Abstract
Paramagnetic gadolinium(III) complexes are widely used to increase contrast in magnetic resonance (MR) images. Contrast enhancement depends on the concentration of the gadolinium complex and on its relaxivity, an inherent property of the complex. Increased relaxivity results in greater image contrast or the ability to detect the contrast agent at a lower concentration. Increasing relaxivity enables imaging of abundant molecular targets.
Relaxivity depends on the structure of the complex, kinetics of inner-sphere and second sphere water exchange, and on the rotational dynamics of the molecule. The latter, and in some cases the former, properties of the complex change when it is bound to its target. All of these properties can be rationally tuned to enhance relaxivitry. In this Microreview we summarize our efforts in understanding and optimizing the relaxivity of contrast agents targeted to serum albumin and to fibrin.
Keywords: gadolinium, relaxivity, serum albumin, fibrin, water exchange
Introduction
Magnetic resonance imaging (MRI) is a mainstay of diagnostic radiology, and a substantial fraction of MRI procedures employ a contrast agent. Commercial contrast agents are discrete Gd(III) complexes of octadentate ligands.[1] Contrast agents are most commonly used in conjunction with T1-weighted MR scans. In T1-weighted imaging, tissue where the T1 is short, e.g. fat, appears bright. The Gd(III) complex serves to shorten the T1 of nearby water protons resulting in areas of signal enhancement corresponding to distribution of the complex.
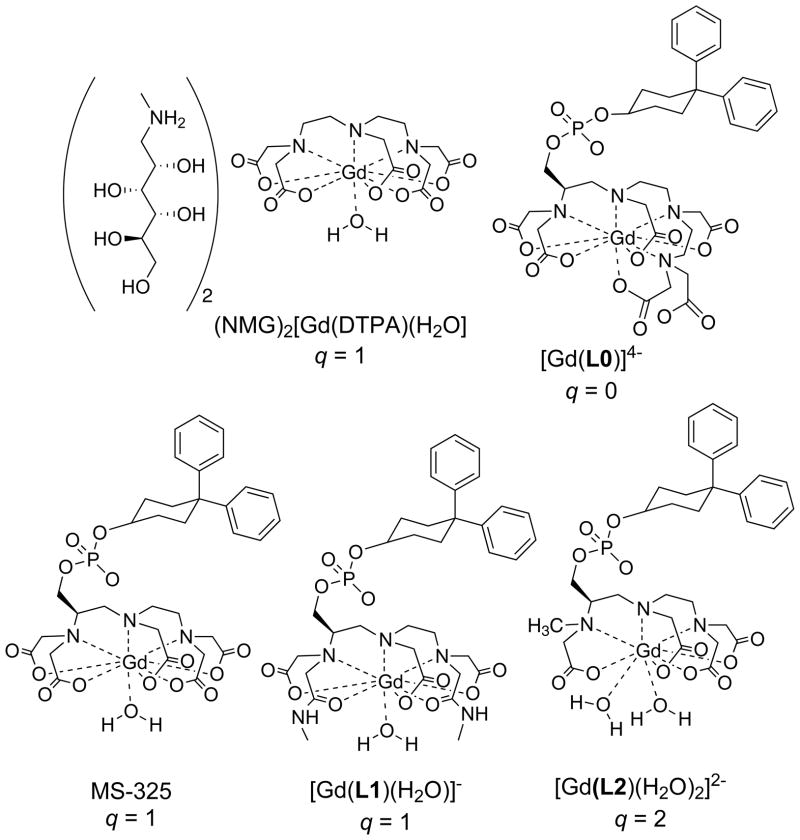
The majority of contrast-enhanced MRI procedures utilize simple hydrophilic complexes that have no active targeting component. For instance NMG2[Gd(DTPA)(H2O)] (Figure 1) has been approved for clinical use since 1988. Although non-specific, these extracellular markers are very useful in identifying and characterizing tumors because of the altered vascular physiology associated with tumors.
Figure 1.
Commercially approved contrast agents NMG2[Gd(DTPA(H2O)] and MS-325, and related serum albumin targeted complexes.
More recently the Gd(DTPA) derivative MS-325 (gadofosveset, Figure 1) has been approved for blood vessel imaging.[2] This complex targets serum albumin in the blood. Non-covalent binding to this blood protein restricts the distribution of the complex to the blood vessels, while the small unbound fraction is continually excreted via the kidneys.[3] Following this initial targeted agent approach, an agent targeting fibrin for imaging blood clots has entered human clinical trials.[4]
Preclinically there are examples of contrast agents targeting collagen for fibrosis imaging,[5] extracellular DNA for necrosis imaging,[6] the folate receptor and the estrogen receptor in some cancers,[7] integrins associated with angiogenesis,[8] and macrophages associated with inflammatory processes[9] to list but a few applications.
An inherent challenge in targeted MR contrast agents is sensitivity of detection. For T1-weighted imaging, the contrast agent must change R1 (R1=1/T1) by an appreciable amount. The intrinsic ability of a complex to change R1 is termed relaxivity, r1 (lowercase r, with units mM−1s−1). To a first approximation, the in vivo R1 is given by equation 1:
| (1) |
Here R10 is the relaxation rate of the tissue in the absence of the contrast agent and [CA] is the concentration of the contrast agent. R10 varies with tissue type and with applied field but is in the range 0.5 – 2 s−1 for most tissues and tumors.[10] Under the most favourable conditions of a low baseline R10 and a very small R1 change of 10%, one still requires 10 μM of contrast agent with a relaxivity of 5 mM−1s−1. In the absence of some sort of amplification mechanism, this means that the target biomolecule must be present at >10 μM. This rules out imaging therapeutic targets like cell surface receptors and enzymes as their concentrations are typically in the low nanomolar range.
To address this sensitivity challenge, two main approaches are used. The first is to employ a targeted nanoparticle that carries a payload of 100’s to 1000’s of paramagnetic ions. This has been successfully employed in a number of preclinical models, e.g. targeting annexin V in apoptosis,[11] macrophage receptors in atherosclerotic plaques,[9a] or integrins associated with angiogenesis,[8b, 8c] to name a few. However nanoparticles are usually restricted to vascular targets; they have a long blood half-life resulting in delays before high target:background ratios are reached; and they often suffer from incomplete elimination of the potentially toxic gadolinium ion.[12] An alternate approach is to use discrete compounds with a targeting moiety such as a peptide conjugated to one or more gadolinium chelates. Discrete complexes can rapidly access the interstitial space and localize at the target of interest. The small size insures rapid elimination through the kidneys. However a key hurdle to overcome is the relatively low sensitivity of these compounds.
We and others have focused on biologically relevant, but abundant targets such as serum albumin,[13] fibrin,[4a, 14] type I collagen,[5] and extracellular DNA associated with necrosis.[6] Analogous to therapeutics, targeted contrast agents require optimization of affinity and specificity for the target biomolecule as well as favorable pharmacokinetic and metabolic stability properties. However since relaxivity is an inherent and tunable property of the compound, we also focus on optimization of relaxivity. In this Microreview we summarize our efforts in understanding and optimizing the relaxivity of contrast agents targeted to serum albumin and to fibrin.
Molecular determinants of relaxivity
Contrast agents act as magnetic catalysts that shorten the relaxation time of bulk water protons. As the paramagnetic complex randomly reorients in solution it creates a fluctuating magnetic field, and this fluctuating field causes relaxation of nearby water protons. This dipolar effect depends on the Gd-H distance (rGdH) to the inverse 6th power and thus is most efficient for water molecules directly coordinated to the Gd(III) ion (inner-sphere water relaxivity, r1IS), but water molecules in the second and outer coordination spheres also provide a significant role in overall relaxivity (r1SS and r1OS).[1, 15] The number of water molecules hydrating the complex (q = number of inner-sphere water ligands) and their mean residency times (τm) also impact relaxivity. The residency time should be long enough to increase the probability of relaxation to occur, but short enough such that the relaxed water molecule undergoes efficient exchange with the bulk. The fluctuating field can be established via three mechanisms: 1) rotational diffusion of the complex (1/τR); 2) thermal transitions of the unpaired electrons of Gd(III) between excited and ground states (1/T1e); and 3) the water molecule approaching and departing the complex, i.e. water exchange (1/τm). The fastest of these processes will dominate. This qualitative description is summarized by equations 2–4 which describe relaxivity for an isotropic system at proton Larmor frequencies above 10 MHz, which is where almost all MRI scanners operate. Here, T1m refers to the T1 of the coordinated water proton, ω refers to the proton Larmor frequency and all other symbols have their usual meanings.
| (2) |
| (3) |
| (4) |
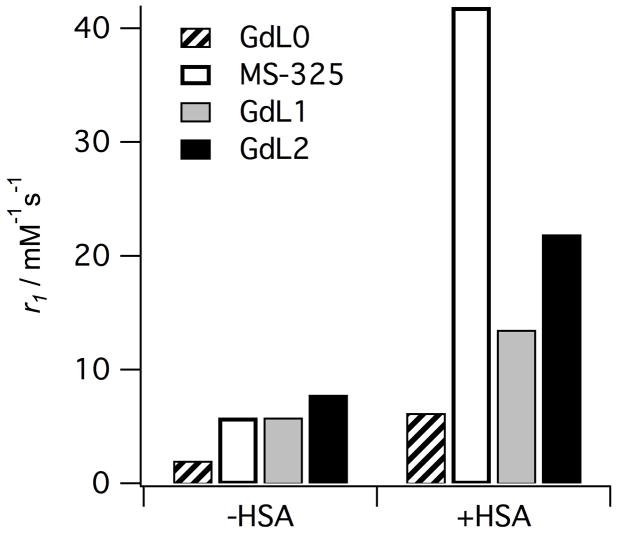
What is apparent from equation 2 is that relaxivity will increase if the number of inner-sphere water molecules (q) increases, or if the term (T1m + τm) is decreased. Figure 2 shows relaxivities for four complexes (Figure 1) measured in the absence and presence of human serum albumin (HSA). In the absence of HSA, we see that relaxivity increases with the number of inner-sphere water molecules, as expected. Note that in the absence of protein that GdL0 has a relaxivity about 1/3 that of MS-325 despite having no inner-sphere water molecule, demonstrating the effect of second- and outer-sphere relaxivity.[2a] Each complex contains the same biphenylcyclohexyl phosphodiester albumin binding group and this results in >85% of the complex bound to HSA under the conditions of the measurement.[1a, 2a, 16] In HSA solution, all the relaxivities increase relative to the values measured in buffer without HSA, but now there are large differences among the compounds and relaxivity no longer correlates with q. It was demonstrated that the differences in relaxivity among HSA-bound MS-325, GdL1, and GdL2 could be attributed to differences in water exchange rates among the complexes.[1a, 2a, 16]
Figure 2.
Relaxivities (r1) of albumin-targeted contrast agents with 0, 1, or 2 inner-sphere water ligands (q) measured in the absence (−HSA) and presence (+HSA) of human serum albumin at 0.47T, 37 °C, pH 7.4.
In the absence of protein, complexes all rotate in solution at about the same rate since they are of similar size. However this rotational rate is on the order of GHz and thus relaxation of water protons at 20 MHz is not very efficient, i.e. T1m is long. Under these conditions, T1m > τm and it is tumbling rate and hydration number that determine relaxivity. Upon binding to HSA, the rotational rate slows down by orders of magnitude and relaxation of the inner-sphere water is much more efficient. Under these conditions, T1m is reduced such that τm > T1m and slow water exchange can limit relaxivity. For GdL1, τm is about 10 times longer than in MS-325.[1a, 2a, 13a] The longer τm has little effect on relaxivity in the absence of protein, but upon binding to HSA, the relaxivity of GdL1 is muted. GdL2 suffers the same problem. It was demonstrated that the water exchange at GdL2 is 16 times slower than at MS-325, and as a result the relaxivity of GdL2 in HSA solution is only about half that of MS-325 despite the former having two inner-sphere water molecules.[16]
The foregoing example illustrates the challenge of optimizing relaxivity. Relaxivity measured in the absence of the target biomolecule is not predictive of the protein-bound relaxivity. The target biomolecule can also be non-innocent in its effect on hydration number and water exchange rate. For instance, it was shown by luminescence lifetime studies and by ENDOR spectroscopy that in some cases a protein side chain can coordinate the Gd(III) ion and displace inner-sphere water resulting in lower relaxivity.[17] It has also been shown that the water exchange rate can be altered when the complex is bound to the protein.[18] Thus it is imperative to optimize relaxivity for the protein-bound complex, and not to assume that molecular factors measured in the absence of protein, like water exchange and hydration number, will be unchanged when the complex is protein bound.
Structure – relaxivity relationships among HSA targeted complexes
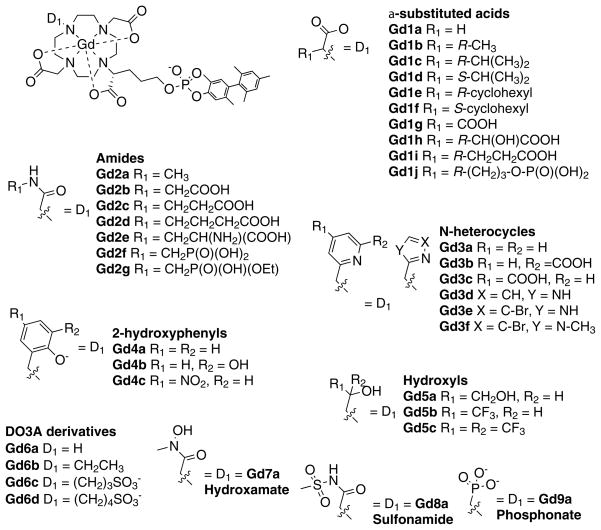
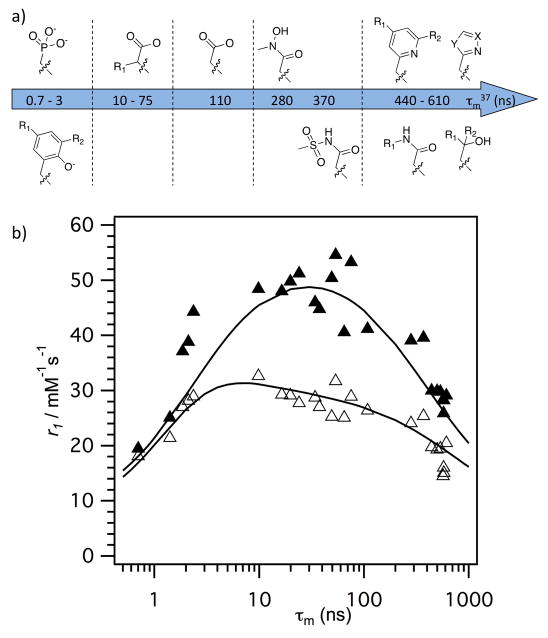
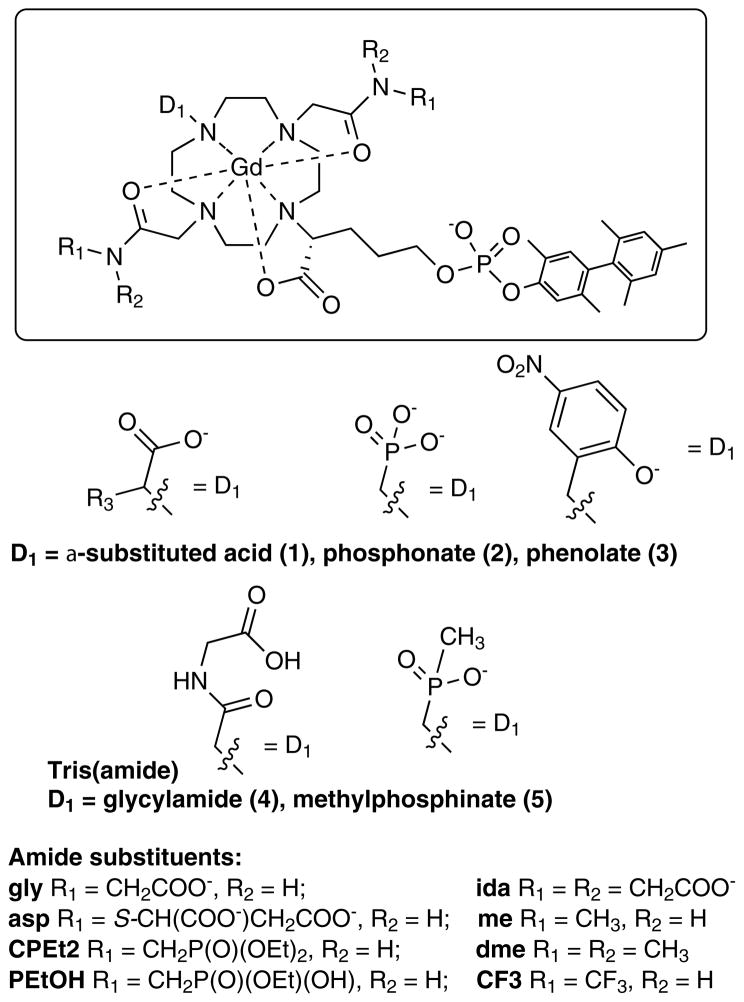
We investigated the effect of the donor group for a series of 38 DOTA derivatives targeted to HSA.[19] DOTA derivatives are known to be kinetically inert with respect to Gd(III) loss.[20] We prepared complexes with a common HSA binding group and varied only one donor group on the ligand (Figure 3). By keeping the HSA binding group constant, we anticipated that the rotational dynamics would be similar for all complexes. This pentamethyl biphenyl moiety imparts high affinity for HSA:[13c] in all complexes, high affinity to HSA was observed with >98% of the complex bound to HSA under the conditions 0.1 mM Gd and 0.67 mM HSA.[19] We measured r1 in the presence and absence of HSA at two field strengths. Variable temperature 1H NMRD and 17O NMR were also used to characterize the complexes. Changing a single donor atom resulted in changes in water exchange rates ranging across 3 orders of magnitude. Donor groups increased water exchange rate in the order: phosphonate > phenolate > α-substituted acetate > acetate > hydroxamate > sulfonamide > amide > pyridyl > imidazole. Relaxivites at 0.47 T, 37 °C, ranged from 12.3 to 55.6 mM−1s−1. Figure 4 shows the observed relaxivities as a function of inner-sphere water exchange rate and demonstrates that there is an optimum range for water exchange, as predicted from theory. The highest relaxivities were observed when the donor group was an α-substituted acetate.
Figure 3.
Library of serum-albumin targeted complexes designed to study the effect of donor atom on relaxivity, inner-sphere water exchange, and electronic relaxation.
Figure 4.
a) Effect of donor group on water residency time (see Figure 3). b) Correlation between observed relaxivity (0.47T = solid symbols; 1.41T = open symbols) and water residency time (τm) at 37 °C for the library of compounds shown in Figure 3, demonstrating that the optimal water residency time is between 10–100 ns at 0.47T but much broader at 1.41T. Solid lines are simulations using the mean values for rotational diffusion and electronic relaxation for all compounds.
Slowing down rotational motion also reveals fast electronic relaxation as limiting relaxivity, see equation 4. Electronic relaxation becomes slower with increasing field strength and is not usually a limiting factor for relaxivity at 1.5T and higher fields. In our study we found that electronic relaxation was slowest for the acetate derivatives, which gave the most symmetric ligand field. As expected, this had the largest positive relaxivity effect at lower fields like 0.47T.
While differences in water exchange rate accounted for most of the observed differences in relaxivity for the compounds in Figure 3, we also noted significant variations in r1 among compounds with the same donor set. For instance in the amide series, the relaxivity of Gd2b with a pendant carboxylate (R1 = CH2COOH) was 30% higher than for the methylamide Gd2a. Increasing the alkyl chain by 1 or 2 carbons (Gd2c or Gd2d) did not result in increased relaxivity, however the methylphosphonate derivatives Gd2f and Gd2g did show this increased relaxivity. We speculated that the increased relaxivity of Gd2b was a result of an increased second-sphere relaxivity wherein the pendant caroboxylate hydrogen bonded to, and stabilized a second sphere water molecule.
The amide pendant groups offered an opportunity to increase 2nd sphere relaxivity, but the amide donors served to slow inner-sphere water exchange and decrease inner-sphere relaxivity. We then sought to determine whether the inner-sphere water exchange rate could be predictably tuned, i.e. if the effect of the amide on water exchange could be offset by adding labilizing donors like phosphonate oxygen. In combination, we attempted to further boost second-sphere relaxivity by varying the amide substituents. Twenty GdDOTA derivatives (Figure 5) were prepared and their relaxivity determined in presence and absence of human serum albumin as a function of temperature and magnetic field.[21] Each compound had a common albumin-binding group and an inner-sphere donor set comprising the 4 tertiary amine N atoms from cyclen, an α-substituted acetate oxygen atom, 2 amide oxygen atoms, an inner-sphere water oxygen atom, and a variable donor group. Each amide nitrogen was substituted with different groups to promote hydrogen bonding with second-sphere water molecules (Figure 5). We found that the reduction of inner-sphere water exchange typical of amide donor groups could indeed be offset by incorporating a phosphonate or phenolate oxygen atom donor in the first coordination sphere, resulting in higher relaxivity. For instance, in the series where the amide was derived from glycine (gly), there was a pronounced effect of donor group. Relaxivity at 0.47T, 37 °C increased from 16.0 to 20.3 to 26.3 to 39.4 to 53.4 mM−1s−1 as D1 was varied from amide (Gd4-gly) to unsubstituted acetate (Gd1a-gly) to α-substituted acetate (Gd1b-gly, τm=290 ns) to phosphonate (Gd2-gly, τm=4.5 ns) to phenolate (Gd3-gly,τm=9.5 ns), respectively. This was clearly an inner-sphere water exchange effect. When D1 was phosphonate, the water exchange rate was too fast at 37 °C and actually limited relaxivity; at lower temperatures relaxivities >70 mM−1s−1 were observed. In general, we found that in a DOTA-like ligand, when two acetate donors were converted to amides, the water exchange rate decreased by a factor of 10. Overall, relativities at 0.47 and 1.4 T, 37 °C, in HSA ranged from 16.0 to 58.1 and from 12.3 to 34.8 mM−1s−1, respectively.
Figure 5.
Library of compounds designed to optimize both inner- and second-sphere relaxivity.
Amide nitrogen substitution for the compounds in Figure 5 was shown to have a profound effect on relaxivity. Pendant phosphonate or carboxylate groups increased relaxivity by as much as 88% compared with the N-methyl amide analog. For instance when D1 was fixed as phosphonate, the relaxivity of the N-methyl derivative was in HSA solution was 27.1, increasing to 39.4 for Gd2-gly, and further to 45.4 mM−1s−1 for Gd2-ida (additional H-bond acceptor). For these complexes it was shown that inner-sphere water exchange did not vary and differences in r1 were attributed to a second-sphere effect. Second-sphere relaxivity alone contributed as much as 24 and 14 mM−1s−1 at 0.47 and 1.4 T, respectively.
Structure – relaxivity relationships among fibrin targeted complexes
Fibrin is a principal component of blood clots and is established as a useful target for imaging thrombosis, implicated in diseases such as coronary artery disease, heart attack, stroke, atrial fibrillation, pulmonary embolism, and deep vein thrombosis.[22] Fibrin is a useful MR target because of its high (μM) concentrations in clots.[23]
Our work in targeting fibrin utilized two different classes of fibrin-binding cyclic disulfide bridged peptides, TN6 (XnCPYGLCXm) and TN7 (XnCXYYGTCXm). They differ in the size of the central ring (TN6: six amino acids; TN7: seven amino acids) as well as the nature of the amino acids. Fibrin affinity was further optimized via unnatural amino acid substitutions and from these peptide leads, different Gd-chelate:peptide conjugates were prepared. It was anticipated that several Gd moities would be required for robust clot detection.
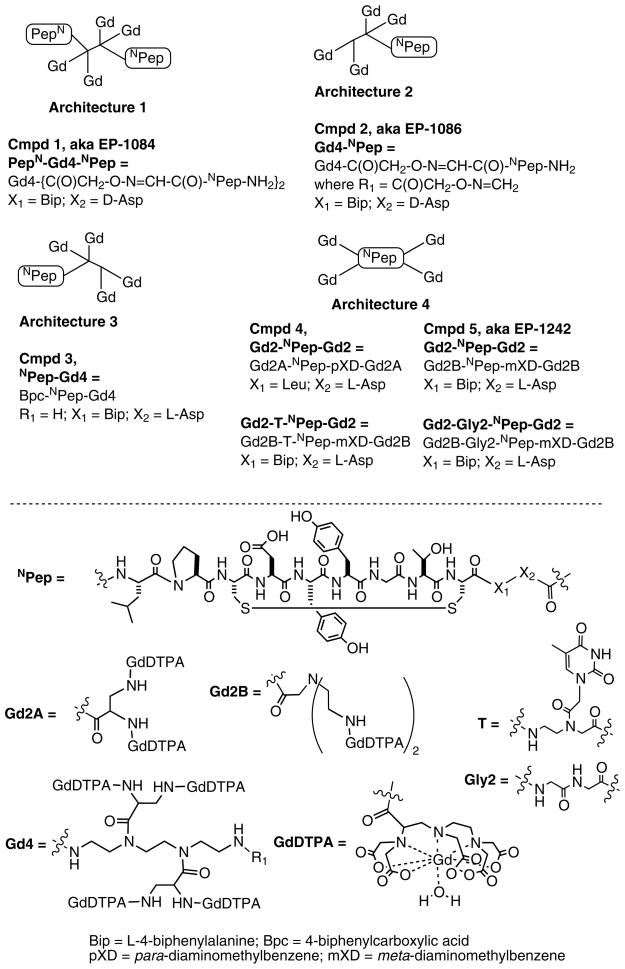
We have reported 7 contrast agents reported based on the TN7 peptide elaborated with 4 GdDTPA moieties, Figure 6.[14a, 14c, 14d] These can be described by four different peptide-chelate conjugate archtypes. Nair et al. used a bifunctional tetrameric GdDTPA scaffold based on triethylenetetraamine (trien) [14a]. The trien scaffold is easily differentiated using the l-(4,4′-dimethyl-2,6-dioxocyclohexylidene) ethyl (Dde) protecting group to selectively protect the primary amines. Two DTPA moieties were attached to the secondary nitrogens by a short diaminopropionate linker to minimize rotational flexibility and enhance relaxivity, while the primary amines were used to introduce the hydroxylamine groups. A serine residue was incorporated at the N-terminus of the peptide and mild periodate oxidation gave an N-terminal a-keto aldehyde. Chemoselective ligation between the hydroxylamine and α-keto aldehyde was carried out at pH 4.6 in water to form oxime bonds, which gave the bivalent conjugate. Complexation with GdCl3 gave EP-1084 (PepN-Gd4-NPep) which showed good fibrin affinity (low Ki value, see Table 1) and specificity, high relaxivity, and positive uptake in an in vivo venous thrombus model. The compact structure of the Gd4 scaffold, coupled with the very short linker between the scaffold and the Gd(DTPA) moiety limited the extent of internal motion within the molecule and resulted in high relaxivity at 0.47 and 1.5T (Table 1).
Figure 6.
Fibrin-targeted TN7 peptide-GdDTPA conjugates designed for imaging blood clots.
Table 1.
Per Gd relaxivities and DD(E) affinities for fibrin-targeted contrast agents shown in Figure 6. Per molecule relaxivities are four times higher. Relaxivities determined in Tris buffer or bound to human fibrin at 37 °C, 1.5 T. Inhibition constants, Ki, to soluble fibrin fragment DD(E). NR=not reported.
| Compound | r1 Tris Buffer | r1 fibrin | % Increase | Ki DD(E) (μM) |
|---|---|---|---|---|
| PepN-Gd4-NPep | 18.2 | 23.1 | 27 | 1.1 |
| Gd4-NPep | 15.5 | 20.7 | 33 | 4.7 |
| NPep-Gd4 | NR | NR | NR | 9.1 |
| Gd2A-NPep-Gd2A | NR | NR | NR | 12.0 |
| Gd2B-NPep-Gd2B | 13.0 | 18.0 | 37 | 4.7 |
| Gd2-Gly2-NPep-Gd2 | 12.4 | 17.8 | 37 | 4.0 |
| Gd2-T-NPep-Gd2 | 13.3 | 28.0 | 114 | 3.5 |
However, subsequent in vivo studies revealed that this compound was being metabolized over the course of the study, and that the gadolinium-containing metabolites no longer bound fibrin. While EP-1084 showed in vivo proof of concept, it was a rather complex molecule and subject to degradation by endogenous peptidases. Zhang et al. addressed the problem of metabolic instability by using the large hydrophilic Gd-chelates to block peptidase activity [14d]. They prepared three different types of conjugates, Gd4-NPep, Bpc-NPep-Gd4 and Gd2-NPep-Gd2, see Figure 6. These compounds, along with the parent peptides were examined in a rat liver homogenate assay. Liver homogenate consists of a mixture of proteases and other enzymes, and represents a harsh challenge to compound stability. The peptides alone, with or without the biphenylalanine (Bip) or D-Asp substitutions, were completely metabolized within the time taken for measurement, t1/2 < 2 min. The bulky hydrophilic Gd(DTPA) tetramer at the N-terminus appeared to block metabolism. This was supported by studies with single peptide analog Gd4-NPep. The half-life of this compound was similar to PepN-Gd4-NPep (10.0 min), suggesting that blockading of the C-terminus might be required for enhanced stability. Bpc-NPep-Gd4 was synthesized with the gadolinium tetramer conjugated directly to the C-terminus and the N-terminus capped with biphenyl carboxamide (Bpc), a group used to block exopeptidase activity. This compound also had a similar half-life in rat liver homogenate (9 min). However capping both the C- and N-termini with a Gd(DTPA) dimeric unit (Gd2A-NPep-Gd2A), resulted in a six- to seven-fold increase in metabolic stability. This suggests that while the D-amino acid and unnatural Bip in Gd4-NPep, and the Bpc in NPep-Gd4, may increase stability, these substitutions are not as effective as using the metal chelate with this specific peptide. While the metabolic stability was improved with compound Gd2A-NPep-Gd2A, unfortunately, its fibrin affinity was significantly reduced (Table 1). However by replacing the N-terminal leucine residue with biphenylalanine to give Gd2B-NPep-Gd2B the fibrin affinity was increased 3-fold and the metabolic stability was retained.
The relaxivity of these compounds was strongly dependent on peptide-chelate architecture. The compact GdDTPA tetramer scaffold used in 1 and 2 resulted in high per Gd relaxivities that increased further when the compounds were bound to fibrin (Table 1). On the other hand, the Gd2-NPep-Gd2 scaffold, while addressing the metabolic stability issue, resulted in lower relaxivity (Table 1) and NMRD analysis suggested that this was due to internal motion that remained after the compound binds to fibrin [14d]. Essentially, the inherent flexibility in the molecule offsets the reduction in tumbling rate upon protein binding.
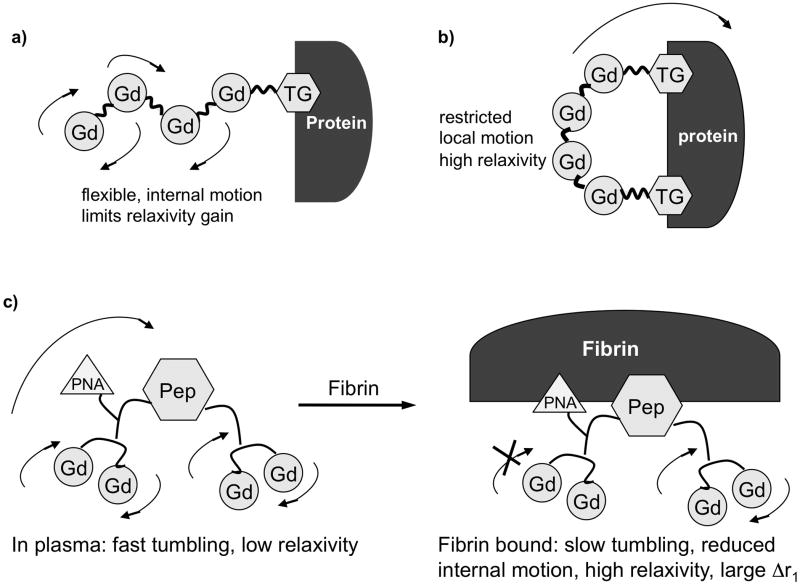
Using a rigid Gd-based multimer such as recently reported,[24] should increase relaxivity, but this relaxivity gain would occur in both the bound and unbound forms of the contrast agent. The ideal targeted agent should have low relaxivity in the unbound state and high relaxivity upon binding to its target. Another approach is to have a flexible complex (low relaxivity) that rigidifies upon binding to the target (high relaxivity). We previously showed that the relaxivity of targeted Gd-based multimers could be increased by introducing two binding moieties that serve to rigidify the molecule upon protein binding. This was successfully applied in a serum albumin-targeted gadolinium tetramer where it was demonstrated that a tetramer with two binding groups resulted in ca. 50% higher relaxivity than the analogous tetramer with one binding group when relaxivity was measured in albumin solution (Figure 7A and 7B).[25] This strategy has also been used with monomeric Gd complexes.[26]
Figure 7.
Strategies for controlling rotational dynamics of mutimeric, targeted contrast agents. A) a single targeting group results in significant internal motion when the complex is bound to the protein, resulting in decreased relaxivity. B) Binding through two sites rigidifies the complex at the target, minimizes internal motion and increases relaxivity. C) Heteroditopic approach to control dynamics: incorporate second, low affinity binding moiety into molecule to anchor complex at the target and increase relaxivity.
The albumin example benefited from the known proclivity of albumin to bind multiple copies of lipophilic anions.[2a, 27] Application of a dual binding approach in the fibrin system is more challenging. The peptides bind to two discrete binding sites per fibrin monomer, but the distance between these two binding sites is not known, and so a dual peptide contrast agent would need to have the linkage between the peptides optimized. This results in a very complex molecule, whose unbound relaxivity will be further increased from the shear size of the molecule (see e.g. PepN-Gd4-NPep, Table 1). We took a heteroditopic approach to this problem. In analogy to the fragment based drug discovery paradigm, we reasoned that there may be additional weak pockets on fibrin near the site where the peptide binds. We screened a series of peptides with further derivatization at the N-terminus and found that insertion of the peptide nucleic acid (PNA) moiety based on thymine showed a modest increase in fibrin affinity.[14c] We prepared the contrast agent Gd2-T-NPep-Gd2 and found that when this compound bound to fibrin, the relaxivity was increased by 50% compared to a control compound lacking the thymine moiety. This suggested that the interaction of the thymine with fibrin served to rigidify the N-terminus of the molecule and enhance relaxivity. Of further distinction was the large increase in relaxivity in going from the unbound to the protein bound state (114% increase at 1.5 T) for the PNA derivative compared to a 30–40% increase for the other compounds in Table 1.
The PNA moiety increases molecular weight by 3% but increases relaxivity by 50% compared to the control Gd2-Gly2-Pep-Gd2. The effect of the PNA group on relaxivity is the equivalent of synthesizing an agent with 6 GdDTPA moieties to achieve equivalent relaxivity. The PNA group has a modest positive impact on fibrin binding but serves to rigidify the N-terminal portion of the molecule upon fibrin binding. Importantly, the PNA group does not increase non-specific protein binding. As a result, relaxivity of Gd2-T-Pep-Gd2 bound to fibrin is more than 50% increased compared to Gd2-Pep-Gd2 while the relaxivity of the two compounds in plasma is comparable. This should result in much greater clot:blood contrast for Gd2-T-Pep-Gd2.
Conclusions
The theoretical understanding of relaxivity is very mature and this is now complimented by rational design principles to create high relaxivity contrast agents. Inner-sphere water exchange rates can be controlled by judicious use of donor groups to increase protein-bound relaxivity. Here we showed that water exchange can be modulated in a predictable manner. The second-hydration sphere can also be tuned to augment relaxivity further. Control of rotational dynamics is a particularly powerful lever for increasing relaxivity. The dual binder strategies described here for serum albumin- and fibrin-targeted agents are generalizable to other targeted contrast agents.
Acknowledgments
This work was supported in part by awards R01EB009062 and R01EB009062-S1 from the National Institute of Biomedical Imaging and Bioengineering and R01CA161221 from the National Cancer Institute.
Biographies

Peter Caravan received his B.Sc. in chemistry from Acadia University in Nova Scotia, Canada, and a Ph.D. degree at the University of British Columbia under the mentorship of Professor Chris Orvig. Following post-doctoral work at the Université de Lausanne with Professor André Merbach, he joined Epix Medical in Cambridge, Massachusetts where he worked on the development of targeted MRI contrast agents. In 2007, he joined the faculty of Radiology at Harvard Medical School and Massachusetts General Hospital where he is currently an Assistant Professor. His research interests include the development of PET, MR, and multimodal imaging probes and their application in the study of cardiovascular disease, fibrosis, and the tumor microenvironment.

Zhaoda Zhang received his B.E. and M.S. in polymer engineering and chemistry at Beijing institute of chemical technology. He received his Ph.D. in organic chemistry with Proessor. Iwao Ojima from SUNY at Stony Brook in 1990. He continued post-doctoral work with Professor Ojima and then with Professor Frances Johnson at Stony Brook. Since 1994 he has worked in the biotech and pharmaceutical industry, conducting discovery research on antisense oligonucleotide therapeutics, MRI contrast agents, and small molecule drugs targeting GPCRs such as S1P1, P2Y2 and mGluR5. He joined MGH as a faculty in 2009 and his current research focuses on developing new MRI contrast agents as well as PET tracers and their applications in functional and molecular imaging.
Footnotes
Dedicated to Chateau Latour on the occasion of the 50th anniversary of their outstanding 1961 vintage.
References
- 1.a) Caravan P. Chem Soc Rev. 2006;35:512. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]; b) Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Chem Rev. 1999;99:2293. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 2.a) Caravan P, Cloutier NJ, Greenfield MT, McDermid SA, Dunham SU, Bulte JW, Amedio JC, Jr, Looby RJ, Supkowski RM, Horrocks WD, Jr, McMurry TJ, Lauffer RB. J Am Chem Soc. 2002;124:3152. doi: 10.1021/ja017168k. [DOI] [PubMed] [Google Scholar]; b) Goyen M, Edelman M, Perreault P, O’Riordan E, Bertoni H, Taylor J, Siragusa D, Sharafuddin M, Mohler ER, 3rd, Breger R, Yucel EK, Shamsi K, Weisskoff RM. Radiology. 2005;236:825. doi: 10.1148/radiol.2363040577. [DOI] [PubMed] [Google Scholar]
- 3.Lauffer RB, Parmelee DJ, Dunham SU, Ouellet HS, Dolan RP, Witte S, McMurry TJ, Walovitch RC. Radiology. 1998;207:529. doi: 10.1148/radiology.207.2.9577506. [DOI] [PubMed] [Google Scholar]
- 4.a) Overoye-Chan K, Koerner S, Looby RJ, Kolodziej AF, Zech SG, Deng Q, Chasse JM, McMurry TJ, Caravan P. J Am Chem Soc. 2008;130:6025. doi: 10.1021/ja800834y. [DOI] [PubMed] [Google Scholar]; b) Vymazal J, Spuentrup E, Cardenas-Molina G, Wiethoff AJ, Hartmann MG, Caravan P, Parsons EC., Jr Invest Radiol. 2009;44:697. doi: 10.1097/RLI.0b013e3181b092a7. [DOI] [PubMed] [Google Scholar]
- 5.a) Caravan P, Das B, Dumas S, Epstein FH, Helm PA, Jacques V, Koerner S, Kolodziej A, Shen L, Sun WC, Zhang Z. Angew Chem Int Ed Engl. 2007;46:8171. doi: 10.1002/anie.200700700. [DOI] [PubMed] [Google Scholar]; b) Caravan P, Das B, Deng Q, Dumas S, Jacques V, Koerner SK, Kolodziej A, Looby RJ, Sun WC, Zhang Z. Chem Commun. 2009:430. doi: 10.1039/b819098d. [DOI] [PubMed] [Google Scholar]
- 6.a) Garanger E, Hilderbrand SA, Blois JT, Sosnovik DE, Weissleder R, Josephson L. Chem Commun (Camb) 2009:4444. doi: 10.1039/b907375b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang S, Chen HH, Yuan H, Dai G, Schuhle DT, Mekkaoui C, Ngoy S, Liao R, Caravan P, Josephson L, Sosnovik DE. Circ Cardiovasc Imaging. 2011;4:729. doi: 10.1161/CIRCIMAGING.111.966374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Konda SD, Aref M, Brechbiel M, Wiener EC. Invest Radiol. 2000;35:50. doi: 10.1097/00004424-200001000-00006. [DOI] [PubMed] [Google Scholar]; b) Konda SD, Wang S, Brechbiel M, Wiener EC. Invest Radiol. 2002;37:199. doi: 10.1097/00004424-200204000-00005. [DOI] [PubMed] [Google Scholar]; c) Wiener EC, Konda S, Shadron A, Brechbiel M, Gansow O. Invest Radiol. 1997;32:748. doi: 10.1097/00004424-199712000-00005. [DOI] [PubMed] [Google Scholar]; d) Gunanathan C, Pais A, Furman-Haran E, Seger D, Eyal E, Mukhopadhyay S, Ben-David Y, Leitus G, Cohen H, Vilan A, Degani H, Milstein D. Bioconjug Chem. 2007;18:1361. doi: 10.1021/bc700230m. [DOI] [PubMed] [Google Scholar]; e) Li MJ, Greenblatt HM, Dym O, Albeck S, Pais A, Gunanathan C, Milstein D, Degani H, Sussman JL. J Med Chem. 2011;54:3575. doi: 10.1021/jm200192y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Mulder WJ, Castermans K, van Beijnum JR, Oude Egbrink MG, Chin PT, Fayad ZA, Lowik CW, Kaijzel EL, Que I, Storm G, Strijkers GJ, Griffioen AW, Nicolay K. Angiogenesis. 2009;12:17. doi: 10.1007/s10456-008-9124-2. [DOI] [PubMed] [Google Scholar]; b) Winter PM, Caruthers SD, Kassner A, Harris TD, Chinen LK, Allen JS, Lacy EK, Zhang H, Robertson JD, Wickline SA, Lanza GM. Cancer Res. 2003;63:5838. [PubMed] [Google Scholar]; c) Winter PM, Morawski AM, Caruthers SD, Fuhrhop RW, Zhang H, Williams TA, Allen JS, Lacy EK, Robertson JD, Lanza GM, Wickline SA. Circulation. 2003;108:2270. doi: 10.1161/01.CIR.0000093185.16083.95. [DOI] [PubMed] [Google Scholar]
- 9.a) Vucic E, Sanders HM, Arena F, Terreno E, Aime S, Nicolay K, Leupold E, Dathe M, Sommerdijk NA, Fayad ZA, Mulder WJ. J Am Chem Soc. 2009;131:406. doi: 10.1021/ja808310u. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Briley-Saebo KC, Shaw PX, Mulder WJ, Choi SH, Vucic E, Aguinaldo JG, Witztum JL, Fuster V, Tsimikas S, Fayad ZA. Circulation. 2008;117:3206. doi: 10.1161/CIRCULATIONAHA.107.757120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Bottomley PA, Hardy CJ, Argersinger RE, Allen-Moore G. Med Phys. 1987;14:1. doi: 10.1118/1.596111. [DOI] [PubMed] [Google Scholar]; b) Rooney WD, Johnson G, Li X, Cohen ER, Kim SG, Ugurbil K, Springer CS., Jr Magn Reson Med. 2007;57:308. doi: 10.1002/mrm.21122. [DOI] [PubMed] [Google Scholar]
- 11.van Tilborg GA, Mulder WJ, Deckers N, Storm G, Reutelingsperger CP, Strijkers GJ, Nicolay K. Bioconjug Chem. 2006;17:741. doi: 10.1021/bc0600259. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi H, Sato N, Kawamoto S, Saga T, Hiraga A, Haque TL, Ishimori T, Konishi J, Togashi K, Brechbiel MW. Bioconjug Chem. 2001;12:100. doi: 10.1021/bc000075s. [DOI] [PubMed] [Google Scholar]
- 13.a) Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ. Inorg Chem. 2007;46:6632. doi: 10.1021/ic700686k. [DOI] [PubMed] [Google Scholar]; b) Caravan P, Greenfield MT, Li X, Sherry AD. Inorg Chem. 2001;40:6580. doi: 10.1021/ic0102900. [DOI] [PubMed] [Google Scholar]; c) Dumas S, Troughton JS, Cloutier NJ, Chasse JM, McMurry TJ, Caravan P. Aus J Chem. 2008;61:682. [Google Scholar]; d) Eldredge HB, Spiller M, Chasse JM, Greenwood MT, Caravan P. Invest Radiol. 2006;41:229. doi: 10.1097/01.rli.0000199293.86956.48. [DOI] [PubMed] [Google Scholar]
- 14.a) Nair S, Kolodziej AF, Bhole G, Greenfield MT, McMurry TJ, Caravan P. Angew Chem Int Ed Engl. 2008;47:4918. doi: 10.1002/anie.200800563. [DOI] [PubMed] [Google Scholar]; b) Uppal R, Catana C, Ay I, Benner T, Sorensen AG, Caravan P. Radiology. 2011;258:812. doi: 10.1148/radiol.10100881. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang Z, Kolodziej AF, Greenfield MT, Caravan P. Angew Chem Int Ed Engl. 2011;50:2621. doi: 10.1002/anie.201007689. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang Z, Kolodziej AF, Qi J, Nair SA, Wang X, Case AW, Greenfield MT, Graham PB, McMurry TJ, Caravan P. New J Chem. 2010;2010:611. doi: 10.1039/b9nj00787c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Bloembergen N, Morgan LO. The Journal of Chemical Physics. 1961;34:842. [Google Scholar]; b) Caravan P, Farrar CT, Frullano L, Uppal R. Contrast Media Mol Imaging. 2009;4:89. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Solomon I. Physical Review. 1955;99:559. [Google Scholar]
- 16.Caravan P, Amedio JC, Jr, Dunham SU, Greenfield MT, Cloutier NJ, McDermid SA, Spiller M, Zech SG, Looby RJ, Raitsimring AM, McMurry TJ, Lauffer RB. Chemistry. 2005;11:5866. doi: 10.1002/chem.200500338. [DOI] [PubMed] [Google Scholar]
- 17.a) Aime S, Gianolio E, Terreno E, Giovenzana GB, Pagliarin R, Sisti M, Palmisano G, Botta M, Lowe MP, Parker D. J Biol Inorg Chem. 2000;5:488. doi: 10.1007/pl00021449. [DOI] [PubMed] [Google Scholar]; b) Zech S, Sun WC, Jacques V, Caravan P, Astashkin AV, Raitsimring AM. ChemPhysChem. 2005;6:2570. doi: 10.1002/cphc.200500250. [DOI] [PubMed] [Google Scholar]
- 18.a) Aime S, Chiaussa M, Digilio G, Gianolio E, Terreno E. J Biol Inorg Chem. 1999;4:766. doi: 10.1007/s007750050349. [DOI] [PubMed] [Google Scholar]; b) Zech SG, Eldredge HB, Lowe MP, Caravan P. Inorg Chem. 2007;46:3576. doi: 10.1021/ic070011u. [DOI] [PubMed] [Google Scholar]
- 19.Dumas S, Jacques V, Sun WC, Troughton JS, Welch JT, Chasse JM, Schmitt-Willich H, Caravan P. Invest Radiol. 2010;45:600. doi: 10.1097/RLI.0b013e3181ee5a9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Jin T, Comblin V, Lopez-Mut A, Merciny E, Desreux JF. Inorg Chem. 1992;31:1095. [Google Scholar]
- 21.Jacques V, Dumas S, Sun WC, Troughton JS, Greenfield MT, Caravan P. Invest Radiol. 2010;45:613. doi: 10.1097/RLI.0b013e3181ee6a49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Aruva MR, Daviau J, Sharma SS, Thakur ML. J Nucl Med. 2006;47:155. [PMC free article] [PubMed] [Google Scholar]; b) Botnar RM, Perez AS, Witte S, Wiethoff AJ, Laredo J, Hamilton J, Quist W, Parsons EC, Jr, Vaidya A, Kolodziej A, Barrett JA, Graham PB, Weisskoff RM, Manning WJ, Johnstone MT. Circulation. 2004;109:2023. doi: 10.1161/01.CIR.0000127034.50006.C0. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Flacke S, Fischer S, Scott MJ, Fuhrhop RJ, Allen JS, McLean M, Winter P, Sicard GA, Gaffney PJ, Wickline SA, Lanza GM. Circulation. 2001;104:1280. doi: 10.1161/hc3601.094303. [DOI] [PubMed] [Google Scholar]
- 23.Spuentrup E, Fausten B, Kinzel S, Wiethoff AJ, Botnar RM, Graham PB, Haller S, Katoh M, Parsons EC, Jr, Manning WJ, Busch T, Gunther RW, Buecker A. Circulation. 2005;112:396. doi: 10.1161/CIRCULATIONAHA.104.529941. [DOI] [PubMed] [Google Scholar]
- 24.Mastarone DJ, Harrison VS, Eckermann AL, Parigi G, Luchinat C, Meade TJ. J Am Chem Soc. 2011;133:5329. doi: 10.1021/ja1099616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Greenfield MT, Spiller M, McMurry TJ, Lauffer RB, Caravan P. Angew Chem Int Ed Engl. 2005;44:6766. doi: 10.1002/anie.200502245. [DOI] [PubMed] [Google Scholar]
- 26.Kielar F, Tei L, Terreno E, Botta M. J Am Chem Soc. 2010;132:7836. doi: 10.1021/ja101518v. [DOI] [PubMed] [Google Scholar]
- 27.Peters TJ. All About Albumin: Biochemistry, Genetics, and Medical Applications. Academic Press; San Diego: 1996. [Google Scholar]