Abstract
Schizophrenia (SCZD) is a heritable developmental disorder. While the molecular mechanism of disease remains unclear, insights into the disorder have been made through a vast array of experimental techniques. Together, MRI brain imaging, pharmacological and postmortem pathological studies have observed decreased brain volume, aberrant neurotransmitter signaling, reduced dendritic arborization and impaired myelination in SCZD. Genome wide association studies have identified common single nucleotide polymorphisms as well as rare copy number variants that contribute to SCZD, while mouse models of candidate SCZD genes show behavioral abnormalities and anatomical perturbations consistent with human disease. The advent of human induced pluripotent stem cells (hiPSCs) makes it possible to study SCZD using live human neurons with a genetic predisposition towards SCZD, even without knowledge of the genes interacting to produce the disease state. SCZD hiPSC neurons show cellular defects comparable to those identified in postmortem human and mouse studies, and gene expression changes consistent with predictions made by GWAS. SCZD hiPSC neurons represent a new tool to look beyond phenotype and begin to dissect the molecular mechanisms of SCZD.
Introduction
Schizophrenia (SCZD) is a neurological disorder characterized by three severe classes of symptoms: positive (hallucinations and delusions), negative (inability to speak, express emotion or find pleasure) and cognitive (deficits in attention, memory and planning) 1, 2. The current treatment regime for SCZD involves chronic treatment with powerful antipsychotics, the strong unpleasant side effects of which often result in cessation of treatment 1. The life expectancy of patients with SCZD is up to ten years less than the general population 3; 40% of schizophrenics suffer from substance abuse 4, 20% are homeless 5 and 10% ultimately commit suicide 6, 7. It is estimated that 1.1% of the population over 18 years of age has SCZD, including 3 million Americans 1.
Human studies of SCZD have relied primarily on brain imaging, pharmacology, postmortem pathology and genetic studies of patient lymphocytes. Animal studies are limited in two important ways: 1) they do not reflect the complex genetic interactions that result in the vast majority of cases of SCZD, and 2) it is difficult to evaluate classical symptoms of SCZD such as hallucinations, delusions and disorganized speech in mice. While many insights have been made through these methods, the molecular and cellular defects that contribute to disease initiation and progression in neurons remain unknown. Recently, a third approach has been described with which to study SCZD 10, 11. Reprogramming of patient fibroblasts to human induced pluripotent stem cells (hiPSCs), followed by hiPSC differentiation to neurons, produces a near limitless source of live human neurons, genetically identical to those present in patients, with which to study this disorder.
This review will summarize the major findings of human and mouse studies of SCZD, and compare these to early reports of SCZD hiPSC neurons.
Insights from Human Studies
Human studies to date have utilized three major approaches: brain imaging, postmortem pathology and genome wide association studies (GWAS).
Observations from brain imaging
Magnetic resonance imaging (MRI) can be used to estimate the volume of various brain regions, though these studies must be interpreted in the context of the high variability of brain measures across individuals 12. While group average differences exist between SCZD patients and controls, anatomic MRI differences are not adequate for diagnosis. For analogy, though men tend to be taller than women on average, height is not sufficient to determine sex; male/female height differences are twice as predictive of gender as most MRI neuroimaging experiments are predictive of a diagnosis of SCZD 13.
In first-episode patients with SCZD, MRI studies consistently observe decreased whole brain volume (and increased ventricular volume) 14–16. Specifically, the greatest decreases are observed in the grey matter of the hippocampus, basal ganglia and thalamus (Figure 1)16–18. In chronically ill patients, the volume loss is most pronounced in the frontal and temporal grey matter areas of the cortex 19, but whether these changes result from disease or long-term treatment with antipsychotic medications is unclear. Longitudinal studies have observed progressive decrease in brain volume and increase in lateral ventricle volume for at least 20 years after the onset of symptoms.
Figure 1.

Average high-resolution magnetic resonance images (MRI scans) showing gray matter loss in adolescents with schizophrenia. Severe loss is observed (red and pink; up to 5% annually) in parietal, motor, and temporal cortices, whereas inferior frontal cortices remain stable (blue; 0–1% loss). Adapted from Thompson et al (2001).
White matter is produced when oligodendrocytes wrap axons in sheaths of myelin, which increases the speed of neurotransmission as well as the timing and synchrony of neuronal firing patterns 20. Beyond the well characterized changes in grey matter, some studies also identify changes in the white matter of the brain in SCZD, particularly in the prefrontal and temporal lobes and corpus collusum 21. Longitudinal brain imaging studies of childhood onset (COS) cases of SCZD have observed both progressive loss of cortical grey matter during adolescence and delayed white matter development 13.
Changes in blood flow and blood oxygenation in the brain are closely linked to neural activity. Functional magnetic resonance imaging (fMRI) measures the change in blood flow as an indicator of brain activity and fMRI comparisons of control and SCZD patients have revealed brain activity changes in the dorsolateral prefrontal cortex. At rest, SCZD patients show cortical hyper-activity and hyper-connectivity between the cortex and hippocampus 22 while during working memory tasks, SCZD patients show reduced activation of the cortex 23. The molecular mechanism of these changes has not been explained.
While brain imaging has associated clinical symptoms to the general brain areas affected, the relationship between brain imaging, cellular pathology and the molecular mechanism of SCZD remains unknown.
Observations from pharmacological studies
The accidental discovery that Chlorpromazine (Thorazine) reduces psychotic symptoms, likely by functioning as an antagonist of dopamine (DA) receptors, provided the basis for the “DA hypothesis” of SCZD. This hypothesis proposed that excessive activation of D2 receptors was the cause of (the positive symptoms of) SCZD. By Positron emission tomography (PET) imaging, researchers determine where given biological molecules, such as DA, bind in the brain. Such work has correlated DA receptor levels with the positive symptoms of SCZD (psychosis and delusions) 25 and strong evidence now links SCZD with increased DA synthesis, DA release, and resting-state synaptic DA concentrations 24. Further support of this hypothesis is the psychosis-inducing effects of DA receptor agonists such as amphetamine.
The DA hypothesis is now believed to be overly simplistic. Among the DA antagonists, Clozapine is uniquely effective for treatment-resistant SCZD 26. This ability may occur through a DA-independent mechanism, a hypothesis supported by anecdotal evidence that Clozapine effectively treats the psychotic symptoms associated with Parkinson’s disease (PD) without exacerbating the tremors, rigidity and bradykinesis caused by the loss of DA neurons in PD 27. Clozapine is an antagonist of D1, D2, D3, and D4 dopamine receptors, α1- and α2-adrenergic receptors, 5-hydroxytryptamine (5-HT) serotonin receptor, H1 histamine receptor, and M1, M2, M3, and M5 receptors; an agonist of the M4 muscarinic receptor28; a modulator of glutamatergic neurotransmission 29 and an inhibitor of GABAA receptor neurotransmission 30; its complex pharmacological properties hint at the roles of others neuronal cell types in SCZD.
Glutamate (GLU)-blocking drugs such as ketamine induce many of the symptoms, including hallucinations and cognitive deficits, associated with SCZD 31, whereas the glutamate receptor (GLUR2/3) agonist LY2140023 has been shown to ameliorate the symptoms of SCZD in recent clinical trials by Eli Lilly 32 (Figure 2). The authors suggest that LY2140023 may work by reducing the presynaptic release of glutamate at limbic synapses where these receptors are expressed.
Figure 2.

Weekly change in clinical assessment of psychosis, measured using the clinical Positive and Negative Symptom Scale (PANSS). Abbreviation: PANSS: positive and negative symptom scale. **p<0.01; ***p<0.001 Adapted from Patil et al (2007).
In summary, pharmacology of SCZD is intricate but evidence supports a role for altered DA and GLU neurotransmitter activity.
Observations from postmortem studies
While brain imaging identified decreased brain volume in SCZD, postmortem studies of brains from patients with SCZD have revealed no widespread neuronal loss or even a glial response to a potential neuronal injury. Instead, pathological studies have observed three major changes in SCZD brain tissue: increased density of pyramidal neurons, aberrations in interneurons, and decreased oligodendrocytes.
In the cortex, pathological changes are consistent with decreased neuronal connectivity in SCZD. Postmortem studies have observed an increased density of cortical pyramidal neurons without changes in cell number 33, 34. Neuronal soma are smaller 34, dendrites are shorter with reduced arborizations 35 and there is reduced dendritic spine density 36,37. The disturbances in prefrontal cognitive functioning in SCZD may be mediated by a process which involves atrophy of neuronal processes or synapses but stops short of actual neuronal loss 33.
Changes in RNA and protein levels of a number of key genes involved in neurotransmission have been observed in postmortem SCZD brain tissue. Glutamate receptor expression is altered in SCZD; expression is decreased in the hippocampus and increased in the cortex 38. Specifically in GABAergic interneurons, one finds decreased GAD67 and calcium-binding proteins in parvalbumin- and calbindin-positive GABAergic neurons. Changes in parvalbumin-positive GABAergic neurons are particularly relevant as they are thought to produce gamma oscillations, which synchronize pyramidal neuron firing, an activity that is impaired in SCZD. It is unclear whether decreased GABAergic inhibitory activity is a cell autonomous cause of SCZD or if it results from decreased glutamatergic input on GABAergic inhibitory neurons in the disease state.
Finally, a number of studies have observed both fewer oligodendrocytes and decreased expression of myelin genes in postmortem SCZD brains. This finding may be consistent with decreases in white matter observed in SCZD.
Genome wide association studies of SCZD
A complex genetic psychiatric disorder, SCZD has a large inherited component with an estimated heritability of 80–85% 39, 40. With tens of thousands of SCZD patients genotyped to date, it is now widely accepted that the complicated heritability of SCZD results from polygenic inheritance of a combination of inherited common polymorphisms and both inherited and de novo rare copy number variations (CNVs). Because a large number of markers collectively account for risk of SCZD, the risk of each one is so small it can be difficult to detect by GWAS. To date, most of the heritable variance of SCZD remains unaccounted for.
Early genetic studies of SCZD focused on traditional pedigree analyses. In a family identified in northern Scotland, more than half of the members suffer from mental illness, generally SCZD, and it was found that a balanced chromosomal translocation (1:11) segregated with disease. The disrupted gene on chromosome 1 was subsequently termed Disrupted-in-Schizophrenia-1 (DISC1)41. The genetic association between Neuregulin-1 (NRG1) and SCZD was first identified in association studies of Icelandic families 42 and subsequently confirmed in follow-up studies in multiple populations in Scotland, Ireland, Netherlands, Taiwan, Korea and China 43–49. Mutations in DISC1 are much more penetrant that those in NRG1, but are also exceedingly rare. While studies of postmortem brain tissue have consistently failed to detect alterations in DISC1 levels in typical SCZD patients, they do observe increased NRG1 in the hippocampus 50 and prefrontal cortex 51, 52 and decreased expression of its receptor ERBB4 53–55, suggesting that NRG1 and ERBB4 may be affected even in the absence of detectable genetic lesions.
Common polygenic variation has been shown to contribute to SCZD. Studies of single nucleotide polymorphisms (SNPs) have estimated that thousands of alleles of very small effect account for nearly 30% of the genetic variance of SCZD 56–58. Much of this common polygenic variation encompasses the major histocompatibility complex region (MHC) region at 6p22–p21 that contains over 200 genes. Though MHC genes have been implicated in immune diseases such as type I diabetes, multiple sclerosis, Crohn's disease, and rheumatoid arthritis, they also contribute to synaptic maturation 59–61; therefore, this genetic evidence should not be presumed to be proof of immune abnormalities in SCZD.
Other common variants now well-implicated in SCZD include TCF4 (transcription factor 4) on chromosome 18q21, zinc finger protein 804A (ZNF804A) on 2q32.1, and neurogranin (NRGN) on 11q24.2 57, 62. TCF4 is a neuronal transcription factor essential for neurogenesis 63, NRGN encodes a postsynaptic protein kinase substrate that binds calmodulin and is enriched in CA1 pyramidal neurons in the hippocampus 64, and ZNF804A is associated with altered neuronal connectivity in the dorsolateral prefrontal cortex 65.
Copy number variants (CNVs) are large deletions or duplications in the genome. So far, only rare (<1% of cases) and large CNVs (>100kb) have been shown to confer high risk of SCZD. SCZD CNVs are highly penetrant and account for up to 20% of SCZD cases. CNVs identified to date disrupt genes, such as ERBB4 66 or NRXN1 67, or regions, including 1q21.1, 15q11.2, 15q13.3, 16p11.2, 22q11.2 66, 68–70. By comparing the DNA of both parents to the SCZD patient (proband), it was shown that CNV mutations frequently occur de novo and are not inherited from either parent. Given that the CNV mutation rate far exceeds the rate for nucleotide substitutions 71 and that current CNV assays detect only the largest CNVs, constituting just 5% of total CNVs, researchers may be underestimating the contribution of CNVs to SCZD 72.
The genetic basis of SCZD does not necessarily conform to classical disease boundaries. Many CNVs that confer high risk of SCZD have been implicated in autism, mental retardation and epilepsy, while many SNPs associated with SCZD are shared with bipolar disorder 66, 68–70. It is likely that numerous disruptions in any number of key neurodevelopmental pathways may be sufficient to produce a diseased state that could ultimately manifest as SCZD.
Insights from Mouse Models
While causal mutations for SCZD have not been identified, several genetic mouse models of SCZD have been developed. Of these, mice with decreased activity of DISC1, NRG1, ERBB4 or the 22q11 genes show behavioral abnormalities and anatomical perturbations that may be relevant to SCZD.
DISC1 mice recapitulate aspects of the SCZD phenotype
DISC1-mutant animals demonstrate behavioral abnormalities such as decreased learning and memory73–75, decreased sociability73, 74, depression73, 74, hyperactivity74, 75 and aggression 74, which are consistent SCZD. Mice with reduced DISC1 activity during development have reduced neurite outgrowth76, reduced cortical migration 76, reduced dendritic complexity73, 75, reduced hippocampal synaptic transmission73, 75 and slightly enlarged ventricles74, but no significant structural defects or signs of neurodegeneration (Figure 3). Conversely, down-regulation of DISC1 in adulthood causes accelerated neural differentiation and increased neural excitability in newborn neurons77. DISC1 seems to function as a molecular scaffold; it interacts with multiple proteins, including the centrosomal protein NUDEL76, 77 required for neurite outgrowth and neuronal migration78, 79, as well as phosphodiesterase 4B (PDE4B)80, a key regulator of cyclic adenosine monophosphate (cAMP), linked to learning, memory and mood81–83. It remains unknown which of these functions is responsible for SCZD pathogenesis.
Figure 3.

Representative phenotypes present in dominant negative DISC1 mouse models of SCZD. Top, increased ventricular volume in dnDISC1 mice. Middle, reduced dendritic arborization in dnDISC1 mice. Bottom, reduced frequency of spontaneous inhibitory postsynaptic currents (IPSPs) in dnDISC1 mice. Adapted from Li et al (2007) and Pletnikov et al (2008).
NRG1 and ERBB4 mice demonstrate role of excitatory glutamatergic input onto GABAergic inhibitory neurons in SCZD
Though mice lacking both copies of either the NRG1 or ERBB4 genes are embryonic lethal due to cardiac defects, heterozygous null NRG1 and ERBB4 animals have behavioral abnormalities such as hyperactivity, increased aggression, and deficiencies in prepulse inhibition, a measure of sensory gating that is abnormal in SCZD 42, 84, 85. While cell layers in the cerebral cortex, hippocampus, and cerebellum develop normally in the mutant mice, there are defects in neurite outgrowth and arborization neuronal migration 86, 87 and impaired synaptic maturation and function 85, 88, 89. Treatment with the antipsychotic drug Clozapine reverses the behavioral and spine defects in these mice 85. ERBB4 is enriched in GABAergic interneurons, while NRG1 is primarily localized at synapses in excitatory glutamatergic neurons. NRG1 increases the number, size and activity of excitatory synapses on GABAergic interneurons 90, 91, renewing support for the hypothesis that SCZD results, at least in part, due to reduced excitatory glutamatergic input onto GABAergic inhibitory neurons.
22q11 models impaired long-range synchrony of neural activity
Mouse models of 22q11.2 represent the first studies of a CNV associated with SCZD; SCZD develops in about 20–25% of individuals with a chromosome 22q11.2 microdeletion. Mice with disruptions of 22q11.2 genes have fewer cortical neurons with slightly smaller spines, altered short- and long-term synaptic plasticity, enhanced neurotransmitter release, altered calcium kinetics in CA3 presynaptic terminals and impaired long-range synchrony between the hippocampus and prefrontal cortex 92–94.
Insights from Olfactory Neural Precursors
Olfactory neural precursors (ONPs) can be expanded following exfoliation of the nasal cavity via a non-invasive method 95. ONPs are capable of self-renewal as well as differentiation to mature electrophysiologically active neurons. ONPs from SCZD patients and controls have been generated and banked 95 to ask whether genetic, structural and/or functional abnormalities are present in SCZD neurons. By similar methods, a second group generated ONPs from control and SCZD patients and performed gene expression comparisons, which identified differences in neurodevelopmental pathways associated with cell migration and axon guidance 96.
ONPs are one source of live human neurons for the study of SCZD. Though differences between SCZD and control ONPs have been identified, it is unclear whether SCZD ONPs will recapitulate all of the neuronal defects present in brain regions such as the cortex or hippocampus. Additionally, ONPs cannot yet be used as a source of cells from the neural lineages specifically implicated in SCZD: glutamatergic neurons, GABAergic neurons, dopaminergic neurons, and oligodendrocytes.
Insights from hiPSC Neurons
Chiang et al first published the generation of hiPSCs from SCZD patients with a DISC1 mutation 11 but did not characterize neurons differentiated from these hiPSCs. We then demonstrated that SCZD hiPSC neurons had defects in neuronal connectivity and gene expression which could be ameliorated following treatment with the antipsychotic Loxapine 10. Specifically, we observed reduced neuronal connectivity, reduced outgrowths from soma, and reduced PSD95 dendritic protein levels, all of which are cellular phenotypes previously described in postmortem SCZD brain tissue as well as animal models of SCZD (Figure 4). Additionally, we observed gene expression differences in SCZD neurons relative to controls, 25% of which had been previously implicated in SCZD, with significant perturbations in genes associated with the WNT pathway, cAMP signaling and glutamate receptor expression. We hypothesize that studies of SCZD hiPSC neurons from an increased number of patients might identify core pathways of genes contributing to SCZD. Pedrosa et al have also generated SCZD hiPSCs from three patients and report that SCZD hiPSC neurons express a number of transcription factors, chromatin remodeling proteins and synaptic proteins relevant to SZCD pathogenesis, independently validating the potential utility of hiPSC neurons in modeling SCZD 97.
Figure 4.
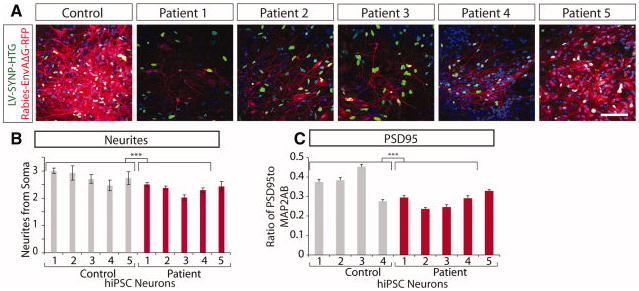
Decreased neuronal connectivity in SCZD hiPSC neurons. A. Representative images of reduced trans-neuronal labeling (red) in SCZD hiPSC neurons relative to controls. B. Graph showing decreased neurites in SCZD hiPSC neurons. C. Graph showing decreased PSD95 protein levels relative to the dendritic marker MAP2AB for control and SCZD hiPSC neurons. Adapted from Brennand et al (2011). Abbreviations: hiPSCs, human induced pluripotent stem cells; Microtubule-associated protein 2AB (MAP2AB); Postsynaptic density protein 95 (PSD95). *** p<0.001
While hiPSC-based studies show exciting promise for the study of SCZD, they remain limited by three types of variability: neuron-to-neuron, hiPSC-to-hiPSC, and patient-to-patient. Interneuron variability is countered by studying more homogeneous populations of hiPSC neurons, generated by directed differentiation protocols to specific neuronal subtypes and subsequent purification by Fluorescence-activated cell sorting (FACS). Inter-hiPSC variation is addressed by comparing multiple hiPSC lines per patient, particularly as it is well established that genetic and epigenetic differences exist between hiPSC lines. Finally, inter-patient variability can be tackled by studying an increased number of patients and controls, and particularly by selecting homogenous patient cohorts characterized by common clinical endophenotypes, pharmacological responses or genetic mutations. Using hiPSC neurons, researchers can now begin to dissect the molecular mechanism of pharmacological response and screen for new drugs to improve cellular phenotypes in SCZD neurons from patients with clear clinical pharmacological non-responsiveness.
Moving Forward: Future hiPSC studies of SCZD
Brain-imaging studies have identified structural changes in SCZD brains, but cannot resolve which neuronal cell types are affected in SCZD. Pharmacological studies have identified a role for DA and GLU; however, chronic antipsychotic treatment alters brain structure and neural activity, confounding studies of human patients affected with SCZD. GWAS studies have yet to account for most of the heritable variance of SCZD. Though animal models have recapitulated aspects of the behavioral and cellular phenotypes of SCZD, they lack the ability to define the complex interacting genetic factors that contribute to disease. hiPSC-based studies will complement brain-imaging, pathological, pharmacological, genetic and animal studies of SCZD.
It will be critical that researchers carefully consider which specific subtypes of neurons should be compared using hiPSC neurons studies. We propose that the field begin to characterize cell-autonomous defects in midbrain DA (mDA) and cortical glutamatergic (cGLU) and GABAergic (cGABA) SCZD hiPSC neurons.
An efficient protocol can now differentiate pluripotent stem cells to populations consisting of approximately 20% DA neurons 98 by recapitulating developmental cues found in the ventral midline when SHH, FGF8 and WNT1 initiate DA differentiation; immature mDA neurons express NURR1, EN1/2 and LMX1A/B, whereas mature mDA neurons also express tyrosine hydoxylase (TH) and aromatic L-amino acid decarboxylase (AADC) 99. Studies using mouse ESCs have shown that NODAL antagonists (LEFTYA) induce expression of the forebrain marker Brain-factor 1 (BF1, FOXG1) and subsequent treatment with WNT antagonists causes regional specification towards cortical fate 100. Genes such as EMX1, FEZF2, and FEZ, are expressed in immature cGLU neurons 101–103, while mature cGLU neurons express CTIP2 and OTX1. 101, 104, 105. Subsequent culture with FGF2 has been shown to increase GABAergic differentiation 106; GABA neurons can be identified by expression of key GABAergic markers, such as Glutamate decarboxylase (GAD65/67), DARPP32, ARPP21, CALBINDIN, or CALRETININ 107, although few if any regional markers of basal ganglia identity have been identified.
Future molecular studies of hiPSC neurons should incorporate SNP, CNV and gene expression data. Studies of quantitative trait loci (eQTLs) will determine how genetic lesions affect gene expression in SCZD neurons. Current eQTL studies can only compare a limited supply of heterogeneous post-mortem brain tissue confounded by variables such as patient treatment history, drug/alcohol abuse and poverty. Ideally, hiPSC-based eQTL studies would produce a renewable supply of more homogeneous cell populations. As hiPSC generation, neuronal differentiation and subtype purification are streamlined and made more efficient, it will become possible to generate any defined neuronal subtypes from hiPSCs generated from hundreds of patients with known genetic backgrounds.
Currently, laborious single cell electrophysiological analysis is the best method to establish the maturity and assess synaptic function of the SCZD hiPSC neurons. Electrophysiological characterization can verify that hiPSC neurons have membrane potentials, undergo induced action potentials and show evidence of spontaneous synaptic activity. However, in order to study functional synaptic activity defects contributing to SCZD, and detect significant effects in heterogeneous patient populations, it will be necessary to increase both the number of patients and neurons that can be studied, We believe that developing more efficient and higher throughput synaptic analyses needs to become a priority of the field.
Beyond neurons, hiPSCs can of course also be differentiated to any other cell type implicated in SCZD, particularly oligodendrocytes 108, 109. Given that consider data suggests that myelin dysfunction contributes to SCZD, comparisons of co-cultures of control and SCZD neurons and oligodendrocytes should reveal whether reduced myelination in SCZD neurons is a cell autonomous effect. Should studies of hiPSC-derived cells reveal aberrant oligodendrocyte activity in SCZD, this may indicate a new point of therapeutic intervention in SCZD.
Moving forward, hiPSC studies must make several critical advances. Future studies should focus on defined neuronal subtypes. More efficient hiPSC generation and neuronal differentiation will ultimately permit eQTL studies of SCZD. More scalable assays of synaptic function will allow characterization of increased numbers of control and patient neurons. New studies will need to recruit better-characterized patient populations with well-defined clinical endophenotypes, pharmacological responses and/or genetic lesions.
Conclusion
It is now time to begin to synthesize the disparate fields of brain imaging, neurobiology and genetics. By generating hiPSC neurons from more and better-characterized patient cohorts, one can now test whether the severity of clinical outcome is predictive of the magnitude of cellular phenotype, if genetic lesions correlate to neuronal gene expression differences or if clinical pharmacological response is predictable by hiPSC neuronal drug response. Beyond correlating genetic lesions to the disease state, we can assay the expression or activity of genes identified through GWAS, both in the specific patient in whom the lesion was identified and across cohorts of SCZD patients. By carefully selecting from patients with well characterized diagnosis, MRI brain scans, genotyping data and clinical treatment history, hiPSCs can be generated specifically from SCZD patients with extreme endophenotypes, for example, patients showing drastically altered brain volumes by MRI or clear treatment resistance. Neurons derived from these patients will allow testing of the genetic causes of each endophenotype. With hiPSCs, we can finally move beyond phenotyping of SCZD patients and begin to develop well-controlled experiments to test the specific molecular and cellular effects of disease and pharmacological treatment and response. It is time to begin correlating clinical, genetic and pharmacological studies in vitro.
Acknowledgments
K.J.B. is supported by a training grant from the California Institute for Regenerative Medicine. The Gage Laboratory is partially funded by CIRM Grant RL1-00649-1, The Lookout and Mathers Foundation, the Helmsley Foundation as well as Sanofi-Aventis.
Footnotes
Author Information
The authors have declared that no competing interests exist.
References
- 1.Association AP. Diagnostic and statistical manual of mental disorders: DSM-IV. Washington, D.C: American Psychiatric Press; 1994. p. 886. [Google Scholar]
- 2.Carpenter WT, Jr, Buchanan RW. Schizophrenia. The New England journal of medicine. 1994;330:681–690. doi: 10.1056/NEJM199403103301006. [DOI] [PubMed] [Google Scholar]
- 3.Hannerz H, Borga P, Borritz M. Life expectancies for individuals with psychiatric diagnoses. Public Health. 2001;115:328–337. doi: 10.1038/sj.ph.1900785. [DOI] [PubMed] [Google Scholar]
- 4.Regier DA, Farmer ME, Rae DS, et al. Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. Jama. 1990;264:2511–2518. [PubMed] [Google Scholar]
- 5.Brown S, Inskip H, Barraclough B. Causes of the excess mortality of schizophrenia. Br J Psychiatry. 2000;177:212–217. doi: 10.1192/bjp.177.3.212. [DOI] [PubMed] [Google Scholar]
- 6.Radomsky ED, Haas GL, Mann JJ, et al. Suicidal behavior in patients with schizophrenia and other psychotic disorders. Am J Psychiatry. 1999;156:1590–1595. doi: 10.1176/ajp.156.10.1590. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell CB, Gottesman Schizophrenia--a high-risk factor for suicide: clues to risk reduction. Suicide Life Threat Behav. 1992;22:479–493. [PubMed] [Google Scholar]
- 8.Wu EQ, Birnbaum HG, Shi L, et al. The economic burden of schizophrenia in the United States in 2002. J Clin Psychiatry. 2005;66:1122–1129. doi: 10.4088/jcp.v66n0906. [DOI] [PubMed] [Google Scholar]
- 9.Murray C, Lopez A. The Global Burden of Disease. Cambridge, MA: Harvard School of Public Health; 1996. [Google Scholar]
- 10.Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang CH, Su Y, Wen Z, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Molecular psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange N, Giedd JN, Castellanos FX, et al. Variability of human brain structure size: ages 4–20 years. Psychiatry Res. 1997;74:1–12. doi: 10.1016/s0925-4927(96)03054-5. [DOI] [PubMed] [Google Scholar]
- 13.Giedd JN, Rapoport JL. Structural MRI of pediatric brain development: what have we learned and where are we going? Neuron. 2010;67:728–734. doi: 10.1016/j.neuron.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vita A, De Peri L, Silenzi C, et al. Brain morphology in first-episode schizophrenia: a meta-analysis of quantitative magnetic resonance imaging studies. Schizophr Res. 2006;82:75–88. doi: 10.1016/j.schres.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Steen RG, Mull C, McClure R, et al. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Br J Psychiatry. 2006;188:510–518. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- 16.Wright IC, Rabe-Hesketh S, Woodruff PW, et al. Meta-analysis of regional brain volumes in schizophrenia. Am J Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- 17.Thompson PM, Vidal C, Giedd JN, et al. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison-Wright I, Glahn DC, Laird AR, et al. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am J Psychiatry. 2008;165:1015–1023. doi: 10.1176/appi.ajp.2008.07101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulshoff Pol HE, Kahn RS. What happens after the first episode? A review of progressive brain changes in chronically ill patients with schizophrenia. Schizophr Bull. 2008;34:354–366. doi: 10.1093/schbul/sbm168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields RD, Stevens-Graham B. New insights into neuron-glia communication. Science (New York, N Y. 2002;298:556–562. doi: 10.1126/science.298.5593.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubicki M, McCarley R, Westin CF, et al. A review of diffusion tensor imaging studies in schizophrenia. J Psychiatr Res. 2007;41:15–30. doi: 10.1016/j.jpsychires.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wexler BE, Bell MD. Cognitive remediation and vocational rehabilitation for schizophrenia. Schizophr Bull. 2005;31:931–941. doi: 10.1093/schbul/sbi038. [DOI] [PubMed] [Google Scholar]
- 23.Whitfield-Gabrieli S, Thermenos HW, Milanovic S, et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 25.Kessler RM, Woodward ND, Riccardi P, et al. Dopamine D2 receptor levels in striatum, thalamus, substantia nigra, limbic regions, and cortex in schizophrenic subjects. Biol Psychiatry. 2009;65:1024–1031. doi: 10.1016/j.biopsych.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane J, Honigfeld G, Singer J, et al. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Arch Gen Psychiatry. 1988;45:789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- 27.Klein C, Gordon J, Pollak L, et al. Clozapine in Parkinson's disease psychosis: 5–year follow-up review. Clin Neuropharmacol. 2003;26:8–11. doi: 10.1097/00002826-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Nord M, Farde L. Antipsychotic occupancy of dopamine receptors in schizophrenia. CNS Neurosci Ther. 2011;17:97–103. doi: 10.1111/j.1755-5949.2010.00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray L, van den Buuse M, Scarr E, et al. Clozapine reverses schizophrenia-related behaviours in the metabotropic glutamate receptor 5 knockout mouse: association with N-methyl-D-aspartic acid receptor up-regulation. Int J Neuropsychopharmacol. 2009;12:45–60. doi: 10.1017/S1461145708009085. [DOI] [PubMed] [Google Scholar]
- 30.Michel FJ, Trudeau LE. Clozapine inhibits synaptic transmission at GABAergic synapses established by ventral tegmental area neurones in culture. Neuropharmacology. 2000;39:1536–1543. doi: 10.1016/s0028-3908(99)00239-7. [DOI] [PubMed] [Google Scholar]
- 31.Krystal JH, Karper LP, Seibyl JP, et al. Subanesthetic effects of the noncompetitive NMDA antagonist ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry. 1994;51:199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- 32.Patil ST, Zhang L, Martenyi F, et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: a randomized Phase 2 clinical trial. Nature medicine. 2007;13:1102–1107. doi: 10.1038/nm1632. [DOI] [PubMed] [Google Scholar]
- 33.Selemon LD, Goldman-Rakic PS. The reduced neuropil hypothesis: a circuit based model of schizophrenia. Biol Psychiatry. 1999;45:17–25. doi: 10.1016/s0006-3223(98)00281-9. [DOI] [PubMed] [Google Scholar]
- 34.Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224. doi: 10.1001/archpsyc.55.3.215. [DOI] [PubMed] [Google Scholar]
- 35.Black JE, Kodish IM, Grossman AW, et al. Pathology of layer V pyramidal neurons in the prefrontal cortex of patients with schizophrenia. Am J Psychiatry. 2004;161:742–744. doi: 10.1176/appi.ajp.161.4.742. [DOI] [PubMed] [Google Scholar]
- 36.Garey LJ, Ong WY, Patel TS, et al. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry. 1998;65:446–453. doi: 10.1136/jnnp.65.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 38.Meador-Woodruff JH, Healy DJ. Glutamate receptor expression in schizophrenic brain. Brain Res Brain Res Rev. 2000;31:288–294. doi: 10.1016/s0165-0173(99)00044-2. [DOI] [PubMed] [Google Scholar]
- 39.Cardno AG, Gottesman Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 40.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 41.Ekelund J, Hovatta I, Parker A, et al. Chromosome 1 loci in Finnish schizophrenia families. Human molecular genetics. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 42.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. American journal of human genetics. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JW, Lee YS, Cho EY, et al. Linkage and association of schizophrenia with genetic variations in the locus of neuregulin 1 in Korean population. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:281–286. doi: 10.1002/ajmg.b.30209. [DOI] [PubMed] [Google Scholar]
- 44.Corvin AP, Morris DW, McGhee K, et al. Confirmation and refinement of an 'at-risk' haplotype for schizophrenia suggests the EST cluster, Hs.97362, as a potential susceptibility gene at the Neuregulin-1 locus. Molecular psychiatry. 2004;9:208–213. doi: 10.1038/sj.mp.4001412. [DOI] [PubMed] [Google Scholar]
- 45.Li T, Stefansson H, Gudfinnsson E, et al. Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Molecular psychiatry. 2004;9:698–704. doi: 10.1038/sj.mp.4001485. [DOI] [PubMed] [Google Scholar]
- 46.Liu CM, Hwu HG, Fann CS, et al. Linkage evidence of schizophrenia to loci near neuregulin 1 gene on chromosome 8p21 in Taiwanese families. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:79–83. doi: 10.1002/ajmg.b.20161. [DOI] [PubMed] [Google Scholar]
- 47.Stefansson H, Sarginson J, Kong A, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. American journal of human genetics. 2003;72:83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang JX, Chen WY, He G, et al. Polymorphisms within 5' end of the Neuregulin 1 gene are genetically associated with schizophrenia in the Chinese population. Molecular psychiatry. 2004;9:11–12. doi: 10.1038/sj.mp.4001436. [DOI] [PubMed] [Google Scholar]
- 49.Bakker SC, Hoogendoorn ML, Selten JP, et al. Neuregulin 1: genetic support for schizophrenia subtypes. Molecular psychiatry. 2004;9:1061–1063. doi: 10.1038/sj.mp.4001564. [DOI] [PubMed] [Google Scholar]
- 50.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto R, Straub RE, Weickert CS, et al. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Molecular psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 52.Chong VZ, Thompson M, Beltaifa S, et al. Elevated neuregulin-1 and ErbB4 protein in the prefrontal cortex of schizophrenic patients. Schizophr Res. 2008;100:270–280. doi: 10.1016/j.schres.2007.12.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hakak Y, Walker JR, Li C, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silberberg G, Darvasi A, Pinkas-Kramarski R, et al. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 55.Sugai T, Kawamura M, Iritani S, et al. Prefrontal abnormality of schizophrenia revealed by DNA microarray: impact on glial and neurotrophic gene expression. Ann N Y Acad Sci. 2004;1025:84–91. doi: 10.1196/annals.1316.011. [DOI] [PubMed] [Google Scholar]
- 56.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huh GS, Boulanger LM, Du H, et al. Functional requirement for class I MHC in CNS development and plasticity. Science (New York, N Y. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulanger LM. MHC class I in activity-dependent structural and functional plasticity. Neuron Glia Biol. 2004;1:283–289. doi: 10.1017/S1740925X05000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nature genetics. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 63.Gulacsi AA, Anderson SA. Beta-catenin-mediated Wnt signaling regulates neurogenesis in the ventral telencephalon. Nat Neurosci. 2008;11:1383–1391. doi: 10.1038/nn.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang FL, Huang KP, Boucheron C. Long-term enrichment enhances the cognitive behavior of the aging neurogranin null mice without affecting their hippocampal LTP. Learn Mem. 2007;14:512–519. doi: 10.1101/lm.636107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esslinger C, Walter H, Kirsch P, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science (New York, N Y. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 66.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science (New York, N Y. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 67.Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Human molecular genetics. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 68.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. The New England journal of medicine. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang F, Gu W, Hurles ME, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W, Zhou Y, Jentsch JD, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pletnikov MV, Ayhan Y, Nikolskaia O, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Molecular psychiatry. 2008;13:173–186. 115. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- 75.Kvajo M, McKellar H, Arguello PA, et al. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamiya A, Kubo K, Tomoda T, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 77.Duan X, Chang JH, Ge S, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sasaki S, Mori D, Toyo-oka K, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Molecular and cellular biology. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shu T, Ayala R, Nguyen MD, et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 80.Millar JK, Pickard BS, Mackie S, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science (New York, N Y. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 81.Morris JA, Kandpal G, Ma L, et al. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Human molecular genetics. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 82.Brandon NJ, Handford EJ, Schurov I, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Molecular and cellular neurosciences. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 83.Ozeki Y, Tomoda T, Kleiderlein J, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li B, Woo RS, Mei L, et al. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barros CS, Calabrese B, Chamero P, et al. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4507–4512. doi: 10.1073/pnas.0900355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopez-Bendito G, Cautinat A, Sanchez JA, et al. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krivosheya D, Tapia L, Levinson JN, et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem. 2008;283:32944–32956. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pitcher GM, Beggs S, Woo RS, et al. ErbB4 is a suppressor of long-term potentiation in the adult hippocampus. Neuroreport. 2008;19:139–143. doi: 10.1097/WNR.0b013e3282f3da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen YJ, Zhang M, Yin DM, et al. ErbB4 in parvalbumin-positive interneurons is critical for neuregulin 1 regulation of long-term potentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21818–21823. doi: 10.1073/pnas.1010669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wen L, Lu YS, Zhu XH, et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1211–1216. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ting AK, Chen Y, Wen L, et al. Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. J Neurosci. 2011;31:15–25. doi: 10.1523/JNEUROSCI.2538-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fenelon K, Mukai J, Xu B, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Earls LR, Bayazitov IT, Fricke RG, et al. Dysregulation of presynaptic calcium and synaptic plasticity in a mouse model of 22q11 deletion syndrome. J Neurosci. 2010;30:15843–15855. doi: 10.1523/JNEUROSCI.1425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sigurdsson T, Stark KL, Karayiorgou M, et al. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benitez-King G, Riquelme A, Ortiz-Lopez L, et al. A non-invasive method to isolate the neuronal linage from the nasal epithelium from schizophrenic and bipolar diseases. J Neurosci Methods. 2011 doi: 10.1016/j.jneumeth.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 96.Matigian N, Abrahamsen G, Sutharsan R, et al. Disease-specific, neurosphere-derived cells as models for brain disorders. Dis Model Mech. 2010;3:785–798. doi: 10.1242/dmm.005447. [DOI] [PubMed] [Google Scholar]
- 97.Pedrosa E, Sandler V, Shah A, et al. Development of Patient-Specific Neurons in Schizophrenia Using Induced Pluripotent Stem Cells. J Neurogenet. 2011 doi: 10.3109/01677063.2011.597908. [DOI] [PubMed] [Google Scholar]
- 98.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development (Cambridge, England) 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- 100.Watanabe K, Kamiya D, Nishiyama A, et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 101.Arlotta P, Molyneaux BJ, Chen J, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 102.Molyneaux BJ, Arlotta P, Hirata T, et al. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 103.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frantz GD, Weimann JM, Levin ME, et al. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weimann JM, Zhang YA, Levin ME, et al. Cortical neurons require Otx1 for the refinement of exuberant axonal projections to subcortical targets. Neuron. 1999;24:819–831. doi: 10.1016/s0896-6273(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 106.Barberi T, Klivenyi P, Calingasan NY, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nature biotechnology. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 107.Aubry L, Bugi A, Lefort N, et al. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keirstead HS, Nistor G, Bernal G, et al. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25:4694–4705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu BY, Du ZW, Zhang SC. Differentiation of human oligodendrocytes from pluripotent stem cells. Nature protocols. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]


