantify the accumulation of the nanoparticles in various organs as a percentage of the intial siRNA dose (Figure 8). The first thing we notice is that the biodistribution profile of free siRNA is quite different than that of either type of complex, suggesting that siRNA remains complexed with the polymer/micelles throughout systemic circulation. If the siRNA became disassociated from the polymer/micelles during circulation, we would expect the biodistribution profiles to resemble that of free siRNA. For both types of complexes and especially for free siRNA, a significant portion of the injected siRNA was found in the gastrointestinal (GI) tract and gall bladder, en route to being removed from the body. However, from the representative PET-CT images shown in Figure S9.1, it is clear that the vast majority of PEGylated polyplexes are immediately (< 90 min) removed from circulation and accumulate in the intestinal cavity whereas there is a diffuse signal from the micelleplexes throughout the entire body, indicating that a subset of the complexes still remain in circulation after 90 minutes. In all cases, no significant amount of radiotracer could be detected after 24 hours post-injection, except in the liver.
Figure 8.
In vivo biodistribution analysis for free siRNA, and PEGylated polyplexes and micelleplexes at N/P 8.
The distribution in the non-excretory tissues more effectively highlights differences in the delivery properties of the PEGylated polyplexes vs. micelleplexes. For instance, after 90 minutes, micelleplexes are found to accumulate more than 3-fold higher than the PEGylated polyplexes in the lung (p = 0.016), the first organ encountered after an intravenous dose. The activities of both the micelleplexes and PEGylated polyplexes in the lung slightly decrease (i.e., by 20 to 30%) after 6 hours, which suggests that the nanoparticles are initially rapidly arrested in but become extravasated rather slowly afterwards from the vascular endothelium of lung tissue.35-37 Previous studies have shown that negatively-charged liposomes accumulate more in the lungs than those that are neutral or positively-charged.37 For both the PEGylated polyplexes and micelleplexes, association with serum albumin (SA) may produce an overall negative in vivo surface charge, which may be a factor contributing to their levels of uptake in the lung. It is most likely some combination of particle size and surface charge that determines the overall levels of accumulation in the lung tissue. Interestingly, free siRNA, although negatively-charged, has very little accumulation in the lungs, most likely because the molecules are too small to be effectively trapped in the vasculature.
After the GI tract and gall bladder, both types of complexes have the next highest accumulation in the liver, with activities that are higher at the 90 minute time point and decrease after 6 hours. The significant degree of uptake in the liver is again attributed to the fact that the association of the complexes with negatively-charged plasma proteins like SA is known to facilitate uptake by non-parenchymal cells within the liver.35 In contrast, levels of free siRNA in the liver increase after 6 hours, which probably reflects the siRNA being degraded by serum nucleases and then scavenged from the bloodstream as waste by the endothelial cells of the liver.38
Notably, in the tumor tissue, micelleplexes accumulate about two to four-fold more efficiently than the PEGylated polyplexes at both 1.5 hours (p = 0.038) and 6 hours (p = 0.001), probably owing to their larger size/bulkier architecture which allows them to be selectively extravasated through blood vessels in the tumor tissue whereby the impaired clearance in the interstitial space contributes to their prolonged retention therein.34 This phenomenon is known as the Enhanced Permeability and Retention (EPR) Effect, and has been shown to be effective for molecules with a size > 45 kDa.39 The PEGylated polyplexes, though of an EPR-capable size after association with SA, are still smaller, and thus probably not as effectively trapped within the tumor tissue as the micelleplexes. In both the PEGylated polyplex and micelleplex cases, the activity in the tumor was about 2-fold higher than in the surrounding muscle tissue, indicating a preference for accumulation in the diseased tissue; the p-values of these comparisons between the tumor and muscle tissues are p = 0.040 (1.5 hours) and 0.002 (6 hours) for the micelleplex case, and p = 0.020 (1.5 hours) and 0.297 (6 hours) for the PEGylated polyplex case. Being able to both discriminate between normal and tumor tissue, and accumulate and be retained at high levels in the tumor, the macromolecular micelleplex particles are clearly the more favorable choice for tumor-targeted delivery.
Conclusions
The objective of this study was to examine some of the potential impacts of changing the architecture of siRNA complexes on their delivery and in vivo biodistribution properties. We have reported the chemistry for a new micelle-based platform for siRNA delivery and have characterized its properties such as binding strength (discussed in Sections S10 and S11 of the SI), enzymatic resistance (Sections S12 and S13), gene silencing, cytotoxicity, cellular uptake and in vivo biodistribution and compared them with conventional polyplex systems. The architectural differences between micelleplexes and the polyplex systems studied did not strongly influence binding affinity or enzymatic resistance. Although the micelleplexes exhibited only a moderate level of cellular uptake, ultimately cellular internalization efficiency did not appear to be the rate-limiting step for achieving gene silencing, since the micelleplexes still mediated a higher level of GAPDH mRNA silencing than either the polyplexes or PEGylated polyplexes. Most likely, the micelle architecture was more favorable for internalization via caveolae-mediated pathways than the other carriers, leading to more efficient delivery of siRNA for gene silencing. However, the fact that folate-conjugation of the micelleplexes did not further enhance their GAPDH silencing efficiency may suggest that endosomal release or other intracellular trafficking steps rather than cellular internalization is the factor ultimately limiting the potency of gene knockdown. Methods to potentially overcome this barrier to efficient gene knockdown are currently being explored for the next generation modification to the micelleplex system.
In conclusion, we have shown that our PEG-PnBA-PDMAEMA micelleplexes are stable and effective siRNA delivery systems and that favorable modifications to architecture and the degree of PEG shielding can be incorporated into polycation-based carriers without loss of function or increased toxicity. The advantages are particularly evident in vivo, where the larger particle size of the micelleplexes clearly provides for improved accumulation and retention in tumor tissue compared to the smaller PEGylated polyplexes due to exploitation of passive-targeting via the EPR effect. We have the additional flexibility to tune the size of the micelleplexes by controlling the molecular weight of the component polymer blocks. This is extremely useful for passive tumor targeting where efficient uptake of nanoparticles depends on how effectively they are designed for navigating the local vasculature, the size and properties of which vary largely by tumor type and location.40-41 All in all, we believe there is value in the fundamental findings of our proof-of-concept design in highlighting architecture as a critical degree of freedom available in designing robust siRNA delivery systems.
Materials and Methods
Materials
The siRNAs used for in vitro experiments were Silencer® Negative Control #1 (catalog number 440421), generously provided by Ambion (Austin, TX), and AlexaFluor 647-tagged, EGFP-specific siRNA (custom synthesis) was purchased from Qiagen (Valencia, CA). siRNA used for in vivo biodistribution was ordered from Samchully Pharm.Co. (Seoul, Korea) with an unmodified antisense strand (5’- UUUCCGUGCUCCAAAACAAAdTdT -3’) and a 5’ amino modifier C6 modification on the 3’ end of the sense strand (5’- UUGUUUUGGAGCACGGAAAdTdT - (CH2)6NH2 -3’). RNase A (ribonuclease protection assay (RPA) Grade) used for the enzymatic degradation study was also purchased from Ambion (Austin, TX), and the RNase inhibitor, RNaseOUT™, used to quench the degradation reaction, and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). A Cell-Titer Non-Radioactive Cell Proliferation (MTT) Assay Kit was purchased from Promega (Madison, WI) for cytotoxicity analyses. Bovine Serum Albumin (BSA) for protein aggregation studies was purchased from USB (Cleveland, OH) and sterile-filtered serum from human male AB plasma was purchased from Sigma Aldrich (Saint Louis, MO) for use in serum stability studies.
Synthesis and Characterization of the Polymers
All polymers used in this study were synthesized by atom transfer radical polymerization (ATRP) using appropriate initiators and catalyst systems. Detailed procedures for the synthesis and characterization of these polymers are described in Section S1 of the SI. Some related information can also be found in our previous publications.13, 42
Micellization of PEG-PnBA-PDMAEMA
Due to the relatively large size of the hydrophobic PnBA block, this polymer is not readily soluble in water. To obtain an aqueous suspension of micelle-like aggregates, a 1% (w/v) solution of PEG-PnBA-PDMAEMA was first prepared by dissolving the polymer in 3 ml of N,N-dimethyl formamide (DMF). The DMF was then removed by solvent exchange against 300 ml of 10 mM Tris-HCl buffer (pH 7.5) under constant stirring using Snakeskin® dialysis tubing (Pierce Biotechnology, Rockford, IL) with a molecular weight cutoff of 3,500 g/mol. The 300 ml of buffer solution was changed every 24 hours during the 72 hour procedure to ensure complete micellization of the polymer and removal of DMF. Finally, a 0.47 % (w/w) aqueous suspension of the micelle-like aggregates was obtained for experimental use. Folate-containing micelles were prepared in by an identical procedure except the starting polymer solution was a 7:1 ratio (by weight) mixture of PEG113-PnBA100-PDMAEMA126 and FOL-PEG113-PnBA100-PDMAEMA113. This 7:1 folate to unmodified copolymers ratio used for preparation of folate-functionalized micelles was the same ratio used in Reference 43. Unlike typical surfactant micelles, these micelle-like aggregates do not exist in a state of equilibrium between unimers and aggregates, but are instead kinetically frozen in a micellar configuration.44 For convenience, though, we will refer to these aggregates as “micelles” hereafter in this paper.
Preparation of Polymer(or Micelle)/siRNA Complexes
250 μM siRNA stock solutions were prepared in either nuclease-free water or siRNA resuspension buffer (100 mM potassium acetate, 30 mM HEPES potassium hydroxide, 2 mM magnesium acetate, pH 7.4) according to the manufacturer's suggested protocol, and stored at -20°C until use. Working concentrations of siRNA (50 μM) were prepared by diluting the stock in 10 mM Tris-HCl buffer (pH 7.5). Stock polymer and micelle solutions (as well as working dilutions) were prepared in 10 mM Tris-HCl buffer (pH 7.5) and stored at 4 °C until use. The polymer/micelle and siRNA stock solutions were prepared at concentrations such that combining the polymer/micelle and siRNA solutions at a volume ratio of “x” would result in an N/P ratio of “x”. N/P refers to the ratio of amine groups on PDMAEMA to phosphate groups on the siRNA backbone. To prepare complexes for analysis using gel electrophoresis, at least 0.75 μl of siRNA (500 ng) was combined with either polymer or micelle solution at the desired N/P ratio. The samples were diluted with TAE (Tris-Acetate-EDTA) buffer, mixed by pipetting vigorously, and allowed to complex quiescently for at least 30 minutes at room temperature. Henceforward, siRNA complexes with PDMAEMA, PEG-PDMAEMA and PEG-PnBA-PDMAEMA will be referred to as “polyplexes”, “PEGylated polyplexes” and “micelleplexes”, respectively.
Atomic Force Microscopy (AFM)
Fluid-state images of micelles and micelleplexes (N/P 8; prepared at a polymer concentration of 0.15 mg/ml) were taken via AFM operating in fluid tapping mode (Nanoscope Dimension 3100, Veeco/Digital Instruments). Samples were prepared by placing 10 μl of the micelle or micelleplex solution onto a negatively-charged substrate. After 2 minutes, excess solution was removed, deionized water was used to rinse the samples twice to remove residual salts and then imaging was performed in 10 mM Tris-HCl buffer (pH 7.5). The negatively-charged, gold-coated substrates were prepared by evaporating titanium and then gold onto a silicon wafer. The coated wafer was then immersed into a 0.001 M solution of 16-mercaptohexadecanoic acid dissolved in ethanol for several days, rinsed with deionized water and then dried with nitrogen. Substrates were kept in an air-tight environment until used for AFM imaging.
Cryogenic Transmission Electron Microscopy (Cryo-TEM)
Cryo-TEM of micelles and micelleplexes (N/P 8; same concentration as in AFM) was performed by depositing 3-4 μl of solution onto copper grids (400 mesh, catalog # G400-cu, Electron Microscopy Sciences). The solutions were then vitrified with liquid ethane and imaged on a CM200 FEI Electron Microscope operating at 200 kV. To achieve appropriate phase contrast, a nominal underfocus of 5-10 μm was used. Digital images were captured on a Gatan UltraScan 4k × 4k camera (model US4000SP) and analyzed in Digital Micrograph software, version 3.10.0.
siRNA Condensation Studies
Using the method described previously, polyplexes, PEGylated polyplexes and micelleplexes were prepared at N/P ratios of 0.5, 1, 2, 5, 8, 10 and 15. A small volume (1 – 4 μl) of gel loading buffer (Blue Juice™, 10×, Invitrogen) was added to the samples before being loaded onto 1% (w/v) agarose gels containing 0.004% (v/v) ethidium bromide (EtBr). The gels were run for 1 hour at 3.4 Volts/cm in TAE buffer, and then imaged on Polaroid 667 film (VWR).
Dynamic Light Scattering (DLS)
A Malvern Zetasizer Nano S instrument was used to measure the hydrodynamic diameters of the polymers and siRNA/polymer complexes. 15 μl solutions of polymers or micelles in 10 mM Tris-HCl buffer or complexes at N/P 8 containing 2.5 μM siRNA were prepared for the measurements and analyzed in a low-volume, quartz cuvette. Hydrodynamic diameters were calculated from the size distribution by volume (generated by the Non-Negatively Constrained Least Squares (NNLS) method for polydisperse samples), provided by the Malvern software, and are reported as the average of 2 independent measurements ± the deviation from the mean; each of these two measurements yielded a result which was actually the average of 10 runs (30 seconds per run).
MTT Assay for Cell Viability
Into a 96-well plate, HeLa cells were seeded at a density of 2 × 104 cells/well in 90 μl of Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and left to grow overnight in a 37 °C, 5% CO2 environment. The next day, the complexes to be analyzed for cytotoxicity were prepared at the desired dilutions in 100 μl of serum and antibiotic-free DMEM and added to cells. Polymer and micelle/siRNA complexes were all prepared at N/P 8 containing siRNA concentrations ranging from 3 nM to 300 nM. The cells were washed with calcium and magnesium-free (CMF) saline 4 hours after transfection and complete growth media (DMEM supplemented with 10% FBS and 1% penicillin streptomycin) was added. Following the 24-hour incubation, a standard MTT assay was performed to determine the percentage of viable cells remaining after exposure to the complexes relative to cells to which only media was added.
Flow Cytometry
Approximately 1 × 106 cells HeLa cells were seeded into 100 mm tissue culture dishes with complete growth media 24 hours prior to the study. The siRNA formulations were prepared according the same methods described above. An anti-EGFP siRNA with a 3’, sense-strand Alexa Fluor 647 label was used in the complex formulations at a final concentration of 30 nM. At the time of transfection, media was removed to wash the cells, and it was then replaced by 5 ml of serum and antibiotic-free media containing the transfection mixtures. The cells were then incubated at 37 ° C for a 4 hour transfection. The medium was then removed from the plate and cells were washed with CMF saline, trypsinized, pelleted by centrifugation for 5 min at 1200 rpm and then resuspended in ice cold PBS and kept on ice. Cellular internalization of various complexes was measured by flow cytometry using a Beckman Coulter FC500 with the FL4 channel for far-red emission, and resulting histograms were analyzed using CXP software. All assays were performed in duplicates. It is important to note that since this flow cytometry experiment could not discriminate between internalized complexes and complexes tightly-bound to the cell membrane, our data is actually a measure of the net cell-associated fluorescence, and not simply uptake.
Endogenous Gene Knockdown Experiments
HeLa cells were seeded in 6-well plates at 3×105 cells/well in DMEM supplemented with 10% FBS, and grown overnight in a 37 °C, 5% CO2, environment. Prior to transfection, cells were observed to be about 60% confluent. Subsequently, the cells were washed with CMF saline and transfection mixtures were added to the appropriate wells in 2 ml of serum and antibiotic-free DMEM. Polymer complexes were prepared at N/P 8 (LF2000 was complexed with siRNA as per the manufacturer's protocol), and the final siRNA concentration in the wells was 30 nM. After 4 hours of transfection, cells were washed once with CMF saline, after which complete DMEM was added for the duration of the experiment. 24 hours post-transfection, the cells were harvested for total RNA using the RNeasy Mini kit (Qiagen) and a reverse transcriptase reaction was performed on a Biometra Tgradient instrument. GAPDH mRNA knockdown levels were measured using SYBR green fluorescent quantification technology in a quantitative, real-time polymerase chain reaction (qRT-PCR; Applied Biosystems 7300 Real Time PCR System) with appropriate primer sets for human GAPDH. As an internal control, GAPDH expression values were normalized to the expression levels of human 18 S mRNA. GAPDH primer sequences: forward: TGACAACTTTGGTATCGTGGA, reverse: CAGTAGAGGCAGGGATGATGT. 18S primer sequences: forward: ACTCTTTCGAGGCCCTGTAAT, reverse: CTCCCAAGATCCAACTACGAG.
Endocytosis Mechanistic Study
PDMP Hydrochloride from Cayman Chemical (Ann Arbor, MI) and Amiloride, Amantadine, Nystatin, Methyl β-Cyclodextrin, and Folic Acid from Sigma Aldrich were freshly diluted with DI water (or 95% ethanol for PDMP HCl and Nystatin) immediately before use. 24 hours prior to the experiment, HeLa cells were seeded at 1 × 106 cells/plate in antibiotic-free DMEM, and in the case of free folate-treated samples, folic acid-free DMEM was used to culture the cells. 100 μl of the inhibitors were added to the 100 mm tissue culture plates containing 4800 μl of serum and antibiotic-free DMEM, and left to incubate for 1 hour prior to the addition of polymer complexes. The final concentration of the endocytosis inhibitors in the plate were 3 mM, 1 mM, 100 μg/ml, 1 mg/ml, 1 mM and 10 μM for Amiloride (Macropinocytosis),45 Amantadine (Clathrin),33 Nystatin (Membrane fusion),45 Methyl β-Cyclodextrin (Caveolar),33 Folic Acid (Folate receptor-mediated)46 and PDMP Hydrochloride (Lipid Raft),45 respectively. After a 1 hour pre-incubation and without washing the cells, 100 μl of polymer/siRNA (AlexaFluor647-tagged) complexes were added to the plates already containing the pharmacological inhibitors and left to incubate for an additional 4 hours at 37 °C, after which the cells were harvested and fixed in 2% paraformaldehyde for analysis of AlexaFluor647 uptake via Flow Cytometry (BD Biosciences FACSCalibur flow cytometer operating with at 630 nm laser). Flow cytometry samples were analyzed in duplicates for each inhibitor.
The values of the optimal inhibitor concentrations used were taken from the respective references cited above, except for the methyl β-cyclodextrin (mβCD) case. The recommended dose of mβCD from Reference 33 was 10 mg/ml, but this dose killed nearly all of the plated HeLa cells and thus was not able to be analyzed via flow cytometry. To determine the appropriate dose, cells were seeded at 100,000 cells/well in a 12-well plate and each well was exposed to a different dose of mβCD. Doses studied were 10, 8, 6, 5, 2.5, 1.0, 0.75, 0.5, 0.25, 0.1, 0.05 and 0 mg/ml mBCD. Any cell death caused by exposure to mBCD was observed visually using optical microscopy at time points 30 min, 1, 2, 3, 4. 5 and 6 hours following addition of the inhibitor. Concentrations above 2.5 mg/ml caused moderate to severe toxicity to HeLa cells, whereas 1.0 mg/ml maintaned a very high level of cell viability (visually, no cells were dead or deformed) over 6 hours. Below 1.0 mg/ml, cells were 100% viable, so 1.0 mg/ml was chosen as a dose potent enoughfor inhibition without toxicity.
In Vivo Biodistribution
Detailed methods for siRNA labeling are provided in Section S9 of the SI. Small animal PET-computed tomography (PET-CT) imaging was performed with an Inveon microPET-CT scanner (Siemens) at Samsung Biomedical Research Institute (Korea). Right before imaging, mice (n = 6 for all samples except the naked siRNA control for which n = 4) were anesthetized with 1% isoflurane breathing tube on a heated (30 °C) pad. 100-200 μCi (microcuries, a unit of radioactivity) of I124-labeled siRNA/polymer (or siRNA/ micelle) complexes was injected via tail vein. Immediately after the micro-PET scan, mice were subjected to a 10-min micro-CT scan, using standard image acquisition parameters. Static micro-PET scans were acquired at 1.5 and 6 hours post-injection with micro-CT scan for anatomical coregistration. To determine temporal changes of tracer concentration in various tissues, ellipsoid or activity-guided, user-defined regions of interest were placed in the region that exhibited organ-characteristic I124 activity as determined by visual inspection. To minimize partial volume effects between tissue types, care was taken not to use overlapped borders between organs. Considering the size of the studied organs, tumors and the spatial resolution of the PET scanner, the partial volume effects are, even if they existed, expected to have a very minor impact on the results of quantitative analysis. Activity concentrations are shown as the percentage of the decay-corrected injected activity divided by the mass of the studied organ (% injection dose per gram or %ID/g).
Cell line and animal experiments
PC9 non-small cell lung adenocarcinoma cell line was kindly provided by Kazuto Nishio (Japan). PC9 cells were cultured and maintained in RPMI 1640 medium (Gibco, USA) containing 1% penicillin-streptomycin and 10% fetal bovine serum. Balb/c-nude mice were purchased from Orient Bio, Inc. (Seongnam, Korea). Exponentially growing 106 PC9 cells in 100 μl of Matrigel (BD Biosciences, USA) were injected subcutaneously into the lower right back of Balb/c-nude mice. Xenograft animal models were used for in vivo experiments, when tumor volumes reached approximately 50-100 mm3. All the animals were maintained in a specific pathogen-free facility at Samsung Biomedical Research Institute at Samsung Medical Center in accordance with institutional guidelines.
Supplementary Material
Acknowledgments
The authors would like to thank the National Science Foundation (CBET-0828574, and DMR-0906567), the Showalter Trust and the Purdue Research Foundation for providing financial support of this research. This work was also supported in part by IUSM/CTR, NIH/NCRR Grant Number RR025761. The GEM Consortium is also acknowledged for fellowship funds to support DJG. The contents of this publication are solely the responsibility of the authors, and do not represent the official views of the funding agencies. We are grateful to Professors Jue Chen, J. Paul Robinson and John A. Morgan at Purdue University for allowing access to various analytical equipment, Professor Stephen P. Beaudoin and Dr. Bich Van Pham for providing thiol-coated AFM substrates and to Dr. Nitin Puri of Ambion (Applied Biosystems) for generously providing some of the siRNA samples used in this work. We also thank the IU Simon Cancer Center Flow Cytometry Facility for access to their flow cytometry equipment.
Footnotes
Supporting Information Available: Detailed procedures of the syntheses of the various polymers used (Section S1), 1H NMR and GPC characterization data (S1), cryo-TEM and AFM histograms (S2), composition information for the siRNA/polymer complexes studied (S3), supplementary information for the DLS technique used (S4), cell viability study (S5), flow cytometry raw data (S6), additional data from the endocytosis mechanistic study (S7), BSA aggregation time study (S8), methods for the in vivo biodistribution study (S9), competing anion assay (S10), ethidium bromide exclusion assay (S11), stability against enzymatic degradation in human serum (S12), and RNAse A degradation results (S13). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dash PR, Read ML, Barrett LB, Wolfert M, Seymour LW. Factors Affecting Blood Clearance and In Vivo Distribution of Polyelectrolyte Complexes for Gene Delivery. Gene Ther. 1999;6:643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Banan M, Puri N. The Ins and Outs of RNAi in Mammalian Cells. Curr. Pharm. Biotechno. 2004;5:441–450. doi: 10.2174/1389201043376643. [DOI] [PubMed] [Google Scholar]
- 4.Gary DJ, Puri N, Won YY. Polymer-Based siRNA Delivery: Perspectives on the Fundamental and Phenomenological Distinctions from Polymer-Based DNA Delivery. J. Control. Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Mishra S, Webster P, Davis ME. PEGylation Significantly Affects Cellular Uptake and Intracellular Trafficking of Non-Viral Gene Delivery Particles. Eur. J. Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 6.Sato A, Choi SW, Hirai M, Yamayoshi A, Moriyarna R, Yamano T, Takagi M, Kano A, Shimamoto A, Maruyama A. Polymer Brush-Stabilized Polyplex for a siRNA Carrier with Long Circulatory Half-Life. J. Control. Release. 2007;122:209–216. doi: 10.1016/j.jconrel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Zelphati O, Uyechi LS, Barron LG, Szoka FC. Effect of Serum Components on the Physico-Chemical Properties of Cationic Lipid/Oligonucleotide Complexes and on Their Interactions with Cells. BBA-Lipid Lipid Met. 1998;1390:119–133. doi: 10.1016/s0005-2760(97)00169-0. [DOI] [PubMed] [Google Scholar]
- 8.Glodde M, Sirsi SR, Lutz GJ. Physiochemical Properties of Low and High Molecular Weight Poly(ethylene glycol)-Grafted Poly(ethylene imine) Copolymers and Their Complexes with Oligonucleotides. Biomacromolecules. 2006;7:347–356. doi: 10.1021/bm050726t. [DOI] [PubMed] [Google Scholar]
- 9.Mullen PM, Lollo CP, Phan QC, Amini A, Banaszczyk MG, Fabrycki JM, Wu DP, Carlo AT, Pezzoli P, Coffin CC, Carlo DJ. Strength of Conjugate Binding to Plasmid DNA Affects Degradation Rate and Expression Level In Vivo. BBA-Gen. Subjects. 2000;1523:103–110. doi: 10.1016/s0304-4165(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Lorenzo C, Barreiro-Iglesias R, Concheiro A, Iourtchenko L, Alakhov V, Bromberg L, Temchenko M, Deshmukh S, Hatton TA. Biophysical Characterization of Complexation of DNA with Block copolymers of Poly(2-(dimethylamino)ethyl methacrylate), Poly(ethylene oxide), and Poly(propylene oxide). Langmuir. 2005;21:5142–5148. doi: 10.1021/la050170v. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg L, Deshmukh S, Temchenko M, Iourtchenko L, Alakhov V, Alvarez-Lorenzo C, Barreiro-Iglesias R, Concheiro A, Hatton TA. Polycationic Block Copolymers of Poly(ethylene oxide) and Poly(propylene oxide) for Cell Transfection. Bioconjugate Chem. 2005;16:626–633. doi: 10.1021/bc049749f. [DOI] [PubMed] [Google Scholar]
- 12.Munier S, Messai I, Delair T, Verrier B, Ataman-Onal Y. Cationic PLA Nanoparticles for DNA Delivery: Comparison of Three Surface Polycations for DNA Binding, Protection and Transfection Properties. Colloid. Surface. B. 2005;43:163–173. doi: 10.1016/j.colsurfb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Effects of the Incorporation of a Hydrophobic Middle Block into a PEG-Polycation Diblock Copolymer on the Physicochemical and Cell Interaction Properties of the Polymer-DNA Complexes. Biomacromolecules. 2008;9:3294–3307. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Wetering P, Cherng JY, Talsma H, Crommelin DJA, Hennink WE. (2-(Dimethylamino)ethyl methacrylate)-Based (Co)polymers as Gene Transfer Agents. J. Control. Release. 1998;53:145–153. doi: 10.1016/s0168-3659(97)00248-4. [DOI] [PubMed] [Google Scholar]
- 15.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Development of a Novel Endosomolytic Diblock Copolymer for siRNA Delivery. J. Control. Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benoit DSW, Henry SM, Shubin AD, Hoffman AS, Stayton PS. pH-Responsive Polymeric siRNA Carriers Sensitize Multidrug Resistant Ovarian Cancer Cells to Doxorubicin via Knockdown of Polo-like Kinase 1. Mol. Pharm. 7:442–455. doi: 10.1021/mp9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong W-H, Sung D-K, Shim Y-H, Bae KH, Dubois P, Park TG, Kim J-H, Seo S-W. Efficient Intracellular siRNA Delivery Strategy through Rapid and Simple Two Steps Mixing Involving Noncovalent Post-PEGylation. J. Control. Release. 2009;138:141–147. doi: 10.1016/j.jconrel.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 18.Zhu CH, Jung S, Luo SB, Meng FH, Zhu XL, Park TG, Zhong ZY. Co-Delivery of siRNA and Paclitaxel into Cancer Cells by Biodegradable Cationic Micelles Based on PDMAEMA-PCL-PDMAEMA Triblock Copolymers. Biomaterials. 2010;31:2408–2416. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 19.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol. Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis ME. The First Targeted Delivery of siRNA in Humans via a Self-Assembling, Cyclodextrin Polymer-Based Nanoparticle: From Concept to Clinic. Mol. Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 21.Nan A. Multifunctional Nanocarrier for Image-Guided Delivery of Bioactive Agents. Nanomedicine. 2007;2:739–743. doi: 10.2217/17435889.2.5.739. [DOI] [PubMed] [Google Scholar]
- 22.Wiewrodt R, Thomas AP, Cipelletti L, Christofidou-Solomidou M, Weitz DA, Feinstein SI, Schaffer D, Albelda SM, Koval M, Muzykantov VR. Size-Dependent Intracellular Immunotargeting of Therapeutic Cargoes into Endothelial Cells. Blood. 2002;99:912–922. doi: 10.1182/blood.v99.3.912. [DOI] [PubMed] [Google Scholar]
- 23.Schiffelers RM, Bakker-Woudenberg IAJM, Snijders SV, Storm G. Localization of Sterically Stabilized Liposomes in Klebsiella Pneumoniae-Infected Rat Lung Tissue: Influence of Liposome Characteristics. BBA-Biomembranes. 1999;1421:329–339. doi: 10.1016/s0005-2736(99)00139-x. [DOI] [PubMed] [Google Scholar]
- 24.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-Specific Knockdown of EWS-FLI1 by Targeted, Nonviral Delivery of Small Interfering RNA Inhibits Tumor Growth in a Murine Model of Metastatic Ewing's Sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 25.Mao SR, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of Poly(ethylene glycol) Chain Length on the Physicochemical and Biological Properties of Poly(ethylene imine)-graft-Poly(ethylene glycol) Block Copolymer/siRNA Polyplexes. Bioconjugate Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 26.Lu TC, Sun J, Chen XX, Zhang PB, Jing XB. Folate-Conjugated Micelles and Their Folate-Receptor-Mediated Endocytosis. Macromol. Biosci. 2009;9:1059–1068. doi: 10.1002/mabi.200900134. [DOI] [PubMed] [Google Scholar]
- 27.Lee H, Son SH, Sharma R, Won YY. A Discussion of the pH-Dependent Protonation Behaviors of Poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) and Poly(ethylenimine-ran-2-ethyl-2-oxazoline) (P(EI-r-EOz)). J. Phys. Chem. B. 2011;115:844–860. doi: 10.1021/jp109151s. [DOI] [PubMed] [Google Scholar]
- 28.van de Wetering P, Zuidam NJ, van Steenbergen MJ, van der Houwen OAGJ, Underberg WJM, Hennink WE. A Mechanistic Study of the Hydrolytic Stability of Poly(2-(dimethylamino)ethyl methacrylate). Macromolecules. 1998;31:8063–8068. [Google Scholar]
- 29.Clark RA, Olsson I, Klebanoff SJ. Cytotoxicity for Tumor-Cells of Cationic Proteins from Human Neutrophil Granules. J. Cell Biol. 1976;70:719–723. doi: 10.1083/jcb.70.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putnam D, Gentry CA, Pack DW, Langer R. Polymer-Based Gene Delivery with Low Cytotoxicity by a Unique Balance of Side-Chain Termini. Proc. Natl. Acad. Sci. USA. 2001;98:1200. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schallon A, Jerome V, Walther A, Synatschke CV, Muller AHE, Freitag R. Performance of Three PDMAEMA-Based Polycation Architectures as Gene Delivery Agents in Comparison to Linear and Branched PEI. React. Funct. Polym. 2010;70:1–10. [Google Scholar]
- 32.Won Y-Y, Sharma Rahul, Konieczny Stephen F. Missing Pieces in Understanding the Intracellular Trafficking of Polycation/DNA Complexes. J. Control. Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrielson NP, Pack DW. Efficient Polyethylenimine-Mediated Gene Delivery Proceeds via a Caveolar Pathway in HeLa Cells. J. Control. Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Fang J, Sawa Tomohiro, Maeda Hiroshi. Factors and Mechanism of “EPR” Effect and the Enhanced Antitumor Effects of Macromolecular Drugs Including SMANCS. In: Maeda, editor. Polymer Drugs in the Clinical Stage. Kluwer Academic/Plenum Publishers; New York: 2003. pp. 29–49. [DOI] [PubMed] [Google Scholar]
- 35.de Wolf HK, Snel CJ, Verbaan FJ, Schiffelers RM, Hennink WE, Storm G. Effect of Cationic Carriers on the Pharmacokinetics and Tumor Localization of Nucleic Acids after Intravenous Administration. Int. J. Pharm. 2007;331:167–175. doi: 10.1016/j.ijpharm.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 36.Hunt CA, Rustum YM, Mayhew E, Papahadjopoulos D. Retention of Cytosine-Arabinoside in Mouse Lung Following Intravenous Administration in Liposomes of Different Size. Drug Metab. Dispos. 1979;7:124–128. [PubMed] [Google Scholar]
- 37.Fidler IJ, Raz A, Fogler WE, Kirsh R, Bugelski P, Poste G. Design of Liposomes to Improve Delivery of Macrophage-Augmenting Agents to Alveolar Macrophages. Cancer Res. 1980;40:4460–4466. [PubMed] [Google Scholar]
- 38.Braet F, Wisse E. Structural and Functional Aspects of Liver Sinusoidal Endothelial Cell Fenestrae: a Review. Comp. Hepatol. 2002;1:1–17. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wester HJ, Kessler H. Molecular Targeting with Peptides or Peptide-Polymer Conjugates: Just a Question of Size? J. Nucl. Med. 2005;46:1940–1945. [PubMed] [Google Scholar]
- 40.Jain RK. Understanding Barriers to Drug Delivery: High Resolution In Vivo Imaging Is Key. Clin. Cancer Res. 1999;5:1605–1606. [PubMed] [Google Scholar]
- 41.Jain RK. Delivery of Molecular and Cellular Medicine to Solid Tumors. Adv. Drug Deliver. Rev. 1997;26:71–90. doi: 10.1016/s0169-409x(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 42.Sharma R, Goyal A, Caruthers JM, Won YY. Inhibitive Chain Transfer to Ligand in the ATRP of n-Butyl Acrylate. Macromolecules. 2006;39:4680–4689. [Google Scholar]
- 43.Yoo HS, Park TG. Folate Receptor Targeted Biodegradable Polymeric Doxorubicin Micelles. J. Control. Release. 2004;96:273–283. doi: 10.1016/j.jconrel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhang LF, Yu K, Eisenberg A. Ion-Induced Morphological Changes in “Crew-Cut” Aggregates of Amphiphilic Block Copolymers. Science. 1996;272:1777–1779. doi: 10.1126/science.272.5269.1777. [DOI] [PubMed] [Google Scholar]
- 45.Lu JJ, Langer R, Chen JZ. A Novel Mechanism Is Involved in Cationic Lipid-Mediated Functional siRNA Delivery. Mol. Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee RJ, Low PS. Folate-Mediated Tumor-Cell Targeting of Liposome-Entrapped Doxorubicin In-Vitro. BBA-Biomembranes. 1995;1233:134–144. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.