Abstract
To better understand the role of bats as reservoirs of Bartonella spp., we estimated Bartonella spp. prevalence and genetic diversity in bats in Guatemala during 2009. We found prevalence of 33% and identified 21 genetic variants of 13 phylogroups. Vampire bat–associated Bartonella spp. may cause undiagnosed illnesses in humans.
Keywords: Bartonella spp., bats, Desmodus rotundus, Guatemala, bacteria, dispatch
Multiple studies have indicated that bats might serve as natural reservoirs to a variety of pathogens, including rabies virus and related lyssaviruses, Nipah and Hendra viruses, Marburg virus, and others (1,2). Bats’ high mobility, broad distribution, social behavior (communal roosting, fission–fusion social structure), and longevity make them ideal reservoir hosts and sources of infection for various etiologic agents. In addition to viruses, bacteria and ectoparasites have been detected in bats (3–5) and can potentially cause human infection (6).
Bartonella spp. have been found in rodents, insectivores, carnivores, ungulates, and many other mammals. Naturally infected hematophagous arthropods, such as fleas, flies, lice, mites, and ticks are frequently implicated in transmitting Bartonella spp (3–5,7). Detection of Bartonella DNA in the saliva of dogs suggests the possibility that Bartonella spp. can be transmitted through biting (8). Increasing numbers of Bartonella spp. have been identified as human pathogens (9,10). However, a mammalian reservoir has not been determined for some newly identified species, such as B. tamiae (9). Extensive surveillance for Bartonella spp. among diverse groups of animals, including bats, has become crucial.
To our knowledge, Bartonella spp. in bats have been studied only in the United Kingdom and Kenya (11,12). To better understand the role of bats as reservoir hosts of Bartonella spp. and their potential risk for infecting humans and animals, we looked for Bartonella spp. in bats in Guatemala, estimated prevalence, and evaluated the genetic diversity of the circulating Bartonella strains.
The Study
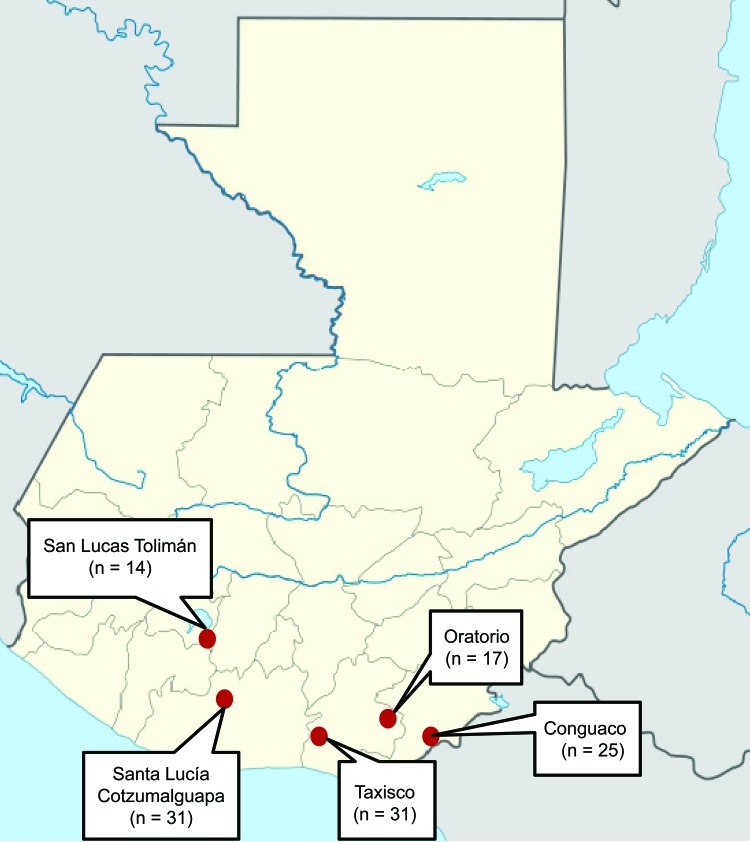
In 2009, a total of 118 bats were collected from 5 sites in southern Guatemala (Figure 1). The bats represented 15 species of 10 genera; the most prevalent (26.3%) species was the common vampire bat (Desmodus rotundus); the other 14 species accounted for 0.8%–12.7% of the bats sampled. Diversity of bats was 6–8 species per site (Table 1). Blood specimens from the bats were collected and cultured for Bartonella spp., according to a published method (12). A total of 41 Bartonella isolates were obtained from 39 (33.1%) of the 118 bats; colonies with different morphologic characteristics were identified from blood of 2 Pteronotus davyi bats. Prevalence of Bartonella spp. in Conguaco (60%, 15/25) was significantly higher than that in Oratorio (11.8%, 2/17), San Lucas Tolimán (14.3%, 2/14), and Taxisco (22.6%, 7/31) but did not differ from that in Santa Lucía Cotzumalguapa (41.9%, 13/31). Bartonella spp. were cultured from 8 bat species. The Bartonella spp. prevalence among Phyllostomus discolor (88.8%, 9/8), P. davyi (70%, 7/10), and D. rotundus (48.4%, 15/31) bats was significantly higher than that among Sturnira lilium (8.3%, 1/12) and Glossophaga soricina (13.3%, 2/15) bats. No Bartonella spp. were found in Artibeus jamaicensis (0/13) and 6 other bat species tested (Table 1).
Figure 1.
Sites of bat collection, showing number of bats collected from each site, Guatemala, 2009.
Table 1. Prevalence of Bartonella spp. in bats from 5 collection sites, Guatemala, 2009.
| Bat species | No. positive/no. cultured |
Overall, no. positive/ no. cultured (%) | ||||
|---|---|---|---|---|---|---|
| Conguaco | Oratorio | San Lucas Tolimán | Santa Lucía Cotzumalguapa | Taxisco | ||
| Artibeus jamaicensis | 0/3 | 0/2 | 0 | 0/3 | 0/5 | 0/13 |
| Artibeus lituratus | 0/1 | 0/1 | 0/1 | 0 | 0 | 0/3 |
| Artibeus toltecus | 1/1 | 0 | 0 | 0 | 0 | 1/1 (100) |
| Carollia castanea | 0 | 0 | 0 | 0/1 | 0 | 0/1 |
| Carollia perspicillata | 0 | 0/2 | 0/3 | 0 | 4/9 | 4/14 (28.6) |
| Desmodus rotundus | 5/7 | 2/4 | 0/1 | 6/12 | 2/7 | 15/31 (48.4) |
| Glossophaga soricina | 1/1 | 0/3 | 1/5 | 0/3 | 0/3 | 2/152 (13.3) |
| Micronycteris microtis | 0 | 0 | 0 | 0 | 1/3 | 1/3 (33.3) |
| Myotis elegans | 0 | 0 | 0 | 0/1 | 0/1 | 0/2 |
| Myotis nigricans | 0 | 0 | 0 | 0 | 0/1 | 0/1 |
| Phyllostomus discolor | 7/8 | 0 | 1/1 | 0 | 0 | 8/9 (88.9) |
| Platyrrhinus helleri | 0 | 0 | 0 | 0/1 | 0 | 0/1 |
| Pteronotus davyi | 0 | 0 | 0 | 7/10 | 0 | 7/10 (70) |
| Sturnira lilium | 1/3 | 0/5 | 0/2 | 0 | 0/2 | 1/12 (8.3) |
|
Sturnira ludovici
|
0/1 |
0 |
0/1 |
0 |
0 |
0/2 |
| Total | 15/25 | 2/17 | 2/14 | 13/31 | 7/31 | 39/118 (33.1) |
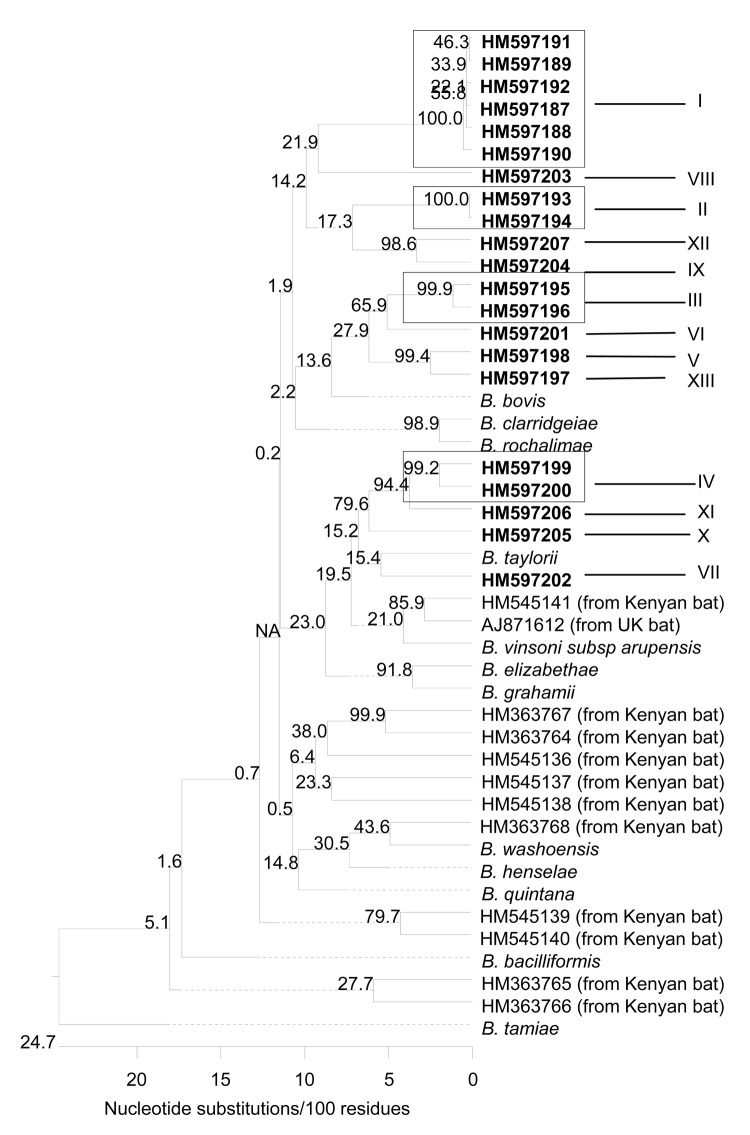
Identity of 41 Bartonella isolates was confirmed by PCR amplification of a specific region in the citrate synthase gene by using primers BhCS781.p (5′-GGGGACCAGCTCATGGTGG-3′) and BhCS1137.n (5′-AATGCAAAAAGAACAGTAAACA-3′). Subsequent sequencing analyses of the 41 isolates revealed 21 genetic variants (Table 2) that clustered into 13 phylogroups (I–XIII) with 6.6%–24.7% divergence. The phylogroups were also distant from any previously described Bartonella species and genotypes identified in bats from the United Kingdom and Kenya (Figure 2). Each phylogroup contained 1–6 variants; similarities within phylogroups were 96.2%–99.7% (Table 2).
Table 2. GenBank accession numbers and distribution of 21 genetic variants of Bartonella spp. in bats from Guatemala, 2009.
| Accession no. | Type strain | Host bat species | No. sequences | Distribution (no. isolates) | Phylogroup |
|---|---|---|---|---|---|
| HM597187 | B29042 | Desmodus rotundus | 1 | D. rotundus (1) | I |
| HM597188 | B29043 | D . rotundus | 3 | D. rotundus (3) | I |
| HM597189 | B29044 | D . rotundus | 2 | D. rotundus (2) | I |
| HM597190 | B29107 | D . rotundus | 1 | D. rotundus (1) | I |
| HM597191 | B29108 | D. rotundus | 3 | D. rotundus (2); C. perspicillata (1) | I |
| HM597192 | B29114 | D. rotundus | 3 | D. rotundus (2); C. perspicillata (1) | I |
| HM597193 | B29102 | Pteronotus davyi | 3 | P. davyi (3) | II |
| HM597194 | B29109 | P. davyi | 1 | P. davyi (1) | II |
| HM597195 | B29119 | D. rotundus | 3 | D. rotundus (3) | III |
| HM597196 | B29122 | D. rotundus | 1 | D. rotundus (1) | III |
| HM597198 | B29116 | Phyllostomus discolor | 2 | P. discolor (2) | V |
| HM597199 | B29126 | Carollia perspicillata | 2 | C. perspicillata (2) | IV |
| HM597200 | B29230 | P. discolor | 1 | P. discolor (1) | IV |
| HM597201 | B29115 | P. discolor | 3 | P. discolor (3) | VI |
| HM597202 | B29110 | Glossophaga soricina | 3 | G. soricina (2); P. davyi (1) | VII |
| HM597203 | B29105 | P. davyi | 3 | P. davyi (3) | VIII |
| HM597204 | B29112 | P. discolor | 2 | P. discolor (2) | IX |
| HM597205 | B29134 | P. davyi | 1 | P. davyi (1) | X |
| HM597206 | B29137 | Sturnira lilium | 1 | S. lilium (1) | XI |
| HM597207 | B29172 | Micronycteris microtis | 1 | M. microtis (1) | XII |
| HM597197 | B29111 | Artibeus toltecus | 1 | A. toltecus (1) | XIII |
Figure 2.
Phylogenetic relationships of the Bartonella spp. genotypes based on partial sequences of the citrate synthase gene detected in bats from Guatemala, Kenya, United Kingdom, and some reference Bartonella spp. The phylogenetic tree was constructed by the neighbor-joining method, and bootstrap values were calculated with 1,000 replicates. A total of 21 Bartonella genotypes, forming 13 Bartonella phylogroups, were identified in the bats from Guatemala. Each genotype is indicated by its GenBank accession number in boldface; the phylogroups are marked by Roman numerals I–XIII.
Of the 13 phylogroups, phylogroups I, IV, and VII were identified in isolates obtained from different bat species (Table 2), suggesting that bats of different species may share the same Bartonella strain; whereas 4 species of bats—C. perspicillata, D. rotundus, P. discolor, and P. davyi—were infected with 2–4 Bartonella strains (Table 2). P. davyi from 2 bats belonged to phylogroups II or VIII.
Conclusions
The high (≈33%) prevalence of Bartonella spp. in bat populations in southern Guatemala might suggest persistent infection of long-lived bats with Bartonella spp., similar to their infection with some viruses (13). Depending on the bat species, Bartonella spp. exhibit high, low, or no infectivity, which may explain the variation in Bartonella spp. prevalence between study sites because the assemblage of bat species differed at each site. Additional studies are needed to illustrate the distribution of Bartonella spp. among the bat fauna in Guatemala and throughout the region.
Further characterization is necessary to verify whether the Bartonella strains representing a variety of distinct phylogroups represent novel Bartonella species. Unlike the discovery in bats in Kenya (12), host specificity of Bartonella spp. was not found in bats in Guatemala. Such lack of specificity may be partly associated with the arthropod vectors that parasitize bats, although we were unable to attempt isolation of agents from the bat ectoparasites. Future studies of bat ectoparasites would enable testing of hypotheses about whether any arthropods may be vectors in the Bartonella spp. transmission cycle and whether ectoparasite specificity contributes to the lack of host specificity observed in this study.
The tendency of some bat species to share roosts, reach large population densities, and roost crowded together creates the potential for dynamic intraspecies and interspecies transmission of infections (14). In accordance with this hypothesis, our finding that co-infection with multiple Bartonella strains in a single bat species, and even in an individual bat, indicate that active interspecies transmission of Bartonella spp. likely occurs among bats in Guatemala. The specificity of ectoparasite arthropod vectors among the bat fauna remains unclear and may contribute to interspecies transmission of Bartonella spp. among bats.
The long life spans of bats (average 10–20 years) may have made them major reservoirs that contribute to the maintenance and transmission of Bartonella spp. to other animals and humans. The bite of the common vampire bat has been long recognized to transmit rabies virus to humans throughout Latin America (2). These bats typically feed on the blood of mammals, including domestic animals and humans (15). Predation of vampire bats on humans is a major problem in Latin America (2). If Bartonella spp. can be transmitted to humans through the bite of bats, the need for further studies with vampire bats is imperative. Bartonella spp.–specific DNA has been detected in ectoparasites collected from bats (3–5). Presumably, if Bartonella spp. are transmitted through a bat ectoparasite vector, some, if not all, bat-associated Bartonella spp. could be transmitted to humans because bats are frequent hosts to a wide variety of ectoparasites, including bat flies, fleas, soft ticks, and mites. However, transmission potential might vary with the degree of synanthropic roosting or foraging behavior within the bat community.
Because an increasing number of Bartonella spp. are being associated with human illness, the need to identify the animal reservoirs of these novel Bartonella spp. and to understand their disease ecology is also increasing. Our study of Bartonella spp. in bats has enlarged our scope of this zoonotic potential as we search for the reservoirs that harbor novel and known Bartonella spp.
Biography
Dr Bai is an associate service fellow in the Bartonella Laboratory, Bacterial Diseases Branch, Division of Vector-Borne Diseases, Centers for Disease Control and Prevention. Her research interests include microbiology, epidemiology, and ecology of zoonotic infectious diseases.
Footnotes
Suggested citation for this article: Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, Turmelle A, et al. Bartonella spp. in bats, Guatemala. Emerg Infect Dis [serial on the Internet]. 2011 Jul [date cited]. http://dx.doi.org/10.3201/eid1707.101867
References
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–45. 10.1128/CMR.00017-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider MC, Romijn PC, Uieda W, Tamayo H, da Silva DF, Belotto A, et al. Rabies transmitted by vampire bats to humans: an emerging zoonotic disease in Latin America? Rev Panam Salud Publica. 2009;25:260–9. 10.1590/S1020-49892009000300010 [DOI] [PubMed] [Google Scholar]
- 3.Loftis AD, Gill JS, Schriefer ME, Levin ML, Eremeeva ME, Gilchrist MJ, et al. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Aragasidae). J Med Entomol. 2005;42:473–80. 10.1603/0022-2585(2005)042[0473:DORBAB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 4.Reeves WK, Loftis AD, Gore JA, Dasch GA. Molecular evidence for novel Bartonella species in Trichobius major (Diptera: Streblidae) and Cimex adjunctus (Hemiptera: Cimicidae) from two southeastern bat caves, U.S.A. J Vector Ecol. 2005.30:339–341. [PubMed]
- 5.Reeves WK, Rogers TE, Durden LA, Dasch GA. Association of Bartonella with the fleas (Siphonaptera) of rodents and bats using molecular techniques. J Vector Ecol. 2007;32:118–22. 10.3376/1081-1710(2007)32[118:AOBWTF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 6.Gill JS, Rowley WA, Bush PJ, Viner JP, Gilchrist MJ. Detection of human blood in the bat tick Carios (Ornithodoros) kelleyi (Acari: Argasidae) in Iowa. J Med Entomol. 2004;41:1179–81. 10.1603/0022-2585-41.6.1179 [DOI] [PubMed] [Google Scholar]
- 7.Roux V, Raoult D. Body lice as tools for diagnosis and surveillance of reemerging diseases. J Clin Microbiol. 1999;37:596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan AW, Maggi RG, Breitschwerdt EB. Bartonella DNA in dog saliva. Emerg Infect Dis. 2007;13:1948–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosoy M, Morway C, Sheff K, Bai Y, Colborn J, Chalcraft L, et al. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from human patients from Thailand. J Clin Microbiol. 2008;46:772–5. 10.1128/JCM.02120-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7. 10.1056/NEJMoa065987 [DOI] [PubMed] [Google Scholar]
- 11.Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ. Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology. 2005;131:489–96. 10.1017/S0031182005008097 [DOI] [PubMed] [Google Scholar]
- 12.Kosoy M, Bai Y, Lynch T, Kuzmin I, Niezgoda M, Franka R, et al. Discovery of Bartonella bacteria in bats from Kenya. Emerg Infect Dis. 2010;16:1875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulkin SE, Allen R. Virus infections in bats. Monogr Virol. 1974;8:1–103. [PubMed] [Google Scholar]
- 14.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–9. 10.1126/science.1188836 [DOI] [PubMed] [Google Scholar]
- 15.Turner DC, Bateson P. The vampire bat: a field study in behavior and ecology. Baltimore: Johns Hopkins University Press; 1975; p. 1–7. [Google Scholar]