Summary
Embryonal rhabdomyosarcoma (ERMS) is an aggressive pediatric sarcoma of muscle. Here, we show that ERMS-propagating potential is confined to myf5+ cells and can be visualized in live, fluorescent transgenic zebrafish. During early tumor growth, myf5+ ERMS cells reside adjacent normal muscle fibers. By late stage ERMS, myf5+ cells are reorganized into distinct regions separated from differentiated tumor cells. Time-lapse imaging of late stage ERMS revealed that myf5+ cells populate newly formed tumor only after seeding by highly migratory myogenin+ ERMS cells. Moreover, myogenin+ ERMS cells can enter the vasculature, whereas myf5+ ERMS-propagating cells do not. Our data suggests that non-tumor propagating cells likely have important supportive roles in cancer progression and facilitate metastasis.
Introduction
Rhabdomyosarcoma (RMS) is a pediatric malignancy that shares common features with skeletal muscle arrested in embryonic development (Xia et al., 2002). The two main subtypes of pediatric rhabdomyosarcoma, embryonal RMS (ERMS) and alveolar RMS (ARMS), differ in their clinical, biological, and molecular characteristics. For example, ERMS and ARMS can be distinguished based on histology and have different long-term prognosis with ERMS patients having better overall outcome than ARMS. These divergent clinical features likely reflect the use of different molecular programs that lead to transformation. For example, we have identified that the RAS pathway is active in a majority of human ERMS (Hettmer et al., 2011; Langenau et al., 2007). By contrast, 85% of ARMS have recurrent chromosomal translocations that juxtapose PAX3 or PAX7 with the forkhead transcription factor (FKHR) (Xia et al., 2002). Finally, it is likely that ERMS and translocation-positive ARMS arise in different cell types that eventually undergo transformation. Keller et al. found that PAX3-FKHR+ ARMS can arise from Myf6 expressing myoblast cells but not dermamyotome or satellite cells that express Pax7 (Keller et al., 2004). By contrast, ERMS can arise from either satellite cells or myoblasts that eventually reinitiate molecular programs found in satellite cells (Rubin et al., 2011). Despite elegant studies defining possible cells of origin in RMS, identification of an ERMS-propagating cell that is required for continued tumor growth in vivo has not been described in mice or humans.
Tumor-propagating cells have been characterized in many malignances, and in some tumors, this potential is confined to a molecularly definable cell population that can be enriched by cell surface markers. For example, in AML a rare CD34+CD38− cell enriches for leukemia-propagating potential while in breast cancer CD44+CD24low/- expression is associated with tumor-propagating potential (reviewed in Dalerba et al., 2007). Molecularly defined, rare CD133+ tumor-propagating cells have also been identified in subset of gliomas and exhibit striking differences in response to nitric oxide and hypoxia inducible factor (HIF) signaling when compared to more differentiated tumors cells (Eyler et al., 2011; Li et al., 2009). Thus, it is likely that many tumors contain hierarchically organized cell subpopulations that retain the capacity to remake tumor and yet give rise to differentiated tumor cell progeny. One might expect that selection would favor the evolution of tumors with high numbers of tumor-propagating cells at a cost of differentiated cell types. Yet paradoxically, in most malignancies, tumor-propagating cells are far less abundant than differentiated tumor cells that are incapable of remaking tumor. These data suggest that differentiated tumor cells may provide important supportive roles in overall growth and maintenance. To date, a role for differentiated, non-tumor-propagating ERMS cells has yet to be fully explored.
Stem cells often reside in distinct niches in normal tissue and their functions are exquisitely controlled by local factors secreted by supporting cells. For example, hematopoietic stem cells (HSCs) have been shown to home to niches within the calvarium that are tightly associated with osteoblasts (Lo Celso et al., 2009). These and other niche-associated cells presumably provide paracrine-signaling factors to recruit and maintain these cells in a specific niche. Unlike other tissues, the muscle stem cell niche is defined by juxtaposition of satellite cells next to differentiated muscle fibers, and their numbers and differentiation capacity are controlled by complex signaling pathways regulated by mature muscle cells (reviewed in Bentzinger et al., 2012). Despite a large body of data defining stem cell niches in normal tissue, few studies have identified tumor-specific niches and/or regions of compartmentalized tumor cell function and less have used microscopic imaging to directly visualize tumor-propagating cells within live animals. In one example, Sipkins et al used a combination of multiphoton and confocal microscopy to image the HSC niche in the calvarium of mice and demonstrated that these sites can attract multiple tumor cell types (Sipkins et al., 2005); however, it is unknown if these malignant cells are capable of reinitiating tumors. In ERMS, as with most solid tumors, it is unknown if tumor-propagating cells reside in distinct regions within the tumor mass and if the more differentiated cells play a role in promoting tumor progression.
Here, we utilize a transgenic zebrafish model of embryonal rhabdomyosarcoma to identify the tumor-propagating cell in this disease and to define the functional consequences of tumor cell heterogeneity within live animals. Because ERMS cell subpopulations can be fluorescent-labeled based on myogenic factor expression, ERMS cell subtypes can be visualized in live animals and the processes of cell growth, division, and local dissemination can be visualized as dynamic processes in live animals. Our data provide an explanation for the large number of non-tumor propagating cells in established cancers and reveals an important supportive role for differentiated tumor cell types in local dissemination and metastasis.
Results
Imaging distinct stages of ERMS growth
Externally-visible ERMS can develop as early as 10 days of life in zebrafish injected with rag2-KRASG12D (Langenau et al., 2007) and >80% of ERMS develop in the tail musculature (n>50). To assess how tumors initiate and evolve in zebrafish ERMS, alpha-actin-GFP transgenic zebrafish were injected at the one-cell stage of development with rag2-dsREDexpress and rag2-KRASG12D (Figure 1A-F), facilitating imaging of ERMS cells in relation to normal muscle. Microinjection of multiple transgenes into one-cell stage animals leads to co-integration and co-expression in animals that develop ERMS (Langenau et al., 2008). This approach provides a robust method to create mosaic transgenic animals with fluorescently-labeled ERMS cell subpopulations (Langenau et al., 2007).
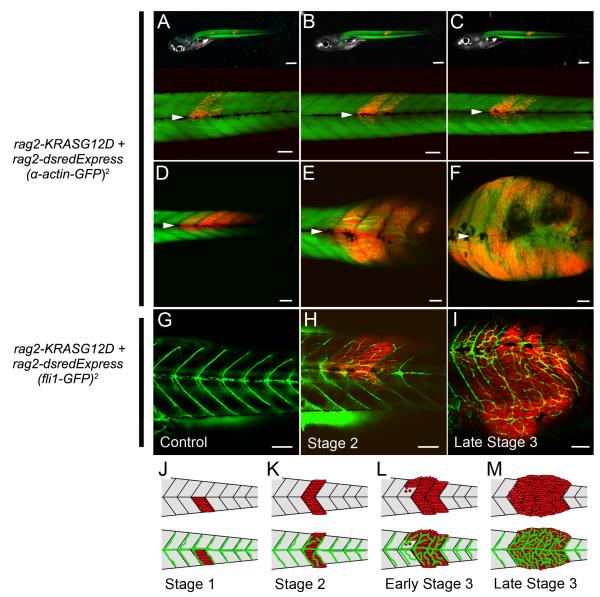
Figure 1. Visualizing distinct stages of embryonal rhabdomyosarcoma growth.
(A-F) rag2-dsRED-labeled ERMS arising in alpha-actin-GFP transgenic zebrafish. (A-C) The same animal imaged at 6, 9, and 12 days post-fertilization (dpf). (D-F) A representative zebrafish where dsRED+ ERMS cells have already bypassed the horizontal myoseptum and migrated into new segments that were previously free of tumor (F, stage 3) at 13, 18, and 24 dpf (D-F, respectively). The horizontal myoseptum is denoted by white arrows in A-F.
(G) fli1-GFP transgenic control animal compared with a rag2-dsRED-labeled ERMS arising in fli1-GFP transgenic zebrafish at early stage 2 (H) or a late stage 3 (I).
(J-M) Schematic of stages of ERMS growth.
Scale bar A-C upper panel 500 μm, A-C lower panel and D-I 100 μm.
See also Movie S1.
Sequential confocal imaging over several days showed that ERMS forms in a choreographed and stereotypical manner (Figure 1A-F, J-M). Specifically, dsRed+ ERMS mononuclear cells arise at the extreme outer borders of the myotome segments and move toward the midline where they are initially unable to bypass the horizontal myosepta - a single cell layer that separates myotome segments (Figure 1A, Stage 1, n=7). After several days, a subset of cells cross the horizontal myosepta and take up residence between normal muscle fibers within the newly colonized myotome segment (stage 2, Figure 1B-C, n=5). Differentiated ERMS cells that express both rag2-dsREDexpress and alpha-actin-GFP can move laterally into neighboring muscle segments by transiting through the collagen matrix of the myoseptum (n=6, Figure 1D-F, Movie S1) or stream into new myotome segments by growing past the edge of myoseptum (n=3, early Stage 3). The collagen matrix of the muscle myoseptum is a cell-impermeable barrier that is the site of muscle attachment in teleost fish and is similar in function to tendons in mouse and humans. Late stage ERMS undergo rapid loss of fibers, breakdown of normal muscle architecture including collagen remodeling, and development of mononuclear tumor cells, reminiscent of the spindle variant of human embryonal rhabdomyosarcoma.
Neovascularization is a hallmark of cancer and an ideal surrogate for assessing tumorigenicity. To assess when KRASG12D-expressing cells are transformed, neovascularization was monitored in fli1-GFP transgenic animals that were injected at the one-cell stage of life with rag2-KRASG12D and rag2-dsREDexpress. Animals were monitored for tumor growth by confocal microscopy beginning at 10 days of life (n=22, Figure 1G-I). ERMS at stage 1 failed to recruit new vasculature (n=0 of 3), but stage 2 and early stage 3 ERMS had begun to recruit new vasculature (n=8 of 8, Figure 1H) with new branches arising from both the intersegmental vessels and vertebral artery. By late stage 3, ERMS developed intricate networks of new vessels (n=11 of 11, Figure 1I). Our imaging studies define distinct stages of ERMS growth and suggest that RAS-expressing cells become fully transformed by stage 2 of tumor development (Figure 1J-M).
Identification of molecularly distinct fluorescent-labeled ERMS cell subpopulations
Previous experiments in zebrafish have identified an ERMS cell subpopulation that had superior tumor-propagating potential when compared to other tumor derived cells. This ERMS-propagating cell was rag2-dsREDexpress+/alpha-actin-negative and expressed high levels of myf5, c-Met, and m-cadherin – markers of satellite and early muscle progenitor cells (Langenau et al., 2007). MYF5 is highly upregulated in human ERMS compared to both translocation positive ARMS and normal muscle (Zibat et al., 2010) and in comparing zebrafish ERMS to normal muscle (Langenau et al., 2007). To directly assess whether myf5 labels distinct ERMS cell subpopulations, myf5-GFP/ myosin light chain 2 -mCherry (mylz2) syngeneic animals were created by four rounds of out crossing to CG1 syngeneic animals (Smith et al., 2010) and injected at one-cell stage with rag2-KRASG12D. myf5-GFP transgenic animals exhibit GFP expression in early somitogenesis and later in satellite cells and early muscle progenitor cells (Chen et al., 2007; Seger et al., 2011) while the mylz2 promoter drives expression in differentiated muscle cells (Ju et al., 2003; Smith et al., 2010). Fluorescent-labeled ERMS cell subpopulations were isolated from double transgenic animals by fluorescence-activated cell sorting (FACS). Reanalysis of sorted cells by FACS confirmed that ERMS contained four distinct populations of cells (purity >87%, and viability >97%).
To verify that discrete fluorescent-labeled ERMS cell subpopulations were molecularly distinct, sorted cell populations were assessed for gene expression differences based on microarray (Figure 2A, Table S1) and real-time PCR (Figure 2B, Figure S1). Gene expression analysis was completed on FACS sorted ERMS cells derived from serially passaged tumors, ensuring that fluorescent-labeled cells were tumor derived. Microarray analysis confirmed that each cell subpopulation exhibited wide differences in gene expression. Subsequent real-time PCR analysis established that the myf5-GFP+/mylz2-negative cells expressed high levels of myf5, cMet, and m-cadherin but not pax7a, pax7b, or differentiated markers (Figure 2B and Figure S1). By contrast, mylz2-mCherry+ ERMS cells expressed high levels of mature muscle markers including myoD, myogenin, troponin I fast-twitch isoform 2 (tnni2a), alpha-actin 1b (acta1b), ventricular myosin heavy chain-like (vmhcl), actin related protein 2/3 complex subunit 5B (arpc5b), carboxypeptidase vitellogenic-like (cpvl), and chemokine (C-X-C motif) receptor 4b (cxcr4b). Finally, the double-negative cell population was comprised of predominantly blood cells that express myeloid-specific peroxidase (mpx) and lymphocyte-specific protein tyrosine kinase (lck). Our data confirms that fluorescent-labeled ERMS cell subpopulations can be prospectively isolated to relative purity following FACS and are molecularly distinct.
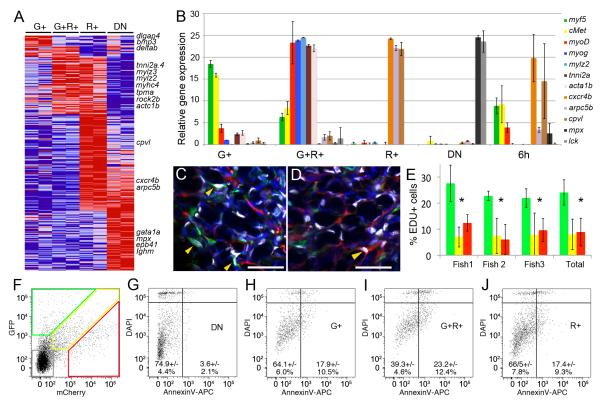
Figure 2. Fluorescent transgenic approaches identify discrete and molecularly definable ERMS cell subpopulations in myf5-GFP/mylz2-mCherry transgenic fish.
(A) Heat map showing differential gene expression between FACS sorted ERMS cell subpopulations isolated from serially-passaged myf5-GFP/ mylz2-mCherry ERMS (microarray log fold-change >1.5). myf5-GFP+/mylz2-mCherry-negative (G+), myf5-GFP+/mylz2-mCherry+ (G+R+), myf5-GFP-negative/mylz2-mCherry+ (R+), and double negative (DN).
(B) Quantitative real-time PCR of sorted ERMS cell subpopulations. Expression values +/− 1 STD.
(C-D) Confocal images of EDU stained sections from serially passaged myf5-GFP+/mylz2-mCherry+ ERMS. Tumor regions with large numbers of either myf5-GFP+ (C) or mylz2-mCherry+ ERMS cells (D). Blue denotes DAPI+ nuclei and white labels EDU+ nuclei. Yellow arrows indicate EDU-labeled cells. Scale bar is 25 μm.
(E) Quantification of EDU-incorporation over a 6 hour EDU pulse. Data for myf5-GFP+/mylz2-mCherry-negative ERMS cells denoted by green bars, myf5-GFP+/mylz2-mCherry+ by yellow, and myf5-GFP-negative/mylz2-mCherry+ cells by red. Three individual tumors shown as well as cumulative data across all tumors (Total). Asterisk denotes p<0.00001 and error bars +/−1 STD.
(F) FACS plot of serially passaged myf5-GFP/mylz2-mCherry ERMS.
(G-J) Gated ERMS cells assessed for DAPI and AnnexinV-APC staining (double negative (DN), G; myf5-GFP+/mylz2-mCherry-negative (G+), H; myf5-GFP+/mylz2-mCherry+ (G+R+), I; myf5-GFP-negative/mylz2-mCherry+ (R+), J). Live cells are shown the DAPI-negative/AnnexinV-negative gates.
Given that FACS could identify unique ERMS cell subpopulations that exhibited wide differences in gene expression, we questioned if these cells also differ in rates of proliferation and cellular turnover. Proliferation was assessed at 6 hours following intraperitoneal injection of EDU into ERMS-affected animals (Figure 2C-E and Figure S1). 24.1 +/− 4.8% myf5-GFP+/mylz2-mCherry-negative cells incorporated EDU over a 6-hour pulse, whereas differentiated ERMS cells that express mylz2-mCherry were far less proliferative (8.9 +/− 5.0%, p<0.00001). By contrast, following 3 day administration of EDU, all fluorescent-labeled ERMS cell sub-fractions exhibited equal proliferative capacity, suggesting that myf5-GFP+/mylz2-mCherry-negative cells divided and differentiated over this time (data not shown). In addition to striking differences in cell proliferation between ERMS cell subpopulations, myf5-GFP+/mylz2-mCherry+ cells had higher levels of apoptotic cellular turnover when compared with myf5-GFP+/mylz2-mCherry-negative and myf5-GFP-negative/mylz2-mCherry+ cells (p<0.01 Fisher exact test, Figure 2F-J). Taken together, our fluorescent transgenic approach identifies unique ERMS cell subpopulations that have different fluorescent reporter expression, divergent gene expression profiles, and varied capacities for proliferation and apoptosis.
myf5-GFP+ cells are the ERMS-propagating cell population
To assess if myf5-GFP transgene expression enriches for ERMS-propagating potential, cells were isolated from transplant animals that developed myf5-GFP+/mylz2-mCherry+ ERMS (Figure 3A-G) and subjected to two rounds of FACS in the presence of propidium iodide or DAPI to isolate highly purified and viable cells (Figure 3H-K, 87.7%-99.7% purity and >98% viability). ERMS cell subpopulations were introduced into CG1 syngeneic recipient animals at limiting dilution (Figure 3L-R), and animals assessed for engraftment from 10 to 120 days post-transplantation (Table 1). All animals developed ERMS before 45 days post-transplantation, confirming that slower cycling ERMS-propagating cell types would not be missed in our analysis. In three ERMS tested, the tumor-propagating activity was confined to the myf5-GFP+/mylz2-mCherry- negative cell subpopulation (Table 1) with an average frequency of 1 in 146 cells capable of reinitiating tumors in recipient animals (range 1 in 87-245, 95% confidence interval). By contrast, only 1 in 4,206 myf5-GFP+/mylz2-mcherry+ cells were capable of inducing tumors (range 1 in 1,550 to 11,409, p=3.38e-15 when compared to ERMS-propagating activity in myf5-GFP+/mylz2-mcherry-negative cells). 0 of 61 animals engrafted disease from terminally differentiated myf5-GFP-negative/mylz2-Cherry+ cells (lower bound for ERMS-propagating potential was 1 in >5,969 cells). In total, we observe a remarkable 28 to >40 fold enrichment of tumor-propagating potential within our myf5-GFP+/mylz2-mCherry-negative cell type when compared with other sorted ERMS cell subpopulations. Similar results were also observed in primary ERMS. Specifically, three primary ERMS were isolated from 20 to 30-day-old larval zebrafish, pooled, and fluorescent-labeled ERMS cell subpopulations isolated by FACS. 3 of 8 animals engrafted disease from 1×102 myf5-GFP+/mylz2-mCherry-negative cells whereas the remaining ERMS cell subpopulations could not transfer disease at this cell dose (0 of 23, purity 83-98% and viability >98.6%, p=0.012 Fisher Exact Test). These results further support our finding that the myf5-GFP+/mylz2-mCherry-negative population is highly enriched for ERMS-propagating activity.
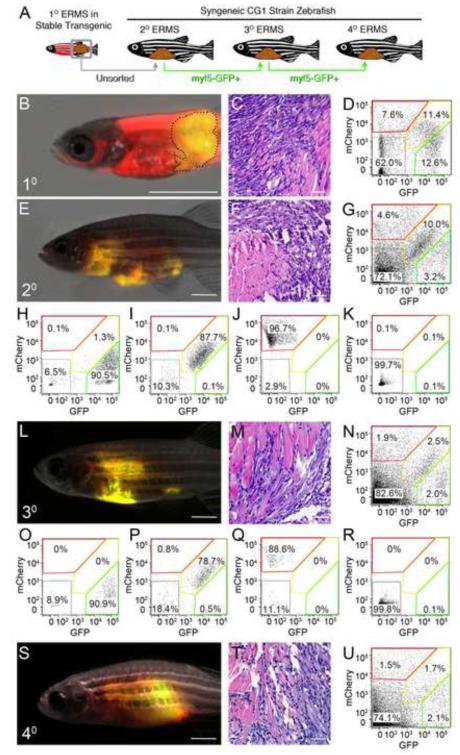
Figure 3. ERMS-propagating cells express myf5-GFP but not the mylz2-mCherry differentiated muscle marker.
(A) Schematic of experimental design.
(B-D) A primary ERMS arising in syngeneic myf5-GFP/mylz2-mCherry transgenic zebrafish (35 dpf). Broken black line denotes tumor area.
(E-G) Fluorescent-labeled ERMS engraft into syngeneic secondary recipient animals when transplanted with unsorted primary ERMS cells.
(H-K) FACS plots of fluorescent-labeled ERMS cells isolated from secondary recipient fish following two rounds of FACS.
(L-R) Transplantation of myf5-GFP+/mylz2-mCherry-negative FACs sorted cells induced ERMS in tertiary transplant animals and (S-U) quaternary recipients. Hematoxylin and eosin stained sections (C,F,M,T) and FACS (D,G,N,U) of primary and serially passaged ERMS. Scale bars equal 2 mm (B, E, L and S) and 100 μm (C, F, M and T).
Table 1. Limiting dilution cell transplantation identifies that myf5-GFP+/mylz2- mCherry-negative cells are the ERMS-propagating cells.
| ERMS #1-2° Transplants | 3° Transplants | ||||
| Cell# | G+ | G+R+ | R+ | Neg | G+ |
| 1000 | 6 of 6 | 2 of 7 | 0 of 6 | 0 of 7 | 6 of 6 |
| 10 | 5 of 9 | 0 of 9 | 0 of 8 | 0 of 10 | 7 of 8 |
| 10 | 0 of 8 | 0 of 8 | 0 of 9 | 0 of 7 | 0 of 10 |
| TPC # | 1 in 140** | 1 in 3461 | NA | NA | 1 in 67 |
| 95% CI | 59-329 | 872-13740 | NA | NA | 31-143 |
| ERMS #2-2° Transplants | |||||
| Cell# | G+ | G+R+ | R+ | Neg | |
| 1000 | 6 of 6 | 0 of 6 | 0 of 6 | 0 of 6 | |
| 10 | 4 of 7 | 2 of 10 | 0 of 10 | 0 of 10 | |
| 10 | 1 of 8 | 0 of 9 | 0 of 10 | 0 of 8 | |
| TPC # | 1 in 109** | 1 in 3495 | NA | NA | |
| 95% CI | 44-270 | 808-15120 | NA | NA | |
| ERMS #3-2° Transplants | |||||
| Cell# | G+ | G+R+ | R+ | Neg | |
| 1000 | 2 of 3 | 0 of 2 | 0 of 3 | 0 of 4 | |
| 10 | 8 of 9 | 0 of 8 | 0 of 8 | 1 of 8 | |
| 10 | 1 of 8 | 0 of 9 | 0 of 9 | 0 of 9 | |
| TPC # | 1 in 159** | NA | NA | 1 in 4840 | |
| 95% CI | 63-401 | NA | NA | 632-37094 | |
p<0.00001
Tumor-propagating cell number (TPC#) and 95% confidence interval (95% CI). Asterisks denote significant differences in TPC# between myf5-GFP+/mylz2-mCherry-negative and double positive ERMS cells (** p<0.00001).
To assess the long-term tumor-propagating potential of the myf5-GFP+/mylz2-mCherry-negative ERMS cells, cells were re-isolated from transplant recipient animals (Figure 3N) and introduced into CG1, syngeneic recipient animals (>78.9% purity and 96% viable, Figure 3O-R). Again, the myf5-GFP+/mylz2-mCherry-negative cell subpopulation was capable of remaking ERMS (Figure 3S-U, Table 1). Histological analysis showed that primary and serially transplanted ERMS arising from myf5-GFP+/mylz2-mCherry negative cell populations have similar morphology and overall proportions of fluorescent-labeled ERMS cell subpopulations (Figure 3C, D, F, G, M, N, T, U).
Visualizing myf5-GFP+ ERMS-propagating cells in vivo
To assess if myf5-GFP+ ERMS cells could be directly visualized in live animals, rag2-dsREDexpress and rag2-KRASG12D were co injected into one-cell stage myf5-GFP transgenic animals and assessed by confocal microscopy. Discrete myf5-GFP+ tumor cells could be readily identified by confocal imaging with a majority of myf5-GFP+ ERMS cells co-expressing both GFP and dsREDexpress (97.5+/−2.9%, n=568 cells counted in 3 animals). Moreover, myf5-GFP+ early muscle progenitor cells from control animals were relatively rare (n=3 animals, 2.3 +/−2.3 cells/ imaging field) whereas myf5-GFP+ cells were abundant in ERMS (n=3 animals, 194.2 +/−23.7 cells/field, T-test p=0.0002). Taken together, our data suggests that a vast majority of myf5-GFP+ cells contained within the boundaries of the ERMS mass are tumor-derived.
To further refine the ERMS cell subpopulations for imaging studies, triple fluorescent transgenic ERMS animals were created by microinjecting myogenin-H2B-RFP, mylz2-lyn-cyan, and rag2-KRASG12D into one-cell stage myf5-GFP transgenic animals (Figure 4A-C). Histone fusion proteins are long-lived and confined to the nucleus, whereas lyn-cyan encodes for membrane localized blue fluorescent protein. Because transgenes co-integrate as concatamers (Langenau et al., 2008), ERMS cells co-express all three transgenes and label distinct tumor cell compartments associated with stages of muscle development (Figure S2). In normal development, myf5 is expressed in satellite cells and early muscle progenitor cells, myogenin is expressed in committed, mid-differentiated muscle myoblasts, and mylz2 is expressed in differentiated myoblasts. Transgenic reporters have been described for all three of these promoters and each drives expression within the correct cellular compartments during normal muscle development (Chen et al., 2007; Du et al., 2003; Ju et al., 2003). Moreover, gene expression studies confirm that these promoters drive correct tissue specific gene expression in ERMS (Figure S2) and additional cell transplantation experiments establish that myf5-GFP+/myogenin-negative cell types exclusively retain ERMS-propagating potential (Figure S2). For example, 4 of 18 animals engrafted ERMS from 1×102 myf5-GFP+/myogenin-H2B-RFP-negative sorted ERMS cells (68.5% purity, 99.8% viable) whereas myogenin-H2B-RFP+ cell types could not induce tumors, irrespective if they expressed myf5-GFP (n=0 of 32, p=0.013, Fisher Exact Test).
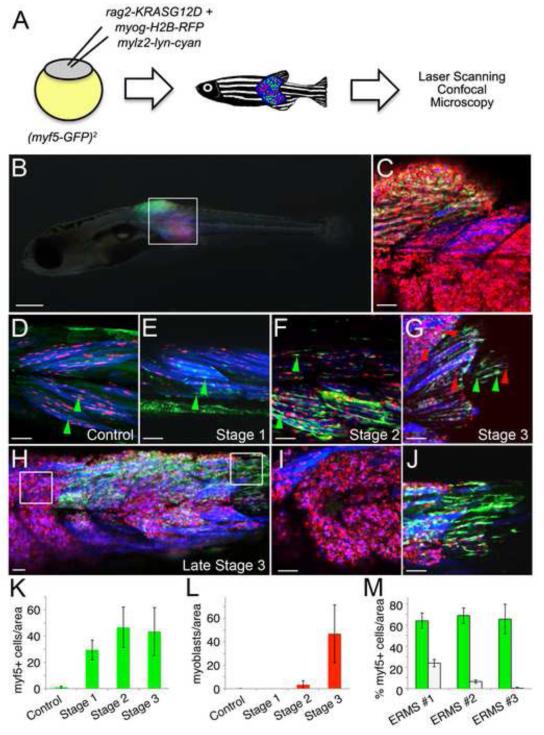
Figure 4. myf5-GFP+ ERMS-propagating cells are dynamically reorganized during tumor growth.
(A) Schematic of the experimental design.
(B) A myf5-GFP transgenic animal injected at the one-cell stage of life with rag2-KRASG12D, myogenin-H2B-RFP, and mylz2-lyn-cyan with triple fluorescent-labeled ERMS at 16 days of life.
(C) A merged confocal image of the boxed region shown in B.
(D) Control myf5-GFP transgenic animal injected with myogenin-H2B-RFP and mylz2-lyn-cyan. myf5-GFP+ muscle precursor cells are denoted by green arrowheads.
(E-G) Representative image of an ERMS-affected zebrafish labeled with myf5-GFP, myogenin-H2B-RFP and mylz2-lyn-cyan at stage 1, 2 and 3, respectively. Green arrowheads denote myf5-GFP+ cells whereas red arrowheads denote mononuclear myogenin-H2B-RFP+ ERMS cells.
(H) Late stage 3 ERMS from a triple fluorescent-labeled animal.
(I-J) Boxed regions in H imaged at higher magnification show regional partitioning of differentiated cells (I) compared with myf5-GFP+ ERMS-propagating cells (J).
(K) Quantification of myf5-GFP+ cells during stages of ERMS growth when compared to control animals.
(L) Quantification of mononuclear myogenin-H2B-RFP+ cells during stages of ERMS growth when compared to control animals.
(M) Quantification of regional compartmentalization of ERMS cells based on differentiation status in late stage 3 tumors (n=3). Green bars denote regions that contain higher percentages of myf5-GFP+ ERMS-propagating cells compared to white bars where myf5-GFP+ cells are less abundant and conversely more differentiated. Error bars in K-M +/− 1 STD. Scale bar is 500 μm (B) and 50 μm (C-J).
See also Figure S2.
Confocal imaging of fluorescent-transgenic ERMS fish that express myf5-GFP/myogenin-H2B-RFP/mylz2-lyn-cyan easily identified myf5-GFP+ cells of which a small subset co-express myogenin-H2B-RFP (Figure 4C). Some myogenin-H2B-RFP+ cells with nuclear fluorescent protein expression fail to express either myf5-GFP or mylz2-lyn-cyan, indicating that these cells are most similar to mid-myoblast stages. Gene expression studies confirm that myogenin-promoter expression drives H2B-flourescent protein expression in a subset of myosin heavy chain expressing muscle cell populations, implying that these represent differentiated cell types (Figure S2). Nearly all mylz2-lyn-cyan positive cells co-express myogenin-H2B-RFP (99.5 %+/− 1%, n=8 ERMS, N>1700 cells counted) reflecting that H2B-flourescent protein expression persists in more differentiated ERMS cells.
myf5-GFP+ cells are reorganized into discrete compartments during late stage tumor growth
We next wanted to define the location of ERMS cell subpopulations during various stages of tumor growth. myf5-GFP transgenic animals were injected with rag2-KRASG12D, myogenin-H2B-RFP, and mylz2-lyn-cyan and imaged by confocal microscopy starting at 10 days of life. Stage 1 ERMS exhibited greatly expanded numbers of myf5-GFP+ cells when compared to control animals and were confined to regions immediately adjacent muscle fibers (Figure 4D-G, K). Mononuclear myogenin-H2B-RFP+ and double positive myogenin-H2B-RFP+/ mylz2-lyn-cyan+ ERMS cells were not observed in stage 1 ERMS; however, they were detected by stage 2 and increased in number as tumors progressed to stage 3 (Figure 4F-I, L). By late stage 3 ERMS, myf5-GFP+ cells lost fiber contacts and began to populate discrete portions of the tumor that were physically separated from more differentiated myogenin-H2B-RFP and mylz2-lyn-cyan expressing ERMS cells (Figure 4H-J, M). The myf5-GFP+ cells were often located within different myotome segments compared to differentiated ERMS cell subpopulations; however, regional partitioning of cells based on differentiation status was also observed within a single myotome segment and in transplanted animals (Figure S2), confirming that compartmentalization did not result from physiological constraints imposed during development but rather was an intrinsic property of ERMS growth.
Human ERMS cells are also compartmentalized based on myogenic factor expression
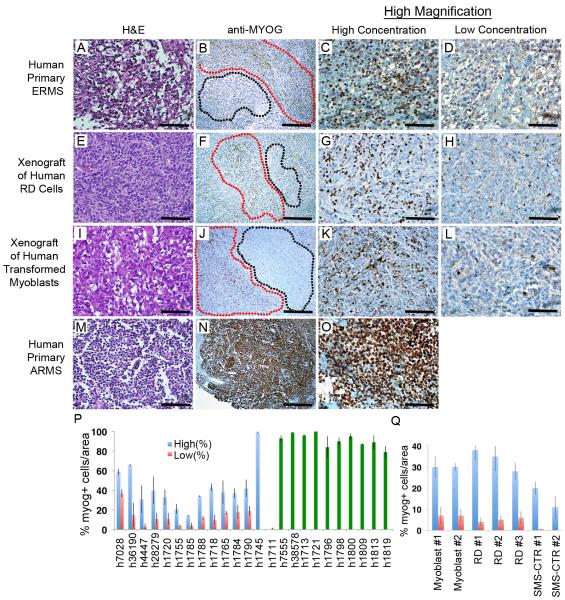
To assess whether human RMS also contain distinct regions of tumor cells based on myogenic factor expression, primary human ERMS and xenografted human ERMS derived from RD and SMS-CTR cell lines (Linardic et al., 2005) were assessed for myogenic marker expression, including Myogenin, PAX7 and MYOD. Distinct regions of high and low Myogenin-expressing cells were seen in a vast majority of primary tumor samples (n=12 of 14, p<0.03, T-statistic) and were present in all xenograft tumors (n=7, six regions/tumor, p<0.02, T-statistic, Figure 5 and Figure S3, Table S2). By contrast most ARMS cells expressed Myogenin and its expression was not confined to specific areas within the tumor mass (n=10, range 79-99%), suggesting that regional partitioning of tumor cells based on Myogenin expression is specific to ERMS. We also stained four primary human ERMS tumors for PAX7 and identified regions of high and low expression in two of the four tumors. In one ERMS sample, expression was diffuse while the other tumor was PAX7 negative, indicating that not all primary ERMS express PAX7. Unfortunately, MYF5 antibodies have not been developed to detect human protein within paraffin embedded sections, precluding analysis of less-differentiated regions contained within the tumors.
Figure 5. Human embryonal rhabdomyosarcoma exhibit regional portioning of cells based on myogenic factor expression.
(A-D) Primary human ERMS.
(E-H) RD human cell lines or (I-L) human RAS-transformed myoblasts introduced into SCID/beige mice.
(M-O) Primary human ARMS. Hematoxylin/Eosin stained sections (A, E, I, M) and anti-myogenin immunohistochemistry performed on adjacent sections (B, F, J, N). Regions containing high numbers of myogenin+ cells are denoted by red outline while regions with low numbers of myogenin+ cells are denoted by black outline (B, F, J). ARMS did not show regional portioning based on myogenin staining (N). Magnified views of areas with high concentrations of Myogenin+ cells (C, G, K, O) or areas with low or absent expression (D, H, L).
(P-Q) Quantification of regional compartments in primary and metastatic human RMS (P) and in mice xenografted with human RD and SMS-CTR ERMS cells and human RAS-transformed myoblasts (myoblasts, Q). Numbers in panel Q denote tumors arising in separate animals. Blue bars denote areas with high percentages of Myogenin+ cells compared to areas with low numbers of cells (red). Green bars denote diffuse and ubiquitous expression of Myogenin within ARMS. Error bars +/− 1 STD. Scale bars are 50 μm (A, C, D, E, G, H, I, K, L, M, O) and 200 μm (B, F, J, N)
myogenin+ ERMS cells are highly migratory and precede the recruitment of myf5+ ERMS-propagating cells into newly colonized areas of growth
Having established that the myf5-GFP-expressing ERMS cell population contains tumor-propagating activity, we wanted to assess if these cells also promote invasive tumor growth. Multiphoton intravital microscopy recordings from myf5-GFP/myogenin-H2B-RFP or myf5-GFP/myogenin-H2B-Amcyan transgenic tumor zebrafish revealed that myf5-GFP+ single-positive ERMS cells were largely stationary and displayed only confined crawling motion (Figure 6A-F, Figure S4, and Movies S2-5). In contrast, myogenin+ ERMS cells were robustly migratory and had the ability to invade across myotome segments through a normally impenetrable collagen matrix. Cells that expressed lower amounts of the H2B-fluorescent fusion protein, were orderly arranged along the direction of muscle fibers, had uniform nuclear shape, and did not show any motility (Movies S2 and S3), suggesting that these were differentiated tumor cells of which a subset had undergone fusion (Figure 6F, right).
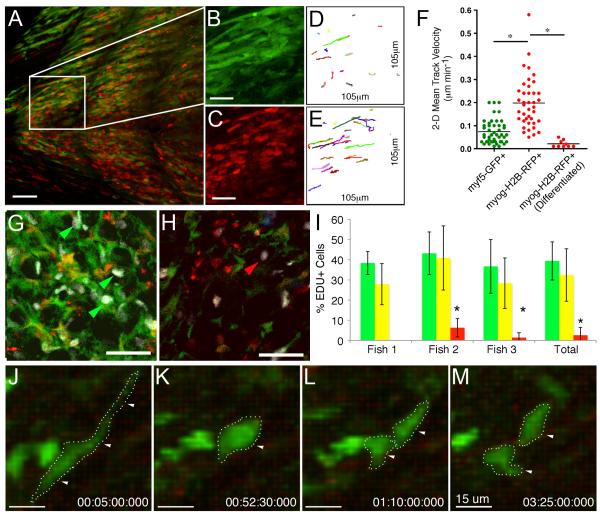
Figure 6. myf5-GFP+ ERMS-propagating cells are slow moving but highly proliferative while myogenin-H2B-RFP+ cells do not divide but are highly migratory.
(A-E) Multiphoton recording of a stage 3 ERMS arising in myf5-GFP/myogenin-H2B-RFP transgenic zebrafish (B, C) Magnified view of the boxed region in panel A showing myf5-GFP+ (B) or myogenin-H2B-RFP+ ERMS cells (C).
(D, E) Tracks of cell movement over the 6.7h observation period. The same areas are shown as in panels B and C, respectively.
(F) Mean track velocities of representative cell types contained within the tumor mass. Asterisk denotes p < 0.001.
(G-H) EDU staining of double transgenic myf5-GFP+/myogenin-H2B-RFP+ primary zebrafish ERMS (35 dpf). Confocal image of a tumor section with high numbers of myf5-GFP+ ERMS cells (G) compared to a section with high numbers of myogenin-H2B-RFP+ cells (H). White denotes nuclei that have incorporated EDU. EDU incorporation into myf5-GFP+ or myogenin-H2B-RFP+ ERMS cells denoted by green or red arrows, respectively.
(I) Quantification of proliferation over the 6 hour EDU pulse. myf5-GFP+/myogenin-H2B-RFP-negative (green bars), double positive (yellow bars), and myf5-GFP-negative/myogenin-H2B-RFP+ (red bars). Error bars +/−1 STD. Asterisk denotes significant differences with p=0.0001.
(J-M) Static images of a myf5-GFP+ ERMS cells dividing. Scales bar is 50 μm (A, G, H), 25 μm (B, C, D and E), and 15 μm (J-M).
ERMS cell subpopulations also differ in their proliferative capacity. Primary ERMS from myf5-GFP+/myogenin-H2B-RFP+ animals were pulsed with EDU for 6 hours and then sectioned and assessed for EDU incorporation (Figure 6G-I). myf5-GFP+ ERMS cells were highly proliferative (39.4+/−9.4%, n=3) whereas myf5-GFP-negative/myogenin-H2B-RFP+ cells rarely proliferated (2.6+/−3.8%, n=3, p=0.0001). In vivo multiphoton imaging of transplant and primary ERMS confirmed that myf5-GFP+/mylz2-negative ERMS-propagating cells are highly proliferative with 27 of 90 GFP+ cells dividing into two daughter cells (n=3 tumors). Multiphoton imaging revealed that resting myf5-GFP+ ERMS cells are elongated (Figure 6J) and then round up in shape just prior to cell division (Figure 6K). Following this dynamic shape change, myf5-GFP+ ERMS cells quickly divide into two GFP-labeled daughter cells (Figure 6L-M). Subsequently, these daughter cells begin to reacquire parental morphology (Movie S6), reminiscent of normal myf5-GFP+ muscle precursors. By contrast, 0 of 90 mylz2+ cells proliferated over this time, irrespective of whether they expressed myf5-GFP.
To further visualize the dynamic movements of ERMS cells in vivo, late stage 3 triple transgenic ERMS affected animals were serially imaged over 16 hours to capture cell movements within ERMS (Figure 7A-H and Movie S7). As was seen in our multiphoton imaging, myf5-GFP+ cells that lack differentiated marker expression move only locally within the tumor and exhibit regional crawling movements whereas myogenin-H2B-RFP+/mylz2-lyn-cyan-negative cells are highly motile and could be easily visualized migrating into adjacent non-affected normal tissue (Figure 7A-D and Movie S7). By contrast, differentiated ERMS cells that express myogenin-H2B-RFP and mylz2-lyn-cyan are largely stationary.
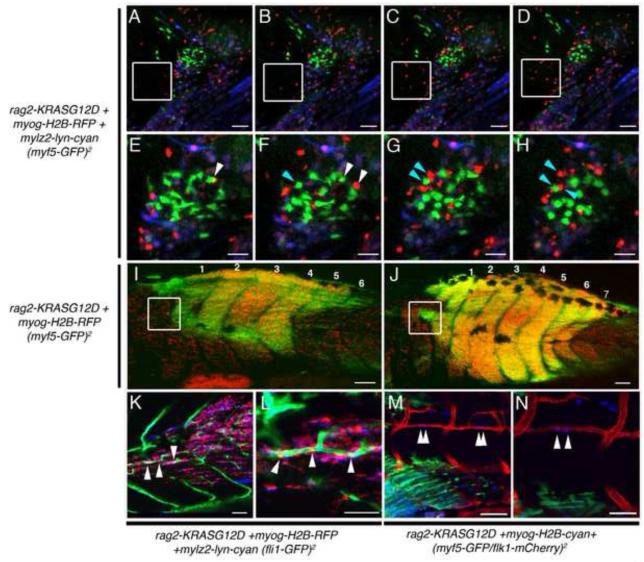
Figure 7. myf5-GFP+ ERMS-propagating cells are recruited to new areas of tumor growth only after seeding by myogenin+ ERMS cells.
(A-H) Time-lapse images of myf5-GFP transgenic animal injected with rag2-KRASG12D, myogenin-H2B-RFP and mylz2-lyn-cyan. Panels are merged image planes taken every hour. myogenin-H2B-RFP+ cells migrate into normal tissues over time (white boxed region). Magnified views of time-lapse images documenting that myf5-GFP+ cells are largely stationary while myogenin-H2B-RFP+ cells are highly migratory and migrate away from GFP+ cells (denoted by arrows in E-H).
(I, J) Serial imaging of a myf5-GFP transgenic animal injected with rag2-KRASG12D and myogenin-H2B-RFP shown at 14 and 17 dpf, respectively. White boxes demark a region that initially contained only myogenin-H2B-RFP+ cells (I) but was later colonized by myf5-GFP+ cells (J).
(K-L) ERMS developing in a fli1-GFP transgenic animal injected with rag2-KRASG12D, myogenin-H2B-RFP, and mylz2-lyn-cyan. (K) Merged z-stacks showing three myogenin-H2B-RFP+ cells associated with and inside fli1-GFP+ vessels, which was confirmed by imaging a single image plane at higher magnification (L, white arrow head).
(M-N) ERMS developing in a flk1-mCherry transgenic animal injected with rag2-KRASG12D and myogenin-H2B-cyan showing four cells entering the vasculature (White arrowheads, M) and a single plane image showing two cells transiting into the vasculature at higher magnification (White arrowheads, N).
Scale bar is 50 μm (A-H, K, L, N) and 100 μm (I, J, M).
To investigate which ERMS cells were the first to migrate into unaffected tissue, serial imaging experiments were conducted over longer observation intervals, focusing on regions that were adjacent to expanding tumor. Serial confocal imaging of fluorescent ERMS fish over several days revealed that myogenin-H2B+ ERMS cells precede the recruitment of myf5-GFP+ cells into newly colonized areas of tumor growth (n=7, Figure 7I, J and Figure S4). Not only do fluorescent-labeled myogenin-H2B+ ERMS cells move locally within the tumor, but they also enter the vasculature (Movie S8). A small portion of myogenin-H2B-RFP+/myf5-GFP-negative cells were associated with vasculature and could invade neovascular beds in fli1-GFP transgenic animals (Figure 7K, L). To verify the fidelity of H2B-fluorescent labeling of ERMS cell sub-fractions and to directly visualize if myf5-GFP+ ERMS cells can enter the vasculature, ERMS was induced in stable transgenic animals that express myf5-GFP/flk-mCherry by co-injecting both rag2-KRASG12D and myogenin-H2b-Amcyan. As was seen using the H2B-RFP transgenic reporter, we find that myogenin-H2B-Amcyan+ cells are highly migratory (Movie S4 and S5), were the first cell type to colonize new areas of tumor growth, and could be observed transiting the vasculature (Figure 7M, N, Movie S5 and S9). By contrast, myf5-GFP+ ERMS cells exhibited reduced motility when compared with myogenin+ ERMS cells and were never observed entering the vasculature (n= 10 animals). Again, slow moving myf5-GFP+ cells were found in newly colonized regions only after initial invasion by myogenin+ ERMS cells.
Discussion
Myf5 as a marker of ERMS-propagating cells
The limiting dilution cell transplantation studies outlined here confirm the existence of a highly purified and molecularly definable ERMS-propagating cell that expresses myf5, m-cadherin, and c-met but not differentiated muscle markers. The myf5-GFP+ ERMS-propagating cell gives rise to all the other differentiated ERMS cells contained within the tumor mass and exhibits enhanced proliferative capacity as assessed by EDU incorporation and direct in vivo cell imaging. These results are in keeping with our previous work showing that ERMS-propagating activity was largely confined to the rag2-dsRED+/alpha-actin-negative ERMS cell population that preferentially expressed myf5 and other activated satellite cell markers (Langenau et al., 2007). However, rag2-dsRED+/alpha-actin-negative ERMS cells exhibited only a modest 3-fold enrichment for tumor-propagating potential when compared to rag2-dsRED+/alpha-actin+ ERMS cells (Langenau et. al., 2007). By contrast, experiments outlined here using new fluorescent transgenic reporter lines and syngeneic zebrafish show that the myf5-GFP+/mylz2-negative ERMS cells exhibit a remarkable 28 to >40-fold enrichment of tumor-propagating potential when compared to other ERMS derived cell populations.
Myf5 is a myogenic regulatory factor related to MyoD and has important roles in muscle development. For example, MyoD/Myf5-deficient mice lack muscle, while deficiencies in only one of these genes does not affect muscle specification (Rudnicki et al., 1993), suggesting important and yet redundant functions of these genes in development. It has also been shown that Myf5 is highly expressed in activated satellite cells and has important roles in postnatal muscle regeneration in response to injury (Cooper et al., 1999; Gayraud-Morel et al., 2009; Ustanina et al., 2007), suggesting that Myf5 may regulate self-renewal in normal muscle satellite cells. Microarray analysis and cross-species comparisons have shown that MYF5 is upregulated in both zebrafish and human ERMS, but not translocation-positive ARMS (Langenau et al., 2007; Zibat et al., 2010), and recent work from Rubin et al has shown that Myf5 is differentially expressed in murine ERMS regardless of which muscle cell subpopulation is initially targeted for transformation (Rubin et al., 2011). These results suggest that Myf5 gene programs are likely re-initiated in transformed cells and may have important roles in driving ERMS growth. By contrast, translocation-positive ARMS fail to express MYF5, precluding MYF5 marker expression as an identifying characteristic of ARMS-propagating cells and raising the interesting possibility that the molecular mechanisms regulating tumor-propagating potential differ between molecular subtypes of rhabdomyosarcoma. Given the critical roles of the Myf5 transcription factor in muscle development and regeneration in mice, it will be important to assess if myf5 is a marker of ERMS-propagating cells or if it plays a regulatory role in ERMS self-renewal and growth.
Regional partitioning of ERMS cells based on differentiation status
Evidence in solid tumors to support a discrete, specialized microenvironment that augments tumor growth and proliferation is now just beginning to emerge. For example, tumor-propagating cells, including those of glioblastomas, have been shown to reside in a vascular niche that promotes both their maintenance and their ability to divide and produce daughter cells capable of inducing tumors (reviewed by Gilbertson and Rich, 2007). In other solid tumors arising in skin, prostate and breast, tumor stromal fibroblasts also serve an essential role in maintaining a favorable microenvironment for tumor growth and expansion. For example, work by Orimo et al has shown that stromal fibroblasts associated with invasive breast carcinoma cells can promote tumor growth and angiogenesis through secretion of SDF-1 (Orimo et al., 2005). In normal muscle, stem cell numbers are exquisitely regulated by paracrine factors like wnt5a (Polesskaya et al., 2003), myostatin (McCroskery et al., 2003), and Notch ligands (Conboy et al., 2003). Many of these factors are secreted by normal fibers that can sense injury and elicit recruitment and expansion of muscle progenitors that are required for regeneration. Thus, mature muscle provides a supportive microenvironment that facilitates homeostatic regulation of muscle stem cells. In our zebrafish ERMS model, we document that myf5-GFP+ ERMS-propagating cells are initially juxtaposed to muscle fibers in an expanded muscle satellite/progenitor cell niche, suggesting that early stage ERMS cells cannot escape the constraints of muscle architecture or are held in check by local secreted factors emanating from normal muscle. By late stages of ERMS growth, ERMS-propagating cells are reorganized and take up residence in defined regions within the tumor mass. Following this regional partitioning of ERMS cells, mid-differentiated myogenin+ ERMS cells show enhanced migratory capability and move away from ERMS-propagating cells from which they had arisen. These mid-differentiated myogenin+ ERMS cells are highly migratory, seed new areas of tumor growth, and cease to move once they turn on differentiated muscle markers including muscle myosin-light chain. Such biologically constrained characteristics of ERMS cells would ensure that tumor-propagating cells remain confined to regionally defined areas and do not compete with differentiated ERMS cell types for local resources including growth factors and oxygen. The extent to which regional partitioning of tumor cells occurs in other solid tumors is unknown; however, assuming this phenomenon is found in diverse cancer types, it will be important to determine if regional partitioning of tumor cells provides protective advantages to tumor-propagating cells, facilitating the retention of a small number of cancer cells that evade treatment and eventually give rise to disease relapse.
A role for differentiated, non-tumor-propagating cells in facilitating tumor growth and metastasis
Myogenin immunohistochemical reactivity found in >80% of RMS cells distinguishes patients with poor clinical outcome (Heerema-McKenney et al., 2008), suggesting that myogenin+ cells have a unique role in RMS progression and metastasis. In our model, myogenin-H2B+ cells arise from myf5-GFP+ ERMS-propagating cells, lack tumor-propagating potential, and are the first cell type to migrate into new areas of tumor growth. A subset of myogenin+ ERMS cells infiltrate blood vessels – a first step toward metastasis- and are also the first to colonize new areas of tumor growth, only to be infiltrated latter by slow-migrating myf5+ ERMS-propagating cells. Our work raises the interesting possibility that differentiated, non-ERMS-propagating cells may create a supportive environment that augments growth and is responsible for local tumor invasion. For example, it is possible that once mid-differentiated myogenin+ cells infiltrate new areas of growth that they secrete factors that recruit slow moving myf5+ ERMS-propagating cells, facilitating tumor spread. Alternatively, it is possible that myogenin+ cells breakdown collagen and cell-cell contacts, acting as trail-blazers to establish migratory tracks that allow slow moving myf5+ ERMS propagating cells to transit into newly forming tumor. Our work also highlights that metastatic capacity and tumor-propagating potential need not be confined to the same tumor cell subpopulations, but rather that local infiltration and metastasis may be facilitated by differentiated, non-tumor-propagating cells. We expect that these same principles may be more broadly applicable to a diversity of cancers, accounting for why tumors retain large numbers of differentiated cell types which themselves are incapable of re-constituting tumor.
Our findings of a myf5+ ERMS-propagating cell population and a myogenin+ migratory population both contributing to tumorigenesis may have profound therapeutic implications. Instead of targeting only tumor-propagating cells for destruction, drug design should also take into account the mechanisms regulating the homeostasis of more differentiated tumor cells and their non-proliferative roles in regulating growth. Moreover, therapies that focus on modulating the differentiation status of ERMS cells should attempt to force the conversion of tumor-propagating cells into cells with terminally differentiated myoblast characteristics that are incapable of re-creating tumor, cannot migrate, and fail to enter into the vasculature.
Experimental Procedures
Study Approval
These studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care under protocol #2011N000127 (zebrafish), the Duke University Institutional Animal Care & Use Committee under protocol A 036-03-02 (mouse), and the Partners Human Research Committee under protocol #2009-P-002756 (human). Samples were obtained from the Pathology Department of Massachusetts General Hospital. Use of decoded, paraffin embedded human tissue samples does not require informed consent.
Animals
CG1-strain (Smith et al., 2010), α-actin-GFP (Higashijima et al., 1997), myf5-GFP (Chen et al., 2007), fli1-GFP (Lawson and Weinstein, 2002), flk1-mCherry (Wang et al., 2010), and mylz2-mCherry transgenic zebrafish (Smith et al., 2010) have been reported previously.
The rag2-KRASG12D, rag2-dsREDexpress, myogenin-H2B-RFP, myogenin-H2B-Amcyan, and mylz2-lyn-cyan constructs were micro-injected into one-cell stage zebrafish singly (rag2-KRASG12D injected into myf5-GFP/mylz2-mCherry syngenic zebrafish, 60 ng/microliter) or as combinations with linearized DNA at a final combined concentration of 120 ng/microliter essentially as described (Langenau et al., 2008).
FACS and ERMS Cell Transplantation
FACS analysis and ERMS cell transplantation were completed essentially as described (Smith et al., 2010; Langenau et al., 2007). Sort gates were placed based on wild-type control fish, and myf5-GFP+, mylz2-mCherry+, or mylz2-lyn-cyan+ ERMS. Dapi, propidium iodide or TOPRO3 was used to isolate viable cells. ERMS tumors were double sorted to obtain pure, viable cell populations. Sort purity was assessed after two rounds of sorting when possible. Following limiting dilution cell transplantation into non-irradiated syngeneic CG1-recipient animals, fish were analyzed for fluorescent tumor engraftment from 10 to 120 days post-transplantation. Tumor-propagating potential was quantified using the Extreme Limiting Dilution Analysis software (http://bioinf.wehi.edu.au/software/elda/). A subset of transplanted fish were sectioned and stained with hematoxylin and eosin to confirm the presence or absence of ERMS.
Immunohistochemistry, EDU and Annexin V staining
Paraffin embedding and sectioning, cryostat sectioning, and immunohistochemical analysis were performed essentially as described (Langenau et al., 2007 and Supplemental Experimental Procedures). EDU staining was performed using the Click-iT Alexa Fluor 647 imaging kit, from Invitrogen. Annexin analysis for apoptotic cells was performed via FACS using annexin V conjugated to Alexa Fluor 647 (Invitrogen).
Gene Expression Analysis-
Total RNA was isolated from 6- and 24-hour post-fertilization AB-strain embryos and FAC sorted ERMS cell subpopulations (TRIzol, GIBCO/BRL) in the presence of glycol blue. Quantitative real-time PCR utilized gene-specific PCR primers (Supplemental Experimental Procedures) and expression was normalized to 18s and ß-actin controls to obtain relative transcript levels using the δδCT method. Relative gene expression was normalized within individual samples and cumulative transcript expression across the four ERMS cell subpopulation was set to 25. Samples were assessed in relation to 6 and 24-hour embryos to ensure that δδCT for any given gene were not lower than 10-fold expression found in normal development. Microarray experiments were completed essentially as described (Fold change cut off >1.5 fold log scale, Langenau et al., 2007). Microarray data has been deposited into the GEO database (GSE32425).
Laser Scanning Confocal Microscopy and Dual Photon Imaging
Larval zebrafish were anesthetized in Tricaine and embedded in a single drop of low melt 1% agarose on a glass bottom petri dish (No 1.5, Mat Tek Corporation). Each petri dish was supplemented with fish water and imaged using an inverted Pascal or LSM510 Zeiss laser scanning confocal microscope or an upright Ultima IV multiphoton microscope (Prairie Technologies). Quantification was completed by counting the total numbers of fluorescent-labeled ERMS cell subpopulations contained in two 250×150 μm areas per animal (Figure 4K-L; control, n=7; Stage 1, n=3; Stage 2, n=4; Stage 3 n=7). Because regional niches can be compartmentalized within a single myotome segment, a smaller area was assessed for total numbers of myf5-GFP+ and myogenin-H2B-RFP+ ERMS cells (50×50 μm2 area, n=6 areas/tumor, Figure 4M).
For cell tracking, sequences of image stacks were transformed into maximum intensity-projected movies using Imaris 7.1 software (Bitplane) and exported as Quicktime movies. Manual 2-D or 3-D tracking was performed using the manual tracking plugin in ImageJ or using Imaris 7.1. Annotation and further processing of movies was completed using ImageJ and Quicktime 7.
Supplementary Material
Significance.
Tumor-propagating potential is not found in all malignant cells, and in most cancers, cells with more differentiated features are largely incapable of remaking tumor and yet comprise a majority of the tumor mass. A role for differentiated malignant cells in tumor growth, including dissemination and metastasis, has not been fully explored. We find that mid-differentiated myogenin-positive ERMS cells lack tumor-propagating potential, yet are responsible for local invasion and can enter the vasculature. Slow moving myf5+ ERMS-propagating cells are recruited to new sites of tumor growth after seeding by differentiated ERMS cells. This finding may explain the clinical observation that Myogenin positivity correlates with poor clinical outcome in human ERMS and suggests that differentiated tumor cells play critical roles in metastasis.
Highlights.
Functional heterogeneity of ERMS cells can be visualized in live zebrafish.
Tumor-propagating potential is restricted to myf5+ ERMS cells.
myogenin+ ERMS cells are highly invasive and colonize new areas of growth.
Metastatic and ERMS-propagating potential reside in distinct cell subpopulations.
Acknowledgements
E.C. and J.S.B are supported by the NIH training grants T32 HL007627 and 5T32CA09216-26, respectively. CL is supported by R01 CA122706 and K12 HD043494. D.M.L. is supported by NIH grants K01 AR055619, 1RO1CA154923, and 1R21CA156056, the Alex’s Lemonade Stand Foundation, the Sarcoma Foundation of America, the American Cancer Society, and the Harvard Stem Cell Institute. I.M.T. is supported by ‘Fundação para a Ciência e Tecnologia’ through fellowship SFRH / BD / 51288 / 2010. We thank Huai-Jen Tsai for myf5-GFP transgenic animals and Clarrisa Henry for critical review of our manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ. Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev Biol. 2007;7:1. doi: 10.1186/1471-213X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112(Pt 17):2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- Du SJ, Gao J, Anyangwe V. Muscle-specific expression of myogenin in zebrafish embryos is controlled by multiple regulatory elements in the promoter. Comp Biochem Physiol B Biochem Mol Biol. 2003;134:123–134. doi: 10.1016/s1096-4959(02)00194-x. [DOI] [PubMed] [Google Scholar]
- Eyler CE, Wu Q, Yan K, MacSwords JM, Chandler-Militello D, Misuraca KL, Lathia JD, Forrester MT, Lee J, Stamler JS, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Tajbakhsh S. Skeletal muscle as a paradigm for regenerative biology and medicine. Regen Med. 2009;4:293–319. doi: 10.2217/17460751.4.2.293. [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- Heerema-McKenney A, Wijnaendts LC, Pulliam JF, Lopez-Terrada D, McKenney JK, Zhu S, Montgomery K, Mitchell J, Marinelli RJ, Hart AA, et al. Diffuse myogenin expression by immunohistochemistry is an independent marker of poor survival in pediatric rhabdomyosarcoma: a tissue microarray study of 71 primary tumors including correlation with molecular phenotype. Am J Surg Pathol. 2008;32:1513–1522. doi: 10.1097/PAS.0b013e31817a909a. [DOI] [PubMed] [Google Scholar]
- Hettmer S, Liu J, Miller CM, Lindsay MC, Sparks CA, Guertin DA, Bronson RT, Langenau DM, Wagers AJ. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A. 2011;108:20002–20007. doi: 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Ju B, Chong SW, He J, Wang X, Xu Y, Wan H, Tong Y, Yan T, Korzh V, Gong Z. Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter. Dev Dyn. 2003;227:14–26. doi: 10.1002/dvdy.10273. [DOI] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Jette CA, Smith AC, Ceol CJ, Bourque C, Look AT, Zon LI. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene. 2008;27:4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–4495. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, Prajapati SI, Abraham J, Arenkiel BR, Chen QR, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Seger C, Hargrave M, Wang X, Chai RJ, Elworthy S, Ingham PW. Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Dev Dyn. 2011;240:2440–2451. doi: 10.1002/dvdy.22745. [DOI] [PubMed] [Google Scholar]
- Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, Luster AD, Scadden DT, Lin CP. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, de Jong JL, Chen AT, Zhou Y, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115:3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kaiser MS, Larson JD, Nasevicius A, Clark KJ, Wadman SA, Roberg-Perez SE, Ekker SC, Hackett PB, McGrail M, Essner JJ. Moesin1 and Vecadherin are required in endothelial cells during in vivo tubulogenesis. Development. 2010;137:3119–3128. doi: 10.1242/dev.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- Zibat A, Missiaglia E, Rosenberger A, Pritchard-Jones K, Shipley J, Hahn H, Fulda S. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29:6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.