Abstract
Clubfoot is a common birth defect characterized by inward posturing and rigid downward displacement of one or both feet. The etiology of syndromic forms of clubfoot is varied and the causes of isolated clubfoot are not well understood. A microduplication of 2.2 Mb on chromosome 17q23.1q23.2 which includes T-box 4 (TBX4), a hindlimb-specific gene, and 16 other genes was recently identified in 3 of 66 families reported as nonsyndromic clubfoot, but additional non-foot malformations place them in the syndromic clubfoot category. Our study assesses whether variation in or around TBX4 contributes to nonsyndromic clubfoot. To determine whether this microduplication was a common cause of nonsyndromic clubfoot, 605 probands (from 148 multiplex and 457 simplex families) with nonsyndromic clubfoot were evaluated by copy number and oligonucleotide array CGH testing modalities. Only one multiplex family (0.68%) that had 16 with clubfoot and 9 with other foot anomalies, had a 350kb microduplication, which included the complete duplication of TBX4 and NACA2 and partial duplication of BRIP1. The microduplication was transmitted in an autosomal dominant pattern and all with the microduplication had a range of phenotypes from short wide feet and toes to bilateral clubfoot. Minimal evidence was found for an association between TBX4 and clubfoot and no pathogenic sequence variants were identified in the two known TBX4 hindlimb enhancer elements. Altogether, these results demonstrate that variation in and around the TBX4 gene and the 17q23.1q23.2 microduplication are not a frequent cause of this common orthopedic birth defect and narrows the 17q23.1q23.2 nonsyndromic clubfoot-associated region.
Keywords: Clubfoot, genetics, TBX4, microduplication, association, malformation
INTRODUCTION
Nonsyndromic clubfoot is a common birth defect characterized by the rigid inward and downward displacement of the foot in equinus deformity [Bakalis et al., 2002; Gurnett et al., 2008]. Calf musculature hypoplasia is a common associated finding. Corrective treatments include serial castings beginning soon after birth, with bracing to maintain the correction. Previously, the majority of true clubfeet would have also required surgical correction; however, improvements in casting have reduced this to 10-20%. Clubfoot can be syndromic, resulting from chromosomal, Mendelian genetic and non-genetic causes, but more commonly occurs as an isolated birth defect [Brewer et al., 1998; Brewer et al., 1999]. Approximately 4,000 newborns in the US and 135,000 worldwide are born with isolated clubfoot annually [Moorthi et al., 2005]. However, the prevalence varies between ethnic groups and males are affected twice as frequently as females [Ching et al., 1969; Moorthi et al., 2005]. Unilateral involvement occurs in slightly less than 50% of cases and the right side is more often affected.
A genetic etiology for isolated clubfoot has been suggested by large population and family studies; segregation analyses suggest a multifactorial etiology with both genetic and environmental factors playing a role [Carter, 1965; Carter, 1969; de Andrade et al., 1998; Rebbeck et al., 1993]. Smoking is the most consistently associated environmental factor with a 2-fold increase in birth prevalence for children born to mothers who smoke during pregnancy [Alderman et al., 1991; Honein et al., 2000]. Smoking in the presence of a positive family history of clubfoot increases the risk twenty-fold, lending support for a role of genes in clubfoot [Honein et al., 2000]. To date, genetic association studies have suggested that apoptotic, homeobox A & D (HOXA and HOXD) and muscle contraction genes contribute to the underlying genetic liability [Ester et al., 2007; Ester et al., 2009; Heck et al., 2005; Weymouth et al., 2011].
A recent study described a 2.2 Mb microduplication of chromosome 17q23.1q23.2 in 3 of 66 families of unknown ethnicity with reportedly isolated clubfoot [Alvarado et al., 2010]. One additional clubfoot family had a deletion involving the same region. While this region contains 17 genes, TBX4 was of particular interest because it has hindlimb-specific expression [Hasson et al., 2010; Menke et al., 2008]. The current study was undertaken to assess whether copy number variation and/or variation in or around TBX4 play a role in clubfoot.
METHODS
Study population and sample preparation
The family-based dataset was comprised of 605 families (148 multiplex families: 112 nonHispanic white (NHW) and 36 Hispanic; 457 simplex families: 211 NHW and 246 Hispanic). Families were recruited after informed consent from clubfoot clinics in Shriners Hospitals for Children in Houston, Los Angeles and Shreveport, Texas Scottish Rite Hospital for Children of Dallas and the University of British Columbia as previously described [Ester et al., 2007; Ester et al., 2009; Heck et al., 2005]. Clinical and radiographic examinations were performed on probands and family members at all centers to exclude syndromic causes of clubfoot. Ethnicity was based on self-report and Hispanic participants were of Mexican descent. After obtaining informed consent, blood and/or saliva samples were collected from affected individuals and family members. DNA was extracted using either the Roche DNA Isolation Kit for Mammalian Blood (Roche, Switzerland) or Oragene Purifier for saliva (DNA Genotek, Inc. Ottawa, Ontario, Canada) following the manufacturer’s protocol.
Copy number variation screening
TaqMan copy number assays were run on all probands with probes, Hs05507252_cn, Hs00361007_cn, Hs06409269_cn, Hs01196629_cn, Hs05521790_cn, Hs05505679_cn and Hs05500071_cn (P/N 4400291, Applied Biosystems, Foster City, California), and analyzed on a 7900HT Sequence Detection System following the manufacturer’s instructions. CopyCaller software was used to calculate copy number values using a maximum likelihood algorithm. Additionally, DNA from two distantly related individuals in family F465, III-10 and V-13, were subjected to chromosomal microarray analysis using an Agilent platform to determine the boundaries of the microduplication. Array Comparative Genomic Hybridization (aCGH) using an array with 180,000 oligonucleotides (Baylor College of Medicine Version 8.1) covering the genome at a resolution of 30 kb was performed. This array has an exon-by-exon coverage of 1714 genes [Boone et al., 2010]. Array CGH was performed using DNA from a gender matched normal individual as reference.
Genotyping and analysis
To determine whether an association existed between the TBX4 region and clubfoot, eight SNPs in and around TBX4 were selected based on heterozygosity in the nonHispanic white population (MAF>0.3) (HapMap CEU dataset - www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?pop=1409), position in or around the gene and extent of linkage disequilibrium (LD) (Table I). Genotyping was performed using TaqMan® Genotyping Assays (Applied Biosystems, Foster City, CA) and detected on a 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol.
Table I.
TBX4 SNP Descriptions
| SNP | Position (bp) |
Alleles | Location | NH W MA F |
HCF |
|---|---|---|---|---|---|
| rs4968564 | 59531816 | C/G | P | 0.2 56 |
0.210 |
| rs3744448 | 59533868 | C/G | E1 (missense) | 0.1 39 |
0.196 |
| rs3744447 | 59534987 | A/C | E2 (synonymous) | 0.1 46 |
0.207 |
| rs3785833 | 59538663 | A/G | I2 | 0.3 50 |
0.366 |
| rs758596 | 59544863 | C/T | I3 | 0.2 55 |
0.190 |
| rs35500387 | 59548328 | C/T | I4 | 0.0 94 |
0.104 |
| rs17601584 | 59552682 | C/T | I4 | 0.2 31 |
0.118 |
| rs3785825 | 59556341 | A/G | I5 | 0.1 80 |
0.101 |
| rs3744438 | 59557600 | A/G | E7 (missense) | 0.1 40 |
0.132 |
| rs3785821 | 59559508 | A/T | I7 | 0.3 24 |
0.403 |
| rs744651 | 59563248 | C/T | D | 0.2 77 |
0.207 |
P = promoter, E = exon, I = intron, D = downstream of gene, MAF = minor allele frequency, HCF = Hispanic corresponding allele frequency
Analysis
For statistical analyses, the data were stratified by ethnicity alone or by family history of clubfoot and ethnicity. Tests for Hardy-Weinberg Equilibrium (HWE) were calculated using SAS (v9.1) and none deviated (p<0.001). Chi-square analysis was performed using SAS to evaluate ethnic differences in allele frequencies. Pairwise linkage disequilibrium values (D’ and r2) were calculated using GOLD [Abecasis and Cookson, 2000].
Linkage and/or association were evaluated using multiple analytic methods to extract the greatest amount of information from the data. Parametric and nonparametric linkage analyses were performed using Merlin [Abecasis et al., 2002]. Linkage parameters were used as described previously [Ester et al., 2009]. Association was tested using Pedigree Disequilibrium Test (PDT), genotype-Pedigree Disequilibrium Test (GENO-PDT) and Association in the Presence of Linkage (APL) [Chung et al., 2006; Martin et al., 2003; Martin et al., 2000].
Sequencing of TBX4 hindlimb enhancers
DNA samples from 95 randomly selected clubfoot probands with a positive family history were sequenced for HLEA and HLEB (hindlimb enhancer A and B), two enhancers that are thought to regulate TBX4 in the developing limb [Menke et al., 2008]. Due to the length of the TBX4 hindlimb enhancers (HLEA: 1323bp, HLEB: 736bp) multiple overlapping primers were used to capture the entire sequence with high confidence base calls (Table III). Sequences were PCR amplified using TopTaq polymerase (Invitrogen) with an annealing temperature of 58oC. Sequencing was done using Sequetech DNA Sequencing Service (Sequetech Corporation) and chromatograms were examined using Sequencher software (Gene Codes Corporation). To examine segregation of the identified variants, sequencing was performed in all family members as described above using HLEA and HLEB primer sets F2R2.
Table III.
Primers for hindlimb enhancer sequencing
| Primer Name | Sequence |
|---|---|
| TBX4_HLEA-F1 | AGTCCTGTTAGAGGCGGACA |
| TBX4_HLEA-R1 | AGACAAGGGCCTCCACCT |
| TBX4_HLEA-F2 | GGGAGAGATCAGCACTCCAG |
| TBX4_HLEA-R2 | CACCTCGGCTGCTTAAGTGT |
| TBX4_HLEA-F3 | CCCTTTGAGCTGAAGGACAG |
| TBX4_HLEA-R3 | GTGGCTCCTCTGGGAGATTC |
| TBX4_HLEB-F1 | TCCCCCAAAGTTATTCACAGA |
| TBX4_HLEB-R1 | CAAGACACTTACAACGGGGATT |
| TBX4_HLEB-F2 | CGGGCCTCCATTTAGAGAA |
| TBX4_HLEB-R2 | TCTGAAAAGCATCACGCTACA |
RESULTS
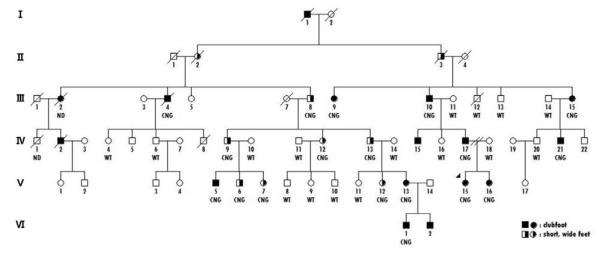
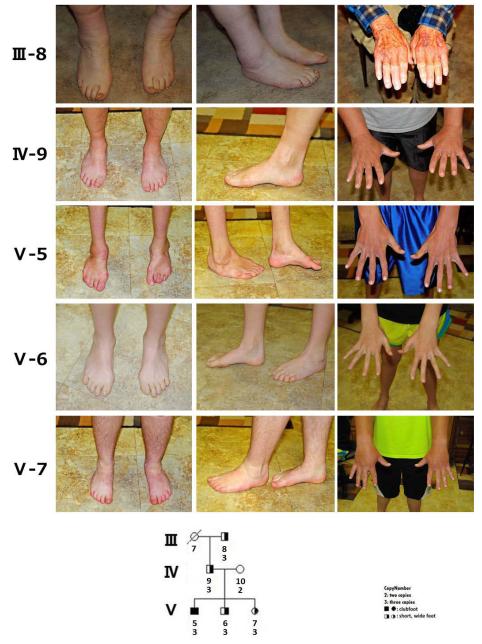
A microduplication was identified in one proband of nonHispanic white origin out of 148 familial cases (0.68%); the microduplication was not present in any of the 457 simplex probands. This proband was from a large family with clubfoot segregating through six generations in an autosomal dominant pattern with variable expressivity (F645) (Fig. 1). Clubfoot was present in 12 individuals who were related through individuals who had unusually short wide feet with toe anomalies (Fig. 2). As shown in Fig. 1, the microduplication (indicated as a CNG (copy number gain) under each pedigree symbol) was present only in individuals with abnormal feet. The foot anomalies in 5 individuals, III-8, IV-9, V-5, V-6, V-7, from three generations with the microduplication are shown in Fig. 2. The phenotypes shown are representative of that seen in the family and range from severe clubfoot residuals to short wide feet with short abnormal toes. The hands were normal in all individuals with the CNG.
Figure. 1. Pedigree of clubfoot family.
Filled circles or squares indicate clubfoot and half filled circles or squares designate short, wide feet. WT indicates wild type or two copies and CNG indicates copy number gain or three copies of the chromosome 17q22.3 region.
Figure. 2. Foot anomalies in individuals with microduplication.
Variability of foot anomalies in microduplication individuals is shown for individuals transmitting the microduplication. Severe clubfoot deformity with muscle hypoplasia is seen in V-5, while his siblings (V-6 and V-7), father (VI-9) and grandfather (III-7) have short, wide feet. The great toes are short in those with foot anomalies and hallux varus is present in V-6. The hands are normal. WT indicates wild type or two copies and CNG indicates copy number gain or three copies of the chromosome 17q22.3 region.
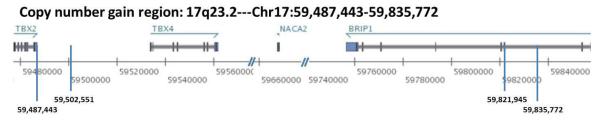
DNA from two relatives, III-10 and V-13, were subjected to chromosomal microarray analysis to define the boundaries of the duplication. As shown in Fig. 3, results of the study showed a copy number gain of chromosome band 17q23.2 spanning at least 0.319 Mb in size from 59502551 to 59821945 and at most 0.348 Mb in size, from 59487443 to 59835772 (HG build 19) and includes complete duplications of TBX4 and NACA2 (nascent polypeptide-associated complex alpha subunit 2) and exons 15 to 20 of BRIP1 (BRCA1 interacting protein C-terminal helicase 1) (NACA2: http://www.genecards.org/cgi-bin/carddisp.pl?gene=NACA2&search=NACA2; BRIP1: http://www.genecards.org/cgi-bin/carddisp.pl?gene=BRIP1&search=BRIP1). The delineated region for the copy number gain is covered by 53 oligonucleotide probes. Although the array used maps to hg18, conversion to hg19 coordinates was done by UCSC conversion for ease of comparison (http://genome.ucsc.edu/cgi-bin/hgLiftOver).
Figure 3. Schematic of chromosome 17q23.2 copy number gain region.
This region, spanning from 59,487,443 to 59,835,772 bps (Genomic coordinates hg19), contains three genes. TBX4 and NACA2 are duplicated; and exons 15 to 20 of BRIP1 are duplicated.
To determine whether variation in TBX4 significantly contributes to isolated clubfoot, we performed linkage and association testing using eight flanking and intragenic SNPs. There was no evidence for linkage in the dataset; these SNPs were uninformative in family 645. No evidence for association was found in the nonHispanic white group. As shown in Table II, only nominal evidence for an association in the Hispanic group was found for rs758596 and rs3785825. No significant alteration of transmission was found by haplotype analysis (data not shown).
Table II.
TBX4 association results
| Hispanics | All Families | Family History | No Family History | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Loc | PDT |
Geno-
PDT |
APL | PDT | APL | PDT | APL |
Geno-
PDT |
APL |
| TBX4 | rs4968564 | P | 0.042 | 0.094 | 0.227 | 0.036 | 0.024 | 0.129 | 0.677 | 0.309 | 0.696 |
| TBX4 | rs758596 | I3 | 0.005 | 0.007 | 0.124 | 0.032 | 0.027 | 0.344 | 0.061 | 0.205 | 0.261 |
| TBX4 | rs35500387 | I4 | 0.884 | 0.893 | 0.091 | 0.552 | 0.769 | 0.812 | 0.198 | 0.301 | 0.040 |
| TBX4 | rs3785825 | I5 | 0.484 | 0.721 | 0.002 | 0.880 | 0.984 | 0.188 | 0.039 | 0.059 | 0.002 |
Loc = location, PDT = Pedigree Disequilibrium Test, Geno-PDT = Genotype Pedigree Disequilibrium Test, APL = Association in the Presence of Linkage
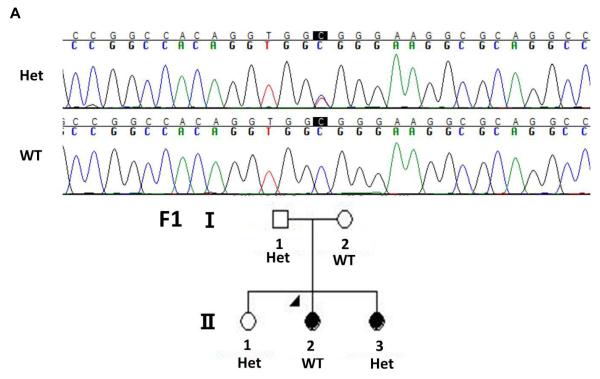
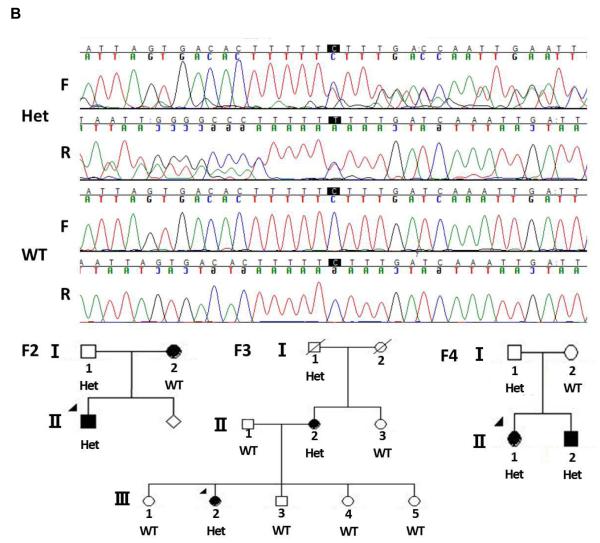
Mutations in limb enhancers have also been shown to cause nonsyndromic limb malformations [VanderMeer and Ahituv, 2011]. To analyze whether mutations in TBX4-associated enhancers could cause clubfoot, we sequenced two known human hindlimb enhancer homologues in the TBX4 genomic region, HLEA and HLEB [Menke et al., 2008], in 95 probands with familial isolated clubfoot. Variants that were identified in the sequences were compared to known human polymorphisms in dbSNP ( http://www.ncbi.nlm.nih.gov/projects/SNP) and known variants were excluded from further analysis. One previously undocumented nucleotide variant was found in the HLEA enhancer, but did not segregate with clubfoot in the proband’s family, suggesting that it is unrelated to the phenotype (Fig. 4A). We found one single base pair deletion in HLEB that was present in three probands. However, as shown in Fig. 4B, this indel also did not segregate with the clubfoot phenotype in the families, suggesting that it is also not associated with clubfoot.
Figure 4. Segregation of hindlimb enhancer variants in clubfoot family members.
A. Chromatogram shows the heterozygote C>T variant in HLEA in Family 1 (F1). This variant does not segregate with the nonsyndromic clubfoot phenotype. B. Chromatograms showing the C deletion in HLEB with both forward and reverse sequences. This polymorphism was present in three families (F2-4) and does not segregate with the nonsyndromic clubfoot phenotype. Het = heterozygote. WT = wild type, F = forward sequence, R = reverse sequence.
DISCUSSION
Nonsyndromic clubfoot is a common orthopedic birth defect that has a multifactorial etiology [Carter, 1965; Carter, 1969] and the genetic underpinnings are just now being delineated. Recently, a microduplication of chromosome 17q23.1q23.2 was found in three probands with a positive family history of clubfoot and was suggested to account for 5% of familial cases of isolated clubfoot [Alvarado et al., 2010]. In the current study, we evaluated 605 probands (148 multiplex and 457 simplex) and identified only one duplication in a single large nonHispanic white family with 16 cases of clubfoot (11 genotyped) and 9 cases (7 genotyped) of other foot anomalies segregating in an autosomal dominant pattern through six generations. Assuming that all individuals with foot anomalies in this family carry the duplication, bilateral clubfoot was the presenting anomaly in 16/25 individuals with the duplication, while the remaining nine people with the duplication had foot anomalies ranging from abnormal toes to short very wide feet. Calf muscle hypoplasia was only present in those with clubfeet. The hands were normal in all individuals with the duplication, which is consistent with the known function of TBX4 as a hindlimb-specific transcription factor [Hasson et al., 2010; Menke et al., 2008].
TBX4 is an important gene in specifying the connective tissue required for muscle and tendon patterning [Hasson et al., 2010]. It is a transcriptional target of paired-like homeodomain 1 (PITX1) and limb initiation is thought to occur through the activation of the Wnt/FGF signaling cascade [Logan and Tabin, 1999; Menke et al., 2008; Takeuchi et al., 2003]. Heterozygous loss-of-function mutations in TBX4 cause small patella syndrome, also known as ischiopatellar dysplasia or Scott-Taor Syndrome (OMIM 147891). Besides small patellae, there is absent, delayed, or irregular ossification of the ischiopubic junctions and increased space between the first and second toes and short fourth and fifth rays of the feet with pes planus. Deletions of 17q23.1q23.2 are well characterized and cause a constellation of anomalies including heart defects and limb anomalies, which do not include clubfoot [Ballif et al., 2010]. In contrast, microduplication of the same region of chromosome 17 is associated with clubfoot and other skeletal anomalies [Alvarado et al., 2010]. The previous study and this study indicate that microduplication of 17q23.1q23.2 can cause a spectrum of foot anomalies ranging from short wide feet to clubfoot. The variable size of the deletion, ranging from 2.2 Mb in the Alvarado et al. families to 350 kb in the present family may determine the extent of other anomalies. Importantly, the Alvarado microduplication families had additional skeletal anomalies involving the pelvis, calcaneous and distal tibial epiphyses. In addition, modifier genes likely influence the severity of the foot anomalies. In the present family, the microduplication involves copy number gain of TBX4 and NACA2 and partial duplication of BRIP1, thus significantly narrowing the critical region for clubfoot. Neither NACA2 nor BRIP1 have a known function in limb development. NACA2 prevents inappropriate targeting of non-secretory polypeptides to the endoplasmic reticulum (ER), most likely by regulating Na/Ca exchange, whereas BRIP1 is required for the maintenance of chromosomal stability [Cantor et al., 2001; Kofuji et al., 1993]. Neither gene has been associated with a developmental anomaly (OMIM). In contrast, TBX4 plays a role in specifying hindlimb formation and is present in the hindlimb by E9.5 in the mouse and continues to be expressed during hindlimb development [Chapman et al., 1996; Naiche and Papaioannou, 2007]. However, it is unclear how overexpression of TBX4 contributes to the foot anomalies observed in this family. Loss of TBX4 during human limb development affects the skeletal elements, and mutations in TBX4 cause a pattern of skeletal abnormalities. While TBX4 plays a role in hindlimb specification, it is not solely responsible for coordinating transcriptional regulation and morphological formation [Chapman et al., 1996; Naiche and Papaioannou, 2007]. PITX1 has recently been shown to also play an important role in hindlimb formation [DeLaurier et al., 2006; Logan and Tabin, 1999]. In this family, the appearance of at least minor foot anomalies in all duplication carriers suggests that overexpression of TBX4 is causing perturbation of foot development.
All four families described in the previous and current study with the 17q23.1q23.2 microduplication are multiplex, i.e. more than one affected family member [Alvarado et al., 2010]. The microduplication was not present in any of our simplex cases, nor in more than 99% of our familial cases. Thus, the microduplication is a rare cause of familial isolated clubfoot and can segregate as an autosomal dominant phenotype. Importantly, our work narrows the critical region for the copy number gain from the 2.2 Mb previously reported, to approximately 350 kb and only three genes. Out of these three genes, TBX4 appears the most likely one responsible for the spectrum of foot anomalies. Moreover, there was no compelling evidence in our dataset for an association between isolated clubfoot and TBX4, nor were any significant variations present in the two known TBX4 hindlimb enhancers sequenced in 95 patients from simplex families. Thus, all familial cases of clubfoot should be tested for this chromosomal duplication but additional studies, such as resequencing of TBX4 in all clubfoot probands, are needed to uncover the cause(s) of isolated clubfoot.
ACKNOWLEDGMENTS
This study was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston. We thank all of the families that kindly participated in this study and made it possible. Thanks to Marie Elena Serna and Rosa Martinez for screening, enrolling and collecting patient samples and to Dr. S. Shahrukh Hashmi for database management. Shriners Hospital for Children and NICHD R01-HD043342-05 supported this work with grants to JTH. NICHD grant number R01HD059862 (N.A.) also supported this research. We would like to thank Dr. Sau-Wai Cheung and the Kleberg Cytogenetics Laboratory at Baylor College of Medicine for their assistance.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Alderman BW, Takahashi ER, LeMier MK. Risk indicators for talipes equinovarus in Washington State, 1987-1989. Epidemiology. 1991;2:289–292. doi: 10.1097/00001648-199107000-00009. [DOI] [PubMed] [Google Scholar]
- Alvarado DM, Aferol H, McCall K, Huang JB, Techy M, Buchan J, Cady J, Gonzales PR, Dobbs MB, Gurnett CA. Familial isolated clubfoot is associated with recurrent chromosome 17q23.1q23.2 microduplications containing TBX4. Am J Hum Genet. 2010;87:154–160. doi: 10.1016/j.ajhg.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalis S, Sairam S, Homfray T, Harrington K, Nicolaides K, Thilaganathan B. Outcome of antenatally diagnosed talipes equinovarus in an unselected obstetric population. Ultrasound Obstet Gynecol. 2002;20:226–229. doi: 10.1046/j.1469-0705.2002.00780.x. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Rosenfeld JA, Traylor RN, Gastier-Foster J, Thrush DL, Astbury C, Bartholomew D, McBride KL, Pyatt RE, Shane K, Smith WE, Banks V, Gallentine WB, Brock P, Rudd MK, Adam MP, Keene JA, Phillips JA, 3rd, Pfotenhauer JP, Gowans GC, Stankiewicz P, Bejjani BA, Shaffer LG. Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet. 2010;86:454–461. doi: 10.1016/j.ajhg.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH, Yang Y, Wiszniewska J, Nowakowska BA, del Gaudio D, Xia Z, Simpson-Patel G, Immken LL, Gibson JB, Tsai AC, Bowers JA, Reimschisel TE, Schaaf CP, Potocki L, Scaglia F, Gambin T, Sykulski M, Bartnik M, Derwinska K, Wisniowiecka-Kowalnik B, Lalani SR, Probst FJ, Bi W, Beaudet AL, Patel A, Lupski JR, Cheung SW, Stankiewicz P. Detection of clinically relevant exonic copy-number changes by array CGH. Human Mutation. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am J Hum Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality-- and tolerance of segmental aneuploidy--in humans. Am J Hum Genet. 1999;64:1702–1708. doi: 10.1086/302410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Carter CO. The inheritance of common congenital malformations. Prog Med Genet. 1965;5:59–84. [PubMed] [Google Scholar]
- Carter CO. Genetics of common disorders. Brit Med Bull. 1969;25:52–57. doi: 10.1093/oxfordjournals.bmb.a070671. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Developmental dynamics : an official publication of the American Association of Anatomists. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ching GH, Chung CS, Nemechek RW. Genetic and epidemiological studies of clubfoot in Hawaii: ascertainment and incidence. Am J Hum Genet. 1969;21:566–580. [PMC free article] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- de Andrade M, Barnholtz JS, Amos CI, Lochmiller C, Scott A, Risman M, Hecht JT. Segregation analysis of idiopathic talipes equinovarus in a Texan population. Am J Med Genet. 1998;79:97–102. doi: 10.1002/(sici)1096-8628(19980901)79:2<97::aid-ajmg4>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- DeLaurier A, Schweitzer R, Logan M. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Developmental biology. 2006;299:22–34. doi: 10.1016/j.ydbio.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Ester AR, Tyerman G, Wise CA, Blanton SH, Hecht JT. Apoptotic gene analysis in idiopathic talipes equinovarus (clubfoot) Clin Orthop Relat Res. 2007;462:32–37. doi: 10.1097/BLO.0b013e318073c2d9. [DOI] [PubMed] [Google Scholar]
- Ester AR, Weymouth KS, Burt A, Wise CA, Scott A, Gurnett CA, Dobbs MB, Blanton SH, Hecht JT. Altered transmission of HOX and apoptotic SNPs identify a potential common pathway for clubfoot. Am J Med Genet A. 2009;149A(12):2745–2752. doi: 10.1002/ajmg.a.33130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnett CA, Boehm S, Connolly A, Reimschisel T, Dobbs MB. Impact of congenital talipes equinovarus etiology on treatment outcomes. Dev Med Child Neurol. 2008;50:498–502. doi: 10.1111/j.1469-8749.2008.03016.x. [DOI] [PubMed] [Google Scholar]
- Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, Mohun TJ, Logan MP. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev Cell. 2010;18:148–156. doi: 10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AL, Bray MS, Scott A, Blanton SH, Hecht JT. Variation in CASP10 gene is associated with idiopathic talipes equinovarus. J Pediatr Orthop. 2005;25:598–602. doi: 10.1097/01.bpo.0000173248.96936.90. [DOI] [PubMed] [Google Scholar]
- Honein MA, Paulozzi LJ, Moore CA. Family history, maternal smoking, and clubfoot: an indication of a gene-environment interaction. Am J Epidemiol. 2000;152:658–665. doi: 10.1093/aje/152.7.658. [DOI] [PubMed] [Google Scholar]
- Kofuji P, Lederer WJ, Schulze DH. Na/Ca exchanger isoforms expressed in kidney. The American journal of physiology. 1993;265(4 Pt 2):F598–603. doi: 10.1152/ajprenal.1993.265.4.F598. [DOI] [PubMed] [Google Scholar]
- Logan M, Tabin CJ. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- Moorthi RN, Hashmi SS, Langois P, Canfield M, Waller DK, Hecht JT. Idiopathic talipes equinovarus (ITEV) (clubfeet) in Texas. Am J Med Genet A. 2005;132:376–380. doi: 10.1002/ajmg.a.30505. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Dietz FR, Murray JC, Buetow KH. A single-gene explanation for the probability of having idiopathic talipes equinovarus. Am J Hum Genet. 1993;53:1051–1063. [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Suzuki T, Kamimura M, Ogura K, Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130:2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- VanderMeer JE, Ahituv N. cis-regulatory mutations are a genetic cause of human limb malformations. Developmental dynamics : an official publication of the American Association of Anatomists. 2011;240:920–930. doi: 10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymouth KS, Blanton SH, Bamshad MJ, Beck AE, Alvarez C, Richards S, Gurnett CA, Dobbs MB, Barnes D, Mitchell LE, Hecht JT. Variants in genes that encode muscle contractile proteins influence risk for isolated clubfoot. Am J Med Genet Part A. 2011;155:2170–2179. doi: 10.1002/ajmg.a.34167. [DOI] [PMC free article] [PubMed] [Google Scholar]