Abstract
Single turnover kinetic studies were conducted using fluorescently labeled HIV Reverse Transcriptase (RT) to evaluate the role of nucleotide-induced changes in enzyme structure in the selectivity against AZT in order to explore why AZT-resistant forms of the enzyme fail to significantly discriminate against AZT. Fluorescent labeling of HIV RT provided a signal to monitor the isomerization from “open” to “closed” states following nucleotide binding. We measured the rate constants governing nucleotide binding and enzyme isomerization for TTP and AZT-triphosphate by the wild-type and AZT-resistant forms of the enzyme containing the thymidine analog mutations (TAMs). We show that the TAMs alter the kinetics of AZT incorporation by weakening ground state nucleotide binding and decreasing the rate of chemistry relative to the wild-type enzyme. However, the slower rate of incorporation of AZT by the TAMs HIV RT is counterbalanced a lower Km resulting from the equilibration of the conformational change step. In contrast, the Km for the wild-type enzyme reflects the balance between rates of binding and incorporation so the conformational change step does not come to equilibrium. These data once again demonstrate that the rate of substrate release, limited by the reverse of the substrate-induced conformational change, is the key determinant of the role of induced-fit in enzyme specificity. Mutations leading to slower rates of incorporation have the unfortunate consequence of lowering the Km value by allowing the conformational change step to come to equilibrium.
HIV Reverse Transcriptase is the RNA- and DNA-dependent DNA polymerase responsible for replication of the Human Immunodeficiency Virus in the first step of the viral life cycle whereby single stranded RNA is copied to form double stranded DNA. The inherent low fidelity of the enzyme results in an incorrect nucleotide incorporation every 104 bases (1) contributing to rapid evolution. HIV infections are treated by combination therapy including nucleoside reverse transcriptase inhibitors (NRTIs) (reviewed in (2)). Once phosphorylated in vivo, the corresponding nucleotide analogs serve as substrates for DNA polymerization and result in chain termination due to the absence of a 3′hydroxyl needed for primer extension. The specificity constant (kcat/Km) governing each nucleotide dictates how efficiently it will be incorporated by the polymerase. Prolonged treatment results in the selection of mutations that confer resistance (3). As a result, the study of polymerase nucleotide specificity is critical to the understanding of underlying mechanisms conferring resistance to facilitate the design of more effective inhibitors.
The contribution of substrate induced conformational changes (induced-fit) on specificity has been highly debated (4). It has been argued on theoretical grounds that a two-step substrate binding model, including ground state substrate binding followed by an enzymatic isomerization preceding catalysis, could not enhance substrate specificity beyond that achievable in a one-step mechanism. However, this argument requires several simplifying assumptions that are not valid if alternate substrates reach different ground states (5), if the conformational change is rate limiting (6), or if substrate binding does not occur at equilibrium (7). Rapid chemical quench flow techniques can be used to measure an apparent nucleotide dissociation constant (Kd,app) and maximum rate of polymerization (kpol) for a given nucleotide to define nucleotide specificity as kcat/Km = kpol/Kd,app. However, attempts to ascribe structural features to changes in kcat and Km leading to nucleotide resistance are problematic because changes in the rate-limiting step invalidates any simple comparison (8).
Development of a fluorescently labeled HIV RT has allowed for measurement of the rates governing an induced-fit model for nucleotide binding (8). This system employs a covalently attached MDCC (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin) fluorophore on the fingers subdomain of the enzyme that reports changes in protein structure from the “open” conformation to the “closed” conformation with nucleotide bound without effecting the kinetics of nucleotide binding and incorporation (8). The fluorescently labeled enzyme has been previously used to determine the specificity constants governing dCTP incorporation and the mechanistic basis for 3TC discrimination, and has resolved the apparent paradox that several nucleoside reverse transcriptase inhibitors appear to bind more tightly (lower Kd,app) to the enzyme than the correct nucleotide when assayed by traditional quench flow methods.
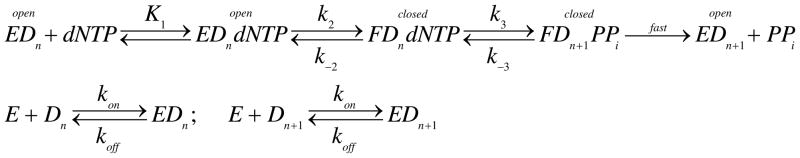
Examination of the polymerase by an induced fit model (Scheme 1) has shown that the relative rates of chemistry (k3) and the reverse of the conformational change (k−2) govern the specificity constant (kcat/Km) for HIV RT (8). When chemistry is fast relative to the conformational change (k3≫k−2) nucleotide binding fails to reach equilibrium and the specificity constant is a function of the rate of formation of the closed enzyme species (kcat/Km = K1k2). Conversely, when the rate of chemistry is slow relative to the rate of the conformational change (k−2≫k3) nucleotide binding reaches equilibrium, and the specificity constant is governed by the product of the equilibrium and the rate of chemistry (K1K2k3). According to this model the relative rates of the conformational change dictate the specificity constant governing nucleotide incorporation for each substrate.
Scheme 1.
Two-step nucleotide binding induced-fit mechanism.
Six mutations (M41L, D67N, K70R, L210W, T215F/Y, and K219Q/E) confer a high level of clinical resistance to AZT in patients who undergo prolonged treatment (9). Previous pre-steady state characterization relying on a single-step nucleotide binding model to determine kpol and Kd,app have indicated that the TAMs do not alter the overall discrimination of AZT (10, 11). It was subsequently discovered that the TAMs function by increasing the rate of ATP-mediated AZT excision (12). Due to recent advances in characterizing the role of the conformational change in nucleotide specificity and discrimination, we have revisited AZT discrimination by the TAMs mutant to investigate if the TAMs alter an induced-fit mechanism. Characterization of this more thorough mechanism of nucleotide binding may find additional contributions to discrimination that were overlooked my traditional analysis. The following work uses a fluorescently labeled MDCC-labeled HIV RT to examine TTP and AZTTP specificity to the enzyme with and without the thymidine analog mutations to examine the underlying mechanistic basis for the minimal AZT discrimination afforded by the TAMs form of the enzyme.
MATERIALS AND METHODS
Expression and Purification of MDCC-labeled HIV Reverse Transcriptase
HIV Reverse Transcriptase was expressed, purified, and labeled according to methods previously described (8). Briefly, p51 C280S and p66 E36C C280S subunits were individually expressed in BL21(DE3) E. coli. Subunits were co-purified in a 1:1 ratio by tandem Q-Sepharose and Bio-Rex 70 columns followed by a single-strand DNA affinity column. Following MDCC labeling, excess MDCC was removed using a Bio-Rex 70 column. The labeled enzyme was assayed by active site titration to determine the concentration, divided into aliquots, and stored at −80°C. The thymidine analogs mutant (TAMs) was produced by introducing six mutations (M41L, D67N, K70R, L210W, T215Y, and K219E) into both p51 C280S and p66 E36C C280S genes using a Quikchange Multi kit (Stratagene). The mutations were confirmed by DNA sequencing, and the TAMs enzyme was expressed, purified, and labeled using the same methods as the wild-type enzyme.
DNA substrates for kinetic studies
The 25/45-mer DNA duplex was prepared after purchasing individual oligonucleotides (25-mer: GCC TCG CAG CCG TCC AAC CAA CTC A; 45-mer: GGA CGG CAT TGG ATC GAC GAT GAG TTG GTT GGA CGG CTG CGA GGC) from Integrated DNA Technologies (1). Oligonucleotides were annealed by mixing 1:1.1 ratio of 25-mer to 45-mer, followed by heating to 95°C for 5 minutes, then allowing to slowly cool to room temperature. The 25-mer oligonucleotide was 5′ 32p labeled for use in the rapid quench flow kinetic assays.
Quench Flow kinetic assays
Chemical incorporation of nucleotide was measured by rapidly mixing a pre-formed enzyme-DNA complex (150 nM MDCC-labeled HIV RT and 100 nM 25/45 DNA) with various concentrations of nucleotide using a KinTek RQF-3 (KinTek Corp). The reaction was stopped by addition of 0.5 M EDTA. Products were separated by 15% denaturing PAGE. All assays were conducted in 50 mM Tris pH 7.5, 100 mM KCl, and10 mM magnesium acetate at 37°C unless otherwise noted. All concentrations are final after mixing.
Stopped Flow kinetic assays
An AutoSF-200x (KinTek Corp) was used to monitor the fluorescence change corresponding to the nucleotide dependent enzyme conformational change. A pre-incubated complex of 200 nM MDCC-labeled HIV RT and 300 nM 25/45-mer was rapidly mixed with various concentrations of nucleotide for 200 ms. The time dependence of the transient change in MDCC fluorescence was recorded by exciting the fluorophore at 425nm and monitoring fluorescence using a 475 nm single band-pass filter with a 25 nm bandwidth (Semrock). To measure the rate of the reverse conformational change (k−2) a 2′3′-dideoxy terminated primer was used. A pre-formed ternary enzyme-DNAdd-dNTP complex (200 nM MDCC-labeled HIV RT, 300 nM 25ddA/45-mer, 1.5 μM nucleotide) was rapidly mixed with 5 μM of an unlabeled enzyme-DNA complex.
Global Analysis of kinetic data
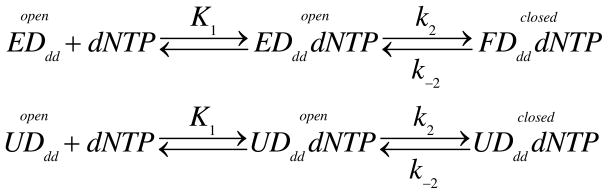
The kinetic data defining nucleotide binding and incorporation were globally fit to a two-step nucleotide binding mechanism shown in Scheme 1 using KinTek Explorer software (KinTek Corp) (13, 14). Values for the kon = 10 μM−1s−1 and koff = 0.2 s−1, the rate constants governing DNA binding and release, respectively, were fixed based upon previous estimates. The smooth curves in each figure panel represent the best simultaneous fit of all of the data according to the model. Nucleotide dissociation rates were measured by competition with unlabeled enzyme and fit according to Scheme 2, where U represents unlabeled enzyme.
Scheme 2.
Measuring the nucleotide dissociation rate by competition
RESULTS
TTP and AZTTP incorporation by the HIV RT-MDCC
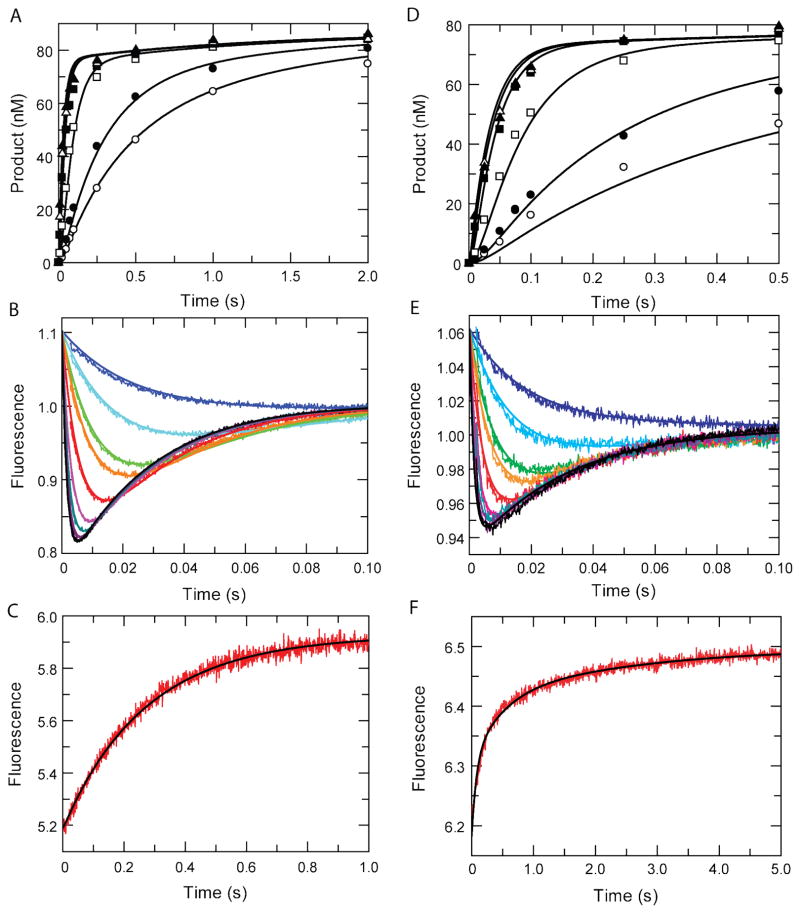
Figure 1A shows the nucleotide concentration dependence of nucleotide incorporation obtained by rapid chemical quench flow methods. In Figure 1B, we show the time dependence of the fluorescence changes measured by stopped-flow methods during nucleotide binding and incorporation at several nucleotide concentrations. Figure 1C shows the measurement of the rate of nucleotide dissociation by competition with unlabeled enzyme. All three experiments were fit globally according to Scheme 1, as illustrated by the smooth curves in each figure, in order to derive the rate constants summarized in Table 1. These data alone could not establish the maximum rate of the conformational change because the rate was too fast to measure at 37°C.
Figure 1. Global fitting of TTP and AZTTP Binding and Incorporation by MDCC-labeled HIV Reverse Transcriptase.
The concentration-dependence of incorporation of TTP (A) and AZTTP (D) was measured using quench-flow methods. Various concentrations of nucleotide (0.25, 0.5, 2, 10, 25, and 100 μM) were mixed with a pre-form enzyme-DNA complex (100 nM 25/45-mer DNA and 150 nM MDCC-labeled HIV RT). The time dependence of the fluorescence change was monitored after mixing TTP (B) or AZTTP (E) with an enzyme-DNA complex. Various concentrations of nucleotide (2, 4, 10, 20, 40, 60, 80, and 100 μM) were mixed with a pre-form enzyme-DNA complex (300 nM 25/45-mer and 200nM MDCC-labeled HIV RT). Fluorescence was observed by excitation of MDCC at 425 nm and monitoring emission with a 475 nm bandpass filter with a 25 nm bandwidth. The reverse rate of the conformational change leading to release of TTP (C) or AZTTP (F) was measured by rapidly mixing a pre-formed enzyme-DNAdd-dNTP complex (200 nM HIV RT-MDCC, 300 nM 25ddA/45-mer, 1.5 μM nucleotide) with a nucleotide trap consisting of 5 μM unlabeled enzyme-DNAdd complex. Nucleotide release from the fluorescent enzyme followed by binding to the trap complex produced the observed increase in fluorescence. Global analysis was performed independently for TTP (A–C) and AZTTP (D–F). Rate constants resulting from global analysis of kinetic data are shown in Table 1.
Table 1.
Kinetic constants governing nucleotide binding and incorporation.
| Enzyme | dNTP | 1/K1 (μM) | k2 (s−1) | k−2 (s−1) | K2 | k3 (s−1) |
|---|---|---|---|---|---|---|
| HIV RT-MDCC | TTP | 310 ± 10 | 2700 ± 100 | 3.9 ± 0.2 | 690 ± 67 | 34.4 ± 0.1 |
| AZTTP | 110 ± 10 | 1500 ± 100 | 15.0 ± 0.5 | 100 ± 10 | 30.2 ± 0.2 | |
| TAMs HIV RT-MDCC | TTP | 1350 ± 30 | 3250 ± 150 | 9.0 ± 0.3 | 360 ± 30 | 45.1 ± 0.1 |
| AZTTP | 470 ± 25 | 1500 ± 100 | 34.4 ± 3.1 | 44 ± 7 | 11.5 ± 0.1 |
Rate constants were derived from global analysis of the data fitted to scheme 1 allowing for a nucleotide included conformational change prior to chemistry. Values for k2 were determined by temperature analysis at lower temperatures to fully resolve the maximum rates. Arrhenius analysis was performed to obtain the maximum rate at 37°C, which is reported above, and was used to constrain the global fit. Global analysis of K1 was performed by locking the initial second-order nucleotide on-rate (k1) to a modest estimate for diffusion-limited collision (100 μM−1s−1). The reverse rate (k−1) was fit globally. The reported ground-state Kd (1/K1) reported above was calculated using the values obtained from global analysis.
To determine the maximum rate of the conformational change (k2) an Arrhenius analysis of the temperature dependence on the decrease of fluorescence upon nucleotide binding was performed (see Supplement). Analysis of the TTP concentration dependence of the rate of the decrease of fluorescence afforded estimates of k2 = 66±2 s−1, 115±4 s−1, and 216±6 s−1 at 10°C, 14°C, and 18°C, respectively (Figure S1). These rates were used to extrapolate to an estimated rate for k2 at 37°C of 2750±100 s−1. This is in accord with a lower limit k2 of 1300s−1 estimated by confidence contour analysis while globally fitting the data shown in Figure 1. The value of k2 derived by extrapolation of the temperature dependence was fixed and all other constants were then derived from the global data fitting. The data at 37°C accurately define the product K1k2, so use of the value of k2 derived from the temperature dependence was only used to estimate K1. Accordingly, a ground state dissociation constant of 1/K1 = 310±10 μM was calculated in fitting the families of curves.
By conventional data analysis, as previously shown (8), the concentration dependence of rate of fast fluorescence change in Figure 1B could be fit to a straight line with a slope of 8.9 μM−1s−1, corresponding to K1k2. This value agrees with the value for K1k2 calculated from the results of global fitting, and provides an additional check of the rates determined by global analysis.
The reverse of the conformational change was determined by monitoring the change in fluorescence from a “closed” ternary complex to the “open” complex when nucleotide disassociates. The absence of a 3′ hydroxyl on the DNA primer allowed for the formation of the “closed” complex while blocking chemistry. The rate of the fluorescence increase observed in Figure 1C, corresponded to the reverse of the conformational change (k−2) during nucleotide release when nucleotide trap is added. This resulted in a k−2 value of 3.9±0.2 s−1, which was well constrained in the global fitting according to confidence contour analysis (13, 14). It should be noted that because we fit the data by simulation with a comprehensive model including the rate of nucleotide binding to the unlabeled enzyme that traps free nucleotide (Scheme 2), it was not necessary to use a large excess of unlabeled enzyme to get a reliable estimate of the rate of release of nucleotide from the labeled enzyme. If the data had been fit by conventional means, it would have been necessary to assume that the binding rate of the free nucleotide to the unlabeled enzyme was much faster than the rate of rebinding to the labeled enzyme. In global fitting with a complete model, which implicitly includes all rate constants, no simplifying assumptions were needed in fitting the data.
Similar experiments were performed to determine the kinetics governing AZTTP binding and incorporation as shown in Figure 1C–E and summarized in Table 1. Arrhenius analysis was performed to determine a maximum rate of the conformational change (k2) of 1500±100 s−1 (figure S2). Nucleotide ground state binding was 3-fold tighter than for TTP, with value of 110±10 μM. The rate of chemistry was 30.2±0.2 s−1, as defined by the rapid chemical quench flow data (Figure 1C) and the slow second phase of the stopped flow data (Figure 1D). The rate of chemistry agrees with previously published reports indicating that rates of incorporation of AZTTP and TTP were comparable (11). Measurement of the reverse of the conformational change (k−2) was fit to a rate of 15.0±0.5 s−1.
TTP and AZTTP incorporation by MDCC-labeled TAMS HIV RT
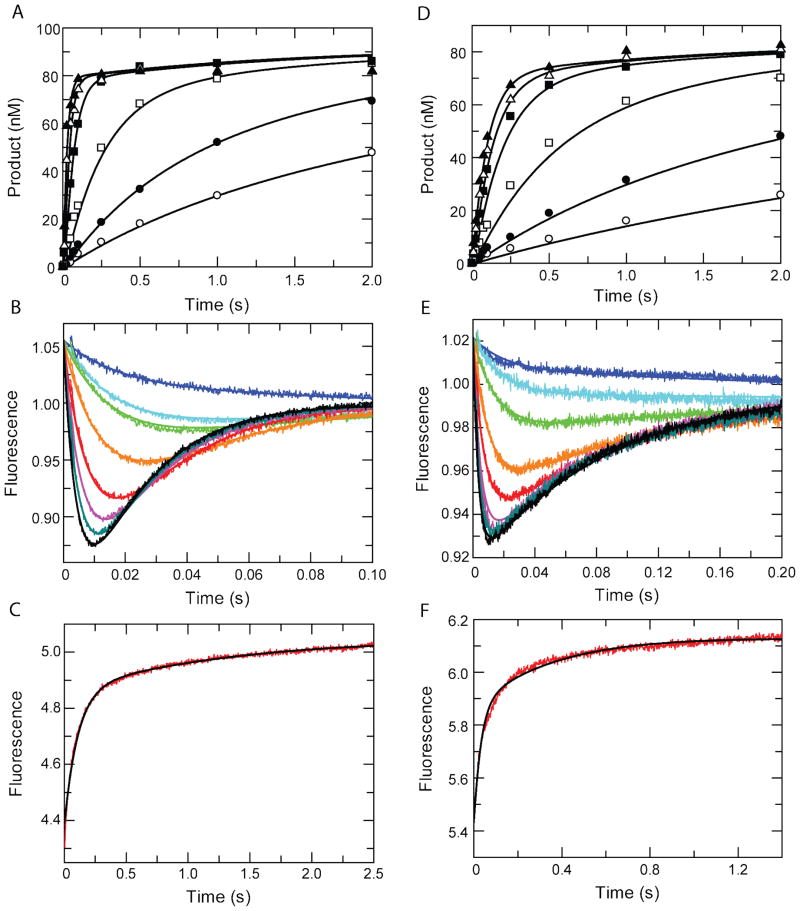
Nucleotide concentration dependence of the rates of incorporation of TTP by the TAMs HIV RT and the slow phase of the stopped flow binding and incorporation data defined a rate of incorporation (k3) of 45.1±0.1s−1 (Figure 2D–E). Temperature dependence of the conformational change rate was performed yielding rates of 91±2 s−1, 191±3 s−1, and 276±9 s−1, for 10°C, 14°C, and 18°C, respectively. The temperature dependence of the rate was fit to the Arrhenius equation to obtain a maximum rate for k2 of 3250±150 s−1 at 37°C (Figure S3). Data contained within the families of curves for the chemical quench flow and stopped flow data were used to constrain a nucleotide ground state binding constant (1/K1) of 1350±30 μM. The reverse rate for the conformational change (k−2) was 9.0±0.3 s−1, as measured by the TTP dissociation experiment (Figure 2F).
Figure 2. Global fitting of TTP and AZTTP Binding and Incorporation by MDCC-labeled TAMs HIV Reverse Transcriptase.
Chemical quench flow concentration dependence of TTP (A) and AZTTP (D). The concentration-dependence of incorporation of TTP (A) and AZTTP (D) by the TAMs HIV RT was measured using quench-flow methods. Various concentrations of nucleotide (0.25, 0.5, 2, 10, 25, and 100 μM) were mixed with a pre-formed enzyme-DNA complex (100 nM 25/45-mer DNA and 150 nM MDCC-labeled TAMs HIV RT). The time dependence of the fluorescence change was monitored after mixing TTP (B) or AZTTP (E) with an enzyme-DNA complex. Various concentrations of nucleotide (2, 4, 10, 20, 40, 60, 80, and 100 μM) were mixed with a pre-formed enzyme-DNA complex (300 nM 25/45-mer and 200 nM MDCC-labeled TAMs HIV RT). Fluorescence was observed by excitation of MDCC at 425nm and observing emission with a 475 nm band-pass filter with a 25 nm bandwidth. The reverse rate of the conformational change for TTP (C) and AZTTP (F) was measured rapidly mixing a pre-formed enzyme-DNAdd-dNTP ternary complex (200 nM TAMs HIV RT-MDCC, 300 nM 25ddA/45-mer, 1.5μM nucleotide) with a nucleotide trap of consisting of 5μM an unlabeled enzyme-DNAdd complex. Nucleotide release from the fluorescent enzyme followed by binding to the trap complex produced the observed increase in fluorescence. Global analysis was performed independently for TTP (A–C) and AZTTP (D–F). Rate constants resulting from global analysis of kinetic data are shown in Table 1.
AZTTP binding and incorporation were also globally fit to a two-step nucleotide binding model. The rate of chemistry (k3) was 11.5±0.1 s−1 (Figure 2D and E). To determine the maximum rate of the conformational change temperature dependence analysis was performed and extrapolated to 37°C to obtain a rate of 1500±100 s−1 (Figure S4). Global analysis resulted in a value of 470±25 μM governing the ground state binding of AZTTP to the “open” enzyme complex. The reverse rate of the conformational change was fit to a value of 34.3±3.1 s−1, as defined by the AZTTP dissociation measurement. Additionally, data to constrain this rate were also observed in the amplitude dependence of the stopped flow binding experiment (Figure 2E) since equilibrium was established prior to chemistry because k−2 was greater than k3. Global fitting results for TTP and AZTTP binding and incorporation are summarized in Table 1.
Determination of specificity constants governing TTP and AZTTP binding and incorporation to wild-type and TAMs HIV RT
The specificity constant (kcat/Km) was calculated as kcat/Km = k1k2k3/(k2k3+k1(k−2+k3)) for each nucleotide and enzyme. The specificity constants governing TTP and AZTTP incorporation by HIVRT-MDCC were 7.5±0.2 μM−1s−1 and 7.7±0.3 μM−1s−1, respectively. These values produced a net discrimination [(kcat/Km)AZTTP/(kcat/Km)TTP] of 1.02±0.08 (Table 2). Overall Kd values for nucleotide binding resulting from a two-step mechanism (Kd,overall = 1/(K1(1+K2))) were 0.4±0.05 μM and 1.1±0.04 μM, respectively. The specificity constants governing TTP and AZTTP incorporation by the TAMs enzyme were 1.9±0.02 μM−1s−1 and 0.8 μM−1s−1, respectively. These values defined the AZTTP discrimination by the TAMs mutant of 2.4±0.05, a modest increase from the wild type enzyme. In addition to reduced specificity constants for TTP and AZTTP, the TAMs enzyme also displayed a weaker overall nucleotide binding affinities of 3.7±0.2 μM and 11.5±0.1 μM, respectively. These values amount to a 10-fold reduction in the overall nucleotide binding interaction by the mutant enzyme. In addition, the TAMs mutations lead to a fourfold reduction in enzyme efficiency due to changes in kcat/Km.
Table 2.
Nucleotide binding, specificity, and discrimination for wild-type and TAMs HIV RT.
| Enzyme | dNTP | Kd,net (μM)a | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1)b | Dc |
|---|---|---|---|---|---|---|
| Wt HIV RT | TTP | 0.4 ± 0.1 | 4.7 ± 0.4 | 33.9 ± 0.2 | 7.5 ± 0.2 | |
| AZTTP | 1.1 ± 0.1 | 3.5 ± 0.4 | 29.3 ± 0.4 | 7.7 ± 0.3 | 1.02 ± 0.08 | |
| TAMs HIV RT | TTP | 3.7 ± 0.2 | 22.5 ± 2 | 44.3 ± 0.4 | 1.9 ± 0.2 | |
| AZTTP | 11.5 ± 0.1 | 14.0 ± 1.2 | 11.1 ± 0.2 | 0.8 ± 0.1 | 2.4 ± 0.05 |
The net Kd as a result of two-step nucleotide binding was calculated as 1/(K1(1+K2)).
kcat/Km was calculated as k1k2k3/(k2k3+(k1(k−2+k3)))
Discrimination was calculated as D = (kcat/Km)AZTTP/(kcat/Km)TTP
DISCUSSION
We have studied the effects of the thymidine analog mutations of HIVRT on nucleotide specificity via a two-step induced fit nucleotide binding model using an engineered enzyme possessing a conformationally sensitive fluorophore. Data were fit globally to a comprehensive model shown in Scheme 1. Fitting data based upon numerical integration of the rate equations with no simplifying assumptions is far more robust and demanding than fitting to equations derived from a simplified form of the model because it requires that the model account for both the rate and amplitude of all reactions simultaneously. It should be noted, however, that the reverse rate of chemistry (k−3) was not determined because available evidence indicates that pyrophosphate (PPi) release is fast, and therefore, there is no information content in the data to define k−3 or k4.
The results indicate that the TAMs modestly improve discrimination against AZT. As reported previously, this small 2.4-fold improvement in discrimination would not be enough to provide the high level of AZT resistance conferred by the TAMs observed clinically (11). Although the overall discrimination against AZT is not greatly effected by the TAMs, the mechanistic basis for this phenomenon is rather intriguing. The ground state binding of TTP and AZTTP to the TAMs mutant is 4.5-fold weaker than the MDCC-labeled wild type enzyme. Although ground-state binding is reduced, AZTTP still binds with a 3-fold stronger affinity than TTP as a result of contributions from conformational change step. This stronger initial ground state interaction may be a result of a favorable interaction between the positively charged 3′azido group of AZTTP and a negatively charged binding pocket. Following ground state binding, both HIVRT and TAMs HIVRT undergo favorable conformational changes (K2) to the “closed” state. The mutations impart a modest increase in the conformational change rate (k2) for TTP, but have no effect on the 1500 s−1 rate of conformational closing when AZTTP is bound.
Arrhenius analysis to determine the maximum rate of the conformational change (k2) at 37°C was also used to examine the thermodynamics of initial ground state binding (K1). Temperature dependence of K1 revealed negative H values between 6.5 and 13.8 kcal/mol indicating initial “ground-state” binding of nucleotide is an exothermic process (Table S1). This is contrasted by the enthalpy of activation values for k2 (linear slopes from figure S1–4) that indicate a weakly endothermic reaction with positive ΔH values of 0.32 ± 0.02 kcal/mol.
The major mechanistic difference between the nucleotide incorporation mechanisms as shown in Table 2 is a function of the kinetic partitioning as a result of the relative rates of chemistry (k3) and the reverse of the conformational change (k−2). Previous applications of an induced-fit model governing nucleotide incorporation for HIVRT and T7 DNA Polymerase have shown that this partitioning allows simplifications to be made when deriving the specificity constant (7, 8). Notably, when the rate of chemistry is much greater than the rate of the reverse conformational change, the specificity constant (kcat/Km) can be simplified to K1k2. This term describes the formation of the closed species, which is then committed to catalysis. Conversely, when k−2≫k3 an equilibrium is established prior to chemistry, which results in a simplified derivation of specificity corresponding to product of the equilibrium constants and the rate of chemistry (K1K2k3). These differences highlight the importance of the relative rates of the conformational change when determining substrate specificity. Previous measurements of k−2 and k3 have shown that each nucleotide favors a given kinetic partitioning, either catalysis or reversal of the conformational change. However, TTP and AZTTP incorporation by the HIVRT-MDCC and TAMs HIVRT-MDCC do not follow this simple trend. The largest observed difference in these rates is only 10-fold when TTP is incorporated by HIVRT-MDCC, resulting in kinetic partitioning between nucleotide incorporation and the “open” state. Because of this partitioning, simplification of the specificity constant to K1k2 or K1K2k3 cannot be used. We observe that kcat/Km values for HIVRT-MDCC incorporation of TTP and AZTTP are equal because the weaker ground state binding of TTP (K1,TTP<K1,AZTTP) is offset by a more favorable conformational change (K2,TTP>K2,AZTTP), and the rates of nucleotide incorporation are equal. A similar analysis of TAMs HIVRT-MDCC provides the mechanistic bases for the modest increase in discrimination. Again we observe weaker ground state binding of TTP to the mutant enzyme than AZTTP. Although TTP binds more weakly to the mutant enzyme, the conformational change to the “closed” state is more highly favored (K2,TTP = 361±31, K2,AZTTP = 43±8) compared to isomerization when AZTTP is bound. This difference in the equilibrium of the conformational change is followed by a faster rate of incorporation for TTP compared to AZTTP. We also note that of the four possible nucleotide/enzyme combinations studied in this work, only AZTTP incorporation by TAMs HIVRT-MDCC has a reverse conformational change rate that is greater than the rate of chemistry (k−2>k3). This allows the two-step binding mechanism to come to equilibrium, leading to a reduced Km value. Thus, it is an unfortunate consequence in that a reduced rate of chemistry allows the binding and conformational change steps to come to equilibrium so that the Km reflects the true Kd,net (Table 2). For the wild type enzyme, the Km is much greater than the true Kd,net because binding and isomerization do not come to equilibrium and the Km is determined by the relative rates of binding and chemistry, Km ~ k3/K1k2.
A similar phenomenon was observed in comparing the kinetic parameters governing the incorporation of 3TC-TP (the triphosphate form of lamivudine) and dCTP (8). The slower rate of incorporation of 3TC-TP allowed the conformational change to come to equilibrium preceding chemistry, leading to a lower Km value than observed for dCTP where the rate of incorporation was faster than the rate of nucleotide release, and the Km value is determined by the ratio of the rate of chemistry divided by the net rate of nucleotide binding. Simple mechanistic conclusions regarding discrimination due to changes in Km are not valid because of changes in the rate constants contributing to the observed Km values.
Modeling of 3TC-TP incorporation by HIV RT required an additional mechanistic step preceding chemistry to account for an additional isomerization observed as a result of a greatly reduced rate of chemistry. This phase was observed in stopped flow experiments measuring the nucleotide induced conformational closing and enzymatic release of nucleotide similar to experiments shown in Figure 2E and F). Like with 3TC-TP, with AZTTP we observe slightly biphasic kinetics of nucleotide dissociation. Although this data can be modeled with an additional isomerization preceding chemistry, we have decided to present the minimal model two-step model that accounts for the experimental data. Additionally, The extra phase observed in Figure 2F may result from the combination of establishing an equilibrium prior to chemistry in the presence of a 2′3′-dideoxy terminated primer.
The absence of discrimination against AZT by the TAMs mutant is likely the result of the high degree of structural homology between the inhibitor and TTP. We reason that because AZT contains a naturally occurring thymine nucleobase and a deoxyribose sugar with a single 3′ azido modification, the inhibitor is structurally similar enough to TTP to not greatly alter the kinetics governing incorporation by either enzyme. Previously we have shown that inhibitors containing more severe modifications in either the sugar ring structure or nucleobase, such as an oxatholane ring substitution and L-stereochemistry of 3TC (8), or 5-fluorination of FTC, result in higher basal and mutant discrimination values than AZTTP, which can be attributed to more severe differences in binding and incorporation kinetics. It is interesting to note that the human mitochondrial DNA polymerase discriminates against AZT by slowing the rate of pyrophosphate release after incorporation (15).
Previous findings have demonstrated that the role of the thymidine analog mutations in AZT resistance is to favor excision a 3′ terminal AZTMP following incorporation. Excision occurs by the mutant enzyme binding pyrophosphate (16) or ATP (17), which then react with the 3′-terminal AZT to form AZTTP or AppppAZT, respectively. This mechanism suggests that the thymidine analog mutation either favor PPi/ATP binding, shift the translocation equilibrium of the DNA to favor the un-translocated state, or both (18, 19). Presumably, the decreases in specificity for both TTP and AZTTP by the TAMs enzyme are an ancillary result of increased AZT excision. Additionally, as few as two of the TAMs (including K215Y/F) can impart resistance to AZT (9), with all six mutation producing a more fit enzyme that is capable of more efficient DNA polymerization and AZT excision. Further studies are needed to explore the effects of the TAMs using the fluorescently labeled enzyme to monitor pyrophosphate or ATP binding during the excision reaction.
The emergence of multiple mutations needed to confer resistance to AZT is evolutionarily much more unlikely than a single mutation, such as M184V or K65R, that result in high levels of analog discrimination against other NRTI’s. One would expect the virus to evolve a discrimination mechanism where the more bulky 3′azido group is sensed by steric interactions thereby leading to increased discrimination, such as in the case with M184V and 3TC. However, the only amino acid residues close enough to interact with the 3′azido group are 111GDAY114, which are highly conserved (20). The emergence of an excision mediated rescue of polymerization conferred by the TAMs may therefore have been evolutionarily favored because discrimination against AZTTP through mutation of 111GDAY114 may have too severely compromised the natural polymerization activity of the enzyme. Further studies are needed to determine how polymerization is effected by mutations in this region.
In conclusion, our data suggest that the TAMs only impart a modest gain in discrimination against AZT by HIV Reverse Transcriptase, a finding that supports previous reports of nucleotide discrimination by the TAMs enzyme. By expanding the kinetic mechanism governing nucleotide binding to include an induced-fit model we have determined the mechanistic bases for this absence of increased discrimination. We have found that the thymidine analog mutations do weaken the ground state binding of AZTTP and also increase the reverse of the conformational change. However, the mutations have only a small effect on the overall kinetic partitioning of the closed species to greatly favor either catalysis of nucleotide release. The result is a decrease in kcat/Km for both TTP and AZTTP, however only subtle changes in AZTTP discrimination. Further studies of the excision reaction using the fluorescently labeled enzyme may provide a more detailed kinetic basis for TAMs resistance to AZT.
Supplementary Material
Acknowledgments
K.A.J. is the President of KinTek Corp., which provided the AutoSF-200x stopped-flow and RQF-3 quench-flow instruments and KinTek Explorer software.
Supported by a grant from the Welch Foundation (F-1604) and the National Institutes of Health (R01GM084741).
Abbreviations
- HIV RT
Human Immunodeficiency Virus Reverse Transcriptase
- TAMs
Thymidine analog mutations
- AZT
3′azido-2′,3′-dideoxythymidine
- AZTTP
3′-azido-2′,3′-dideoxythymidine-5′-triphosphate
- TTP
Thymidine triphosphate
- 3TC
2′,3′-dideoxy-3′-thiacytidine
- MDCC
7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin
Footnotes
Supporting Information Available. Figures S1–4. Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 2.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deval J, Courcambeck J, Selmi B, Boretto J, Canard B. Structural determinants and molecular mechanisms for the resistance of HIV-1 RT to nucleoside analogues. Curr Drug Metab. 2004;5:305–316. doi: 10.2174/1389200043335478. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 5.Post CB, Ray WJ., Jr Reexamination of induced fit as a determinant of substrate specificity in enzymatic reactions. Biochemistry. 1995;34:15881–15885. doi: 10.1021/bi00049a001. [DOI] [PubMed] [Google Scholar]
- 6.Herschlag D. The role of induced fit and conformational changes of enzymes in specificity and catalysis. Bioorganic Chemistry. 1988;16:62–96. [Google Scholar]
- 7.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellinger MW, Johnson KA. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc Natl Acad Sci U S A. 2010;107:7734–7739. doi: 10.1073/pnas.0913946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 10.Lacey SF, Reardon JE, Furfine ES, Kunkel TA, Bebenek K, Eckert KA, Kemp SD, Larder BA. Biochemical studies on the reverse transcriptase and RNase H activities from human immunodeficiency virus strains resistant to 3′-azido-3′-deoxythymidine. J Biol Chem. 1992;267:15789–15794. [PubMed] [Google Scholar]
- 11.Kerr SG, Anderson KS. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;36:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 12.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson KA, Simpson ZB, Blom T. FitSpace Explorer: An algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic Acids Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arion D, Parniak MA. HIV resistance to zidovudine: the role of pyrophosphorolysis. Drug Resist Updat. 1999;2:91–95. doi: 10.1054/drup.1999.0076. [DOI] [PubMed] [Google Scholar]
- 17.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 18.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu X, Das K, Han Q, Bauman JD, Clark AD, Jr, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.