Abstract
CUG binding protein 1 (CUGBP1) regulates multiple aspects of nuclear and cytoplasmic mRNA processing, with implications for onset of myotonic dystrophy. CUGBP1 harbors three RRM domains and preferentially targets UGU-rich mRNA elements. We report on crystal structures of CUGBP1 RRM1 and tandem RRM1/2 domains bound to RNAs containing tandem UGU(U/G) elements. Both RRM1 in RRM1-RNA and RRM2 in RRM1/2-RNA complexes use similar principles to target UGU(U/G) elements, with recognition mediated by face-to-edge stacking and water-mediated hydrogen bonding networks. The UG step adopts a left-handed Z-RNA conformation, with the syn guanine recognized through Hoogsteen edge-protein backbone hydrogen-bonding interactions. NMR studies on the RRM1/2-RNA complex establish that both RRM domains target tandem UGUU motifs in solution, while filter-binding assays identify a preference for recognition of GU over AU or GC steps. We discuss the implications of CUGBP1-mediated targeting and sequestration of UGU(U/G) elements on pre-mRNA alternative-splicing regulation, translational regulation and mRNA decay.
Human CUG-binding protein 1 (CUGBP1) is a founding member of the CUGBP1 and ETR-3-like factors (CELF) family of RNA binding proteins that have been implicated in the regulation of pre-mRNA splicing, and in the control of mRNA translation and deadenylation (Barreau et al., 2006). CUGBP1 has also recently been identified as a decay-promoting factor associated with several short-lived mRNAs (Vlasova et al., 2008).
CUGBP1 is one of the key proteins whose function in both alternative splicing and translational regulation is altered in myotonic dystrophy type 1 (DM1), a multi-systemic disease affecting skeletal muscle, heart and the central nervous system (Ranum and Cooper, 2006). In DM1, expansion of CUG repeats within the 3′-UTR of the DMPK mRNA, results in hyperphosphorylation and stabilization of CUGBP1, which is normally downregulated in adult tissues (Kalsotra et al., 2008; Kuyumcu-Martinez et al., 2007). In turn, increased levels of CUGBP1 leads to aberrantly spliced products in which embryonic and neonatal rather than adult splicing patterns are retained in DM tissues (Osborne and Thornton, 2006; Ranum and Cooper, 2006). Expression of mis-spliced mRNAs encoding muscle-specific chloride channel (CIC-1), insulin receptor (IR), and cardiac troponin T (cTNT), result in disease symptoms such as myotonia and insulin resistance. In addition to triggering aberrant splicing, CUGBP1 has also been reported to enhance translation of the cdk inhibitor p21 (Timchenko et al., 2001) and the transcriptional regulator myocyte enhancer factor 2A (MEF2A) in DM1 cells (Timchenko et al., 2004). Mouse models, which exhibit both splicing defects (Ho et al., 2005) and altered translational regulation (Timchenko et al., 2004), support the muscle pathogenic role of increased CUGBP1 in DM1. Moreover, elevated CUGBP1 levels have recently been demonstrated in DM1 mouse models that express CUG repeats in the context of the DMPK transgene (Mahadevan et al., 2006).
CELF proteins from diverse species bind preferentially to GU-rich sequence elements within mRNA (Vlasova and Bohjanen, 2008), including U/G-rich intronic motifs (Charlet et al., 2002; Philips et al., 1998; Savkur et al., 2001), as well as to 3′-UTR U-purine rich motifs of c-jun AU-rich elements (ARE) (Paillard et al., 2002), c-mos EDEN (Paillard et al., 2003), and to 3′-UTR GRE in human papillomavirus 16 (Goraczniak and Gunderson, 2008). In addition, CELF proteins target 3′-UTR GU-rich regulatory elements (GRE) represented by a conserved 11-nucleotide (nt) UGUUUGUUUGU sequence found in a number of human transcripts that exhibited rapid mRNA decay (Vlasova et al., 2008), Mutation of G nucleotides to C disrupted CUGBP1 binding to GREs both in vitro and in vivo (Goraczniak and Gunderson, 2008; Vlasova et al., 2008). Systemic evolution of ligand exponential enrichment (SELEX) also revealed that CUGBP1 (Marquis et al., 2006) and CUGBP2 (Faustino and Cooper, 2005) bound selectively to GU-rich RNA with the preference for UGU and UGUU-rich motifs, respectively. Recent data on microarray-monitored expression of 24,426 alternative splicing events in different human samples identified clusters of UGUGU motifs associated with CELF, along with five other clusters of short (4–7 nucleotides) cis-regulatory motifs enriched near the cassette exons which map to trans-acting regulators, such as PTB, Fox, Muscleblind, TIA-1 and hnRNP F/H (Castle et al., 2008). A recent study on the regulation of TNF mRNA stability, indicates that CUGBP1 principally recognizes sequences that flank the AU-rich element (ARE), which contain multiple non-consecutive UGU motifs, rather than the ARE itself (Zhang et al., 2008).
CUG-BP1 and the other CELF members all contain three highly conserved RNA recognition motifs (RRMs). RRM1 and RRM2 (designated RRM1/2) are adjacent to each other at the N-terminus, while RRM3 is located near the C-terminus (Barreau et al., 2006). RRM2 and RRM3 are separated by a 160–230 (211 in CUGBP1) residues divergent linker. It was suggested that RRM1 and RRM2 are necessary for specific binding to the CUG repeats (Timchenko et al., 1999), and on this basis were initially structurally characterized by NMR in the free state (Jun et al., 2004). The NMR solution structure of a CUGBP1 RRM1/2 construct (PDB ID 2DHS) revealed that both RRM1 and RRM2 adopt the same βαββαβ topology (Figure 1A) in which the four β-strands form an anti-parallel β-sheet packed against two α-helices, a typical RRM fold (Maris et al., 2005). The linker between the CUGBP1 RRM1 and RRM2 exhibited higher conformational flexibility than other regions in the NMR structure. Most commonly, individual RRM’s recognize 3 to 4 nucleotides of single-stranded RNA via their β-sheet (Maris et al., 2005). A recent solution NMR structure established how CUGBP1 RRM3 specifically recognizes the UGU trinucleotide segment of the bound (UG)3 RNA through extensive stacking and hydrogen-bonding interactions within the pocket formed by the β-sheet and the conserved N-terminal extension (Tsuda et al., 2009).
Figure 1. Sequence Alignment and Secondary Structures of RRM Domains of CUGBP1 and ITC Analysis of RRM-RNA Complex Formation.
(A) Structure-based sequence alignment of human CUGBP1 RRM1, RRM2 and RRM3 domains generated with ESPript (http://espript.ibcp.fr/ESPript/ESPript/help.php). The conserved RNP2 and RNP1 segments of the RRM sequences are in black boxes. Asterisks denote residues that form hydrogen bonds with RNA bases via their side chains; triangles indicate aromatic and aspartate residues involved in base-stacking with RNA bases; and circles mark residues interacting with RNA via their backbone functional groups.
(B–D) Isothermal titration calorimetry (ITC) binding curves for complex formation between tandem CUGBP1 RRM1/2 and GUUGUUUUGU 10-nt RNA (panel B), for complex formation between CUGBP1 RRM1 and UUGUU 5-nt RNA (panel C), and for complex formation between CUGBP1 RRM2 and UUGUU 5-nt RNA (panel D).
A thermogram as a result of titration is shown in the top panel, and a plot of the total heat released as a function of the molar RNA/protein ratio is shown in the bottom panel. Solid lines indicate non-linear least-squares fit to a theoretical titration curve using Microcal software, with ΔH (binding enthalpy kcal mol−1), KD (association constant), and n (number of binding sites per monomer) as variable parameters.
(E) Tabulation of energetic parameters for RRM-RNA recognition from ITC binding curves.
Here we address how individual and tandem RRM domains of CUGBP1 target GU-rich regulatory mRNA elements. To further elucidate principles underlying sequence-specific RNA recognition by the RRM domains of CUGBP1, we have solved the crystal structures of individual RRM1 and tandem RRM1/2 domains bound to 12 and 13-nucleotide RNAs containing a pair of UGU(U/G) motifs. The structures of these complexes highlight how the UGU(U/G) segment is bound by the RRM1 domain in the RRM1-RNA complex and by RRM2 in the RRM1/2-RNA complex through a novel recognition of the guanine of the UGU trinucleotide, as well as through canonical base stacking interactions of the last two (UU or UG) bases with conserved aromatic residues and amino acid side chains. The bound UG segment in both complexes adopts an unanticipated left-handed Z-helical conformation. While NMR-based solution studies demonstrate that both RRM1 and RRM2 contribute to binding of a 12-nucleotide target RNA containing a pair of UGUU motifs, crystallographic studies show that only RRM2 binds this RNA, while RRM1 is involved in crystal packing interactions. Finally, filter-binding assays identify a preference of CUGBP1 RRM domains for recognition of GU over either AU or GC steps.
RESULTS
Choice of Protein and RNA Constructs for CUGBP1 Complexes
Crystallization trials were undertaken on three CUGBP1 fragments containing tandem RRM1 and RRM2 (RRM1/2 residues 1-187, 14-187 and 17-187) and various synthetic RNAs of 12- and 13-nt length containing either a pair of UGUU or UGUGUG sequence motifs, which were either adjacent to each other or separated by a U or UU linker. The crystals were obtained for the RRM1/2 construct comprising residues 14-187 in complexes with 12-nt RNA sequences, GUUGUUUUGUUU, GUUGUUUUUGUU (complexes 1 and 2, respectively), and 13-nt UGUGUGUUGUGUG sequence (complex 3). All three sequences formed complexes with RRM1/2 as verified by polyacrylamide electromobility shift assays. RRM1/2 also showed a well-dispersed 15N-HSQC NMR spectrum, allowing complex formation to be monitored on addition of above 12- and 13-nt sequences.
In addition, crystallization trials were undertaken on a CUGBP1 RRM1 construct (residues 14-100) in complex with 12-mer GUUGUUUUGUUU (complex 4).
Binding Affinities of Single Versus Tandem CUGBP1 RRM-UGUU Interactions
We have monitored the binding affinities by isothermal titration calorimetry (ITC) of individual RRM1 and RRM2 domains of CUGBP1 to UUGUU 5-nt RNA and tandem RRM1/2 domains to GUUGUUUUGUUU containing a pair of UUGUU steps. The dissociation constant (KD) values are 0.65 μM for the RRM1/2-GUUGUUUUGUUU complex (Figures 1B,E), 29 μM for the RRM1-UUGUU complex (Figures 1C,E) and 45 μM for the RRM2-UUGUU complex (Figures 1D,E).
Crystallization and Structure Determination of CUGBP1 Complexes
The crystals obtained for CUGBP1 RRM1/2-RNA complexes 1, 2 and 3 all belong to the I222 space group with similar unit cell parameters and contain one complex in the asymmetric unit, with crystallographic data and refinement statistics listed in Table 1. The structures of RRM1/2 in complexes 1 and 2 were determined by multi-wavelength anomalous dispersion (MAD) phasing on Se atoms using selenomethionine-labeled protein. The structure of complex 3 was solved by molecular replacement using the refined structure of complex 1 determined at 1.85 Å resolution, as a search model (see Supplemental Experimental Procedures). The data collection and refinement statistics for all three complexes are listed in Table 1. All three structures are very similar and contain one RRM1/2 molecule and either 5 (UUGUU segment) of 12 RNA nucleotides in complexes 1 and 2, or 6 (UUGUGU segment) of 13 RNA nucleotides in complex 3 with the remaining RNA segment disordered in each structure. While the bound U7-U8-G9-U10-G11-U12 sequence can be unambiguously identified in complex 3, there are two options for the bound UUGUU sequences in the crystal structures of complexes 1 and 2. To determine which of these two motifs is found in the crystal structures, we have replaced U2 and U8, one at a time, in the GUUGUUUUUGUU sequence, with 5-bromouridines. The electron density maps for the Br atoms in the two structures obtained with the derivative RNA sequences clearly indicate that both BrU2-U3-G4-U5-U6 and BrU8-U9-G10-U11-U12 can be bound by the protein (Supplemental Figures S1A and S1B). To simplify the structural description of the complexes, we numbered the bound RNA motifs as U2-U3-G4-U5-U6 in the crystal structures of complexes 1 and 2. The experimental multiple anomalous dispersion (MAD) electron density map for the bound U3-G4-U5 RNA segment and contacting amino acid residues of complex 1 is shown in Figure S1C.
Table 1.
Data Collection and Refinement Statistics
| Complex 1 | Complex 2 | Complex 3 | Complex 4 | |
|---|---|---|---|---|
| CUGBP1 RRM | RRM1/2 | RRM1/2 | RRM1/2 | RRM1 |
| RNA sequence | GUUGUUUUGUUU | GUUGUUUUUGUU | UGUGUGUUGUGUG | GUUGUUUUGUUU |
| Data collection | MAD | MAD | Native | Native |
| Space group | I222 | I222 | I222 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 47.6, 70.0, 132.2 | 47.4, 70.1, 131.7 | 47.7, 70.0, 132.6 | 59.2, 62.1, 122.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-1.85 | 50-1.9 | 40-2.2 | 40-2.75 |
| Rsym or Rmerge | 3.6 (36.9)a | 4.2 (36.6) | 12.2 (42.9) | 11.4 (37.6) |
| I/σI | 36.7 (3.6) | 36.3 (3.7) | 11.5 (3.0) | 13.1 (4.6) |
| Completeness (%) | 99.6 | 99.7 | 98.2 | 99.8 |
| Redundancy | 3.8 | 3.8 | 5.5 | 5.3 |
| Refinement | ||||
| Resolution (Å) | 20-1.85 | 20-1.9 | 20-2.2 | 20-2.75 |
| No. reflections | 18,231 | 16,774 | 10,839 | 11,775 |
| Rwork/Rfree | 20.4/24.5 | 20.7/24.7 | 20.1/27.1 | 18.3/26.3 |
| No. residues | ||||
| Protein | 175 | 175 | 175 | 344 |
| RNA | 5 | 5 | 6 | 20 |
| Water | 100 | 98 | 64 | 29 |
| B-factors | ||||
| Protein | 31.5 | 33.6 | 31.4 | 32.0 |
| RNA | 35.1 | 34.9 | 40.8 | 48.3 |
| Water | 37.3 | 40.0 | 35.9 | 26.7 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.010 | 0.007 |
| Bond angles (°) | 1.3 | 1.3 | 1.5 | 1.0 |
Values in parentheses are for highest-resolution shell.
The CUGBP1 RRM1-RNA complex 4 crystals belong to the P212121 space group and contain two RNA and four RRM1 molecules in the asymmetric unit, with the structure solved by molecular replacement. The crystallographic data and refinement statistics are listed in Table 1, while the simulated annealing omit map for the bound U8-G9-U10 RNA segment in complex 4 is shown in Figure S1D.
The crystal structures of RRM1/2-RNA and RRM1-RNA complexes are shown in Figure 2, with details of intermolecular contacts between these two complexes compared in Figures 2 to 5.
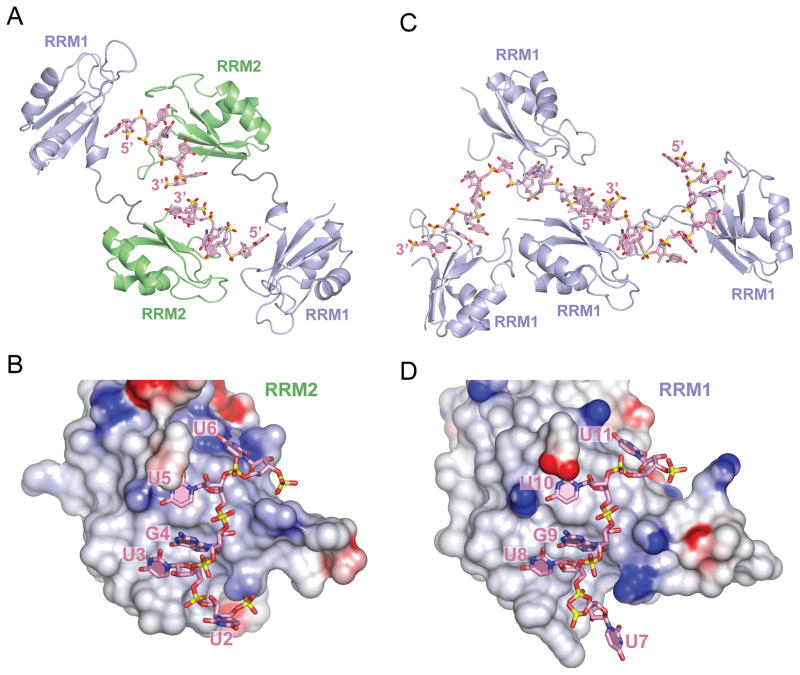
Figure 2. Overall Structures of CUGBP1 RRM1/2-RNA and RRM1-RNA Complexes.
(A) Ribbon and stick representation of two crystallographically related CUGBP1 RRM1/2 molecules bound to UGUU segments of two molecules of RNA GUUGUUUUGUUU RNA (sequence 1). The RRM2 domain (green) interacts with the U3-G4-U5-U6 segment, while RRM1 domain (blue) interacts with the U2 of the symmetry related RNA molecule. RNA strands are colored pink, with the backbone phosphorous atoms colored yellow. Nitrogen, oxygen and phosphate atoms are colored dark-blue, red and yellow in the RNA structure.
(B) An electrostatics surface view of RRM2 bound to U2-U3-G4-U5-U6 in the RRM1/2-RNA complex (RNA sequence 1) generated using the GRASP and PyMol programs. Basic and acidic regions of the protein appear in blue and red, with the intensity of the color being proportional to the local potential. The U3-G4-U5-U6 segment in pink (stick representation) contacts RRM2, while U2 is flipped out and directed away from RRM2.
(C) Ribbon and stick representation of four CUGBP1 RRM1 molecules bound to two molecules of UGUGUGUUGUGUG RNA (sequence 4) in the crystallographic asymmetric unit of the complex. Each RRM1 interacts with either U3-G4-U5-U6 or U8-G9-U10-U11 segment of the RNA. Protein and RNA are color coded as in panel A.
(D) An electrostatics surface view of RRM1 bound to U7-U8-G9-U10-U11 in the RRM1-RNA complex (RNA sequence 4). The U8-G9-U10-U11 segment in pink contacts RRM1, while U7 is flipped out and directed away from RRM1.
See also Figure S1.
Figure 5. Superposition of UG steps in RRM-RNA complexes of CUGBP1 with CG steps in Z-helical RNA.
(A) Stereo-view of the superposed structures of U3-G4 in complex with CUGBP1 RRM1/2 and C3-G4 of dUr(CG)3 Z-RNA helix in complex with ADAR1 Zα domain (PDB code 2GXB, chain E). Similar conformations of the sugar-phosphate backbones and the guanines (syn) in U3-G4 (pink) and C3-G4 (cyan) are highlighted. A water molecule mediates interaction between the guanine amino group and the phosphate oxygen of U5/C5 in the two structures. RNA intramolecular and base-pair hydrogen bonds are colored in grey, while protein-RNA intermolecular hydrogen bonds are in orange. Residues of CUGBP1 RRM2 interacting with U3 phosphate, G4 base and G4 2′-OH are colored in green; residues of ADAR1 Zα domain interacting with C3 phosphate and G4 2′-OH are colored cyan. The pseudorotation angles of the sugar rings are 153° (C2′-endo) for U3 and 18° (C3′-endo) for G4 in the CUGBP1 complex, while they are 162° (C2′-endo) for C3 and 17° (C3′-endo) for G4 for the ADAR1 complex.
(B) Stereo-view of the superposed structures of U8-G9 in complex with CUGBP1 RRM1 and C3-G4 of dUr(CG)3 Z-RNA helix in complex with ADAR1 Zα domain. Similar conformations of the sugar-phosphate backbones and the guanines (syn) in U8-G9 (pink) and C3-G4 (cyan) are highlighted.
See also Table S1.
Structure and intermolecular Contacts in the CUGBP1 RRM1/2-RNA Complex
The overall structure of CUGBP1 RRM1/2 bound to the UUGUU segment of the 12-nt GUUGUUUUGUUU sequence (complex 1), and crystal packing of two complex molecules related by two-fold crystallographic symmetry, are shown in Figure 2A. Within each polypeptide chain, the RRM1 (blue) and RRM2 (green) domains do not interact with each other, and the interdomain linker adopts an extended conformation. The two RRM domains have the same β1α1β2β3α2β4 topology (Figures 1A and 2A) with the two α-helices packed against a four-stranded β-sheet (pairwise Cα rmsd between RRM1 and RRM2 of 0.84 Å for 70 matching residues, Figure 1A), as observed in the RNA-free RRM1/2 structure determined by NMR (PDB ID 2DHS) in solution (Jun et al., 2004). The backbone conformations of the RRM’s are retained between the NMR structure of RRM1/2 in the free state and the x-ray structures in the RNA-bound state, with the largest differences observed for the loops between β2 and β3 of RRM1 and between α2 and β4 of RRM2. In addition, the relative orientation of the two RRMs, as well as conformation of the interdomain linker (residues 97-107), are different between the NMR-based solution (free state) and x-ray (RNA-bound state) structures. Crystal packing interactions could contribute to the observed differences between solution and crystalline states.
CUGBP1 RRM2 contacts U3-G4-U5-U6 of the 5-nt RNA segment within the 12-mer sequence via its β-sheet surface and the two loops, β1α1 and α2β4 (Figures 2B and stereo view in 3A), whereas U2 is flipped out and interacts with the RRM1 of the symmetry related molecule (Figures 2A, 2B and S2). The bound U3-G4-U5-U6 segment is surrounded by positively charged patches on the surface of the β-sheet and the β1α1 loop of the RRM2 (Figure 2B). Positioning of the first three nucleotides of the bound UGU(U/G) motif are virtually identical in all three complexes, whereas the fourth base, U in complexes 1 and 2, and G in complex 3, shows the largest conformational variations (Figure S3).
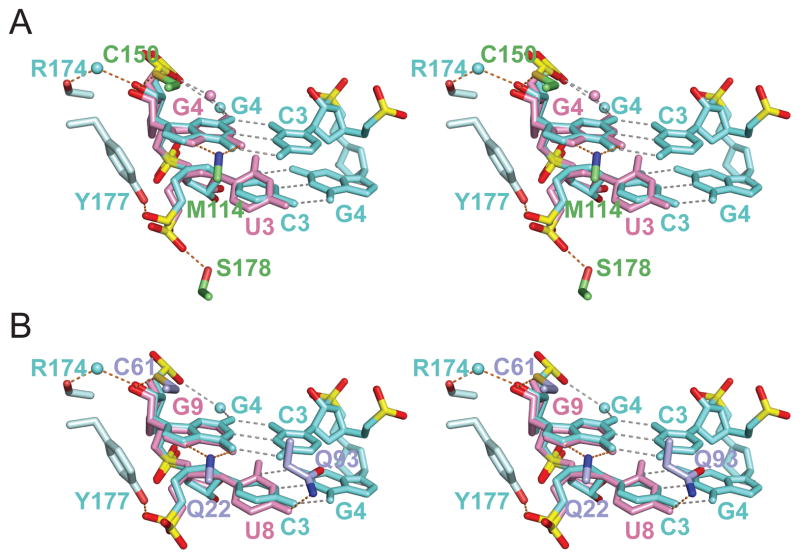
Figure 3. Protein-RNA Intermolecular Interactions in the CUGBP1 RRM1/2-RNA and RRM1-RNA Complexes.
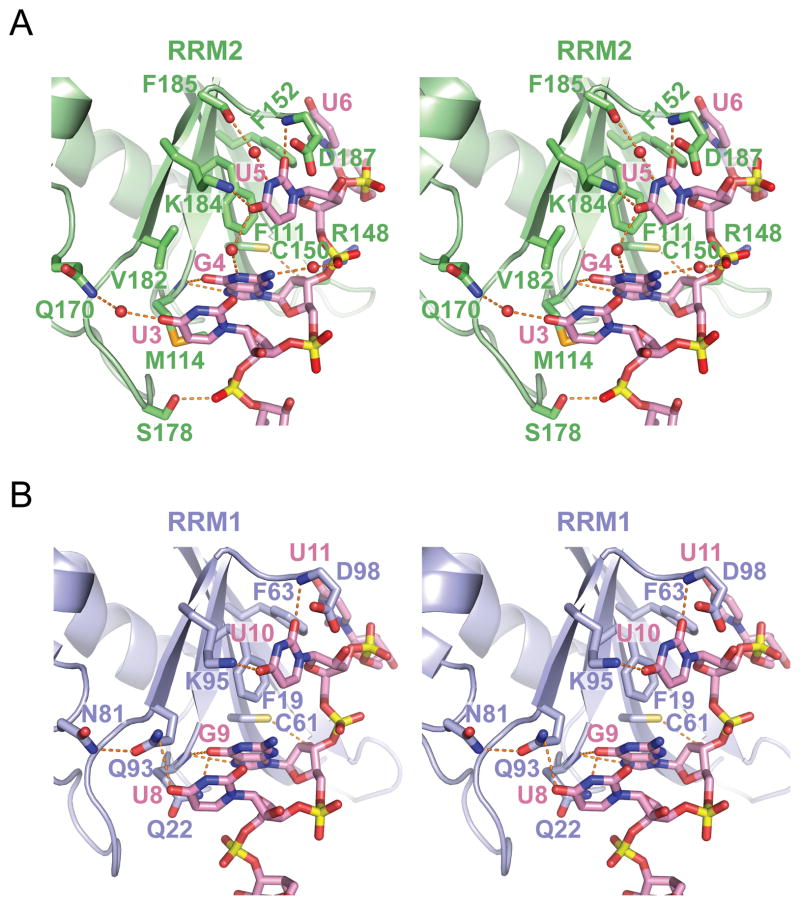
(A) Stereo-view of the protein-RNA interface highlighting intermolecular contacts between U3-G4-U5-U6 segment of GUUGUUUUGUUU RNA (sequence 1) and RRM2 in the RRM1/2-RNA complex. Stacking interactions of U5 base with Phe residue, as well as hydrogen-bonding contacts involving U3, G4 and U5 are highlighted.
(B) Stereo-view of the protein-RNA interface highlighting intermolecular contacts between U8-G9-U10-U11 segment of UGUGUGUUGUGUG RNA (sequence 4) and RRM1 in the RRM1-RNA complex. Stacking interactions of U10 base with Phe residue, as well as hydrogen-bonding contacts involving U8, G9 and U10 are highlighted.
See also Figure S2.
Sequence-specific intermolecular recognition involving U3 (Figures 3A in stereo and 4A), G4 (Figures 3A in stereo and 4B) and U5 (Figure 3A in stereo and 4C) in the RRM1/2-RNA complex 1 are mediated by a network of direct and water-mediated hydrogen bonds and base-aromatic amino acid stacking interactions (Auweter et al., 2006), as outlined in detail in the Supplemental Information. Specifically, the Watson-Crick edges of U3, U5 and U6 are involved in a total of three direct and two water-mediated hydrogen bonds. The Watson-Crick and Hoogsteen edge of G4 are involved in two direct and one water-mediated hydrogen bonds. While U3 and U5 adopt anti glycosidic torsion angles, G4 adopts an unanticipated syn torsion angle in the complex.
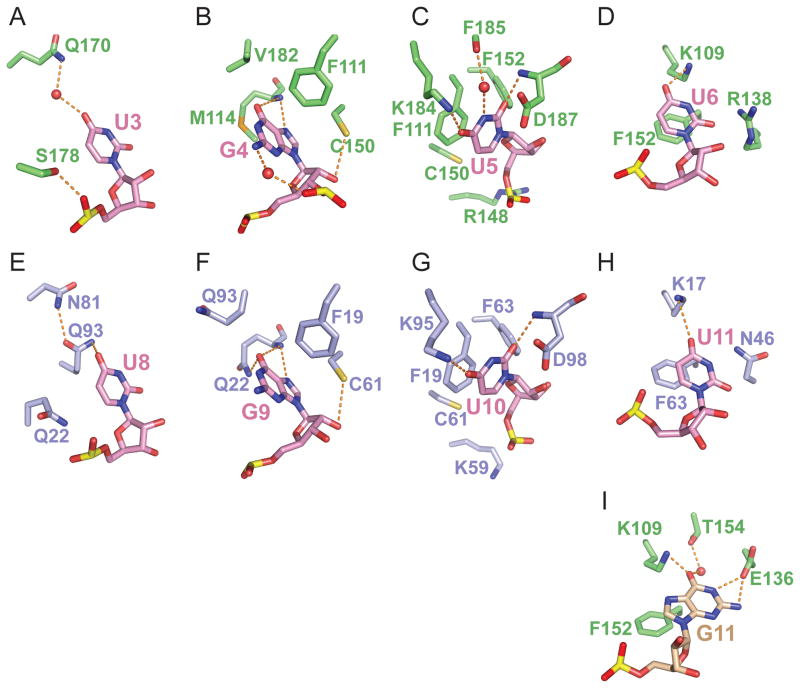
Figure 4. Intermolecular Recognition of individual nucleotides in CUGBP1 RRM1/2-RNA and RRM1-RNA Complexes.
(A–D) Intermolecular hydrogen bonds associated with recognition of U3-G4-U5-U6 by RRM2 in the RRM1/2-RNA complex (GUUGUUUUGUUU RNA sequence 1). The Watson-Crick edges of U3 (panel A), U5 (panel C) and U6 (panel D) are involved in three direct and two water-mediated hydrogen bonds. The Watson-Crick and Hoogsteen edges of G4 (panel B) are involved in two direct and one water-mediated hydrogen bonds.
(E–H) Intermolecular hydrogen bonds associated with recognition of U8-G9-U10-U11 by RRM1 in the RRM1-RNA complex (UGUGUGUUGUGUG RNA sequence 4). The Watson-Crick edges of U8, U10 and U11 are involved in four direct hydrogen bonds. The Watson-Crick and Hoogsteen edges of G4 are involved in three direct hydrogen bonds.
(I) Intermolecular hydrogen bonds associated with recognition of G11 in the RRM1/2-RNA complex (UGUGUGUUGUGUG RNA sequence 3). The Watson-Crick edge of G11 is involved in three direct and one water-mediated hydrogen bonds.
See also Figure S3.
Interestingly, the U2 base bound by the symmetry related RRM1 (Figure S2) appears to occupy a position equivalent to U5 bound by RRM2 (Figure 3A). Details of the intermolecular contacts between U2 and RRM1 are outlined in the Supplemental Information.
The U6 base is only partially defined in the electron density map of complex 1, while the G11 base, that occupies this position in complex 3, is fully-defined in the electron density map. Details of the intermolecular contacts involving U6 (and G11) and RRM2 (Figures 3A in stereo view and 4D,I) are outlined in the Supplemental Information.
Structure and intermolecular contacts in the CUGBP1 RRM1-RNA Complex
The overall crystal structure of CUGBP1 RRM1 bound to the 12-nt GUUGUUUUGUUU sequence (complex 4) is shown in Figure 2C. The crystallographic asymmetric unit is composed of four RRM1 molecules bound to two 12-nt RNAs, with each of the four RRM1 domains contacting either U3-G4-U5-U6 or U8-G9-U10-U11 segments of the two 12-nt RNAs. Both UGUU tetranucleotide segments are similarly positioned on RRM1 and are surrounded by basic patches on the surfaces of the β-sheet and the β1α1 loop (U8-G9-U10-U11 shown in Figure 2D).
We observe similar intermolecular contacts between U8-G9-U10-U11 and the RRM1 domain in the RRM1-RNA complex and U3-G4-U5-G6 and the RRM2 domain in the RRM1/RRM2-RNA complex (compare Figures 2B with 2D, stereo views 3A with 3B, 4A–D with 4E–H).
CUGBP1 RRM1- and RRM2-bound U-G RNA Steps Adopt Left-handed Z-helices
The sugar-phosphate backbones of the U3-G4 step in the CUGBP1 RRM1/2-RNA complex shares unanticipated structural features with that reported for the Z-RNA helix. Thus, superposition of U3(anti)-G4(syn) with the C3(anti)-G4(syn) step of the Z-RNA-ADAR1 Zα complex (Placido et al., 2007), highlights their striking structural similarity (stereo pair in Figure 5A). Similarly, the U8(anti)-G9(syn) step in the CUGBP1 RRM1-RNA complex also adopts a Z-RNA helical conformation (stereo pair in Figure 5B).
NMR Chemical Shift Mapping of RNA-binding Surface of CUGBP1 RRM Domains
Similarities in the mode of RNA recognition by RRM1 in the CUGBP1 RRM1-RNA complex (Figures 2D and 3B) and RRM2 in the CUGBP1 RRM1/2-RNA complex (Figures 2B and 3A) suggests that tandem RRM domains should recognize a pair of UGU(U/G) segments within the RNA sequence. Nevertheless, we did not observe simultaneous binding of RRM1 and RRM2 to adjacent UGU(U/G) motifs in the 12–13 nt RNAs in the crystal structures of complexes 1–3 (Figure 2A), presumably due to crystal packing interactions.
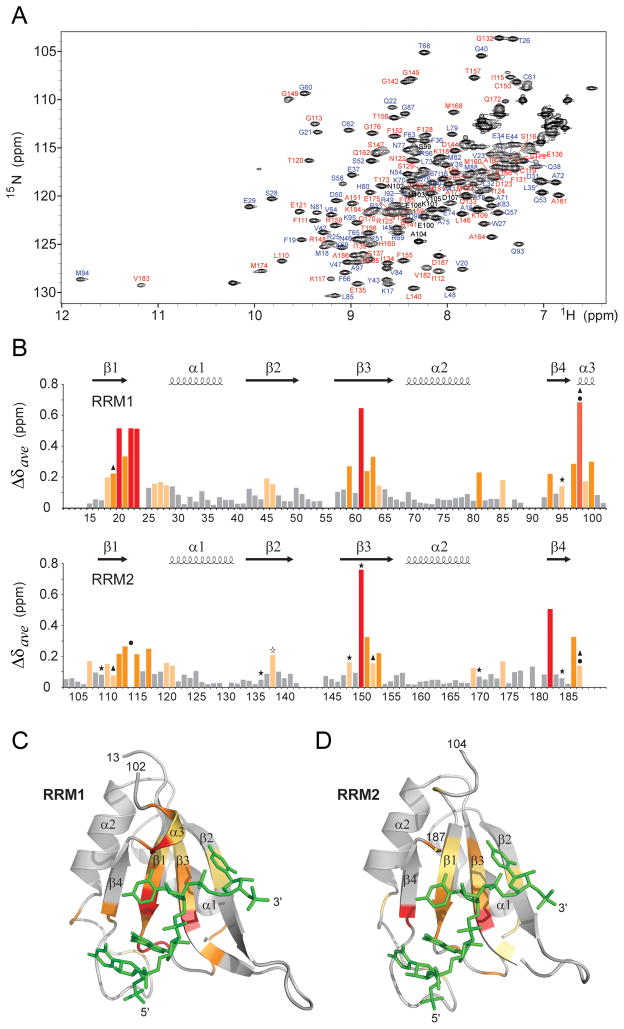
To determine whether both RRM1 and RRM2 are involved in RNA binding in solution, we performed studies of CUGBP1 RRM1/2 (14–187) in the free state and bound to GUUGUUUUUUGU RNA (complex 2) by NMR spectroscopy. We could assign 97% of backbone residues (excluding prolines) in a 1H-15N-HSQC spectrum of the complex (RRM1 and RRM2 residues are labeled in blue and red, respectively; Figure 6A). The largest weighted 1H/15N chemical shift changes on complex formation map to β1, β3, and β4 strands, as well as to the β1α1 loop, α3-helical segment of the interdomain linker, and C-terminus residues (Figure 6B). These changes when mapped onto the backbone structure of RRM1 (Figure 6C) and RRM2 (Figure 6D), indicate that RNA binds to the same surface on each domain. Phe19/Phe111, Gln22/Met114, Cys61/Cys150, Phe63/Phe152, Gln93/Val182 and Asp98/Asp187 residues, that mediate intermolecular contacts in the crystal structures of RRM1/2-RNA complexes 1–3 (Figure 3A) and of RRM1-RNA complex 4 (Figure 3B), are located ether within or adjacent to these regions (Figure 6B). Smaller complexation shifts are rationalized in the Supplemental Information.
Figure 6. Resonance Assignments and Chemical Shift Changes in NMR Spectra of CUGBP1 RRM1/2-RNA Complex upon Binding of GUUGUUUUUGUU RNA (complex 2).
(A) 1H-15N HSQC spectrum of RRM1/2 bound to GUUGUUUUUGUU RNA (sequence 2) in 20 mM Na-Hepes, pH 7.0, 100 mM NaCl, 1mM DTT at 25 °C. Amino acid assignments are listed adjacent to the resolved cross peaks, with those from RRM1 in blue, from RRM2 in red and from the interdomain linker in black.
(B) Histogram outlining the magnitude of the average chemical shift perturbation of the 15N and 1H backbone amide resonances of the CUGBP1 RRM1/2 on complex formation with RNA. The average chemical shift difference Δδave between the free and RNA-bound forms of RRM1/2 was calculated using a correlation: , where ΔδH is the chemical shift of amide proton and ΔδN is the chemical shift of amide nitrogen. Protein secondary structure elements are indicated on the top. Asterisks denote residues that form hydrogen bonds with RNA bases via their side chains; triangles indicate aromatic and aspartate residues involved in base-stacking with RNA bases; and circles mark residues interacting with RNA via their backbone functional groups in the crystal structure of the complex.
(C) The average amide chemical shift perturbations mapped onto the backbone trace of the RRM1 domain bound to UGUU motif (stick representation in green) in the crystal structure of the RRM1-RNA complex (complex 4).
(D) The average amide chemical shift perturbations mapped onto the backbone trace of the RRM2 domain bound to UGUU motif (stick representation in green) in the crystal structure of the RRM1/2-RNA complex (complex 1).
Residues that undergo changes in average amide chemical shift are color-coded as follows: red (>5-fold above average), orange (>2-fold above average) and yellow (between 1.2 to 2-fold above average) in panels B, C and D.
See also Figure S4.
NMR Relaxation Times of CUGBP1 RRM1/2 in Free and RNA Bound States
Transverse and longitudinal relaxation rate ratios (R2/R1) were measured to obtain information about the dynamics of the CUGBP1 RRM1 and RRM2 domains in the absence and presence of GUUGUUUUUGUU 12-mer RNA (Figure S4 and Supplemental Information). The estimated rotational correlation times are consistent with partial rigidification of the tandem RRM domains upon formation of the RNA complex.
Impact of G-U Motif Substitutions on Binding Affinity in CUGBP1 Complexes
To assess RNA-binding specificity of CUGBP1 RRM1/2 construct, we measured apparent equilibrium dissociation constants (KD) using nitrocellulose filter binding assay with GUUGUUUUUGUU RNA dodecamer (complex 2) containing base substitutions at G4, U5, G10 and U11 positions. RRM1/2 binds this RNA with 1:1 stoichiometry as determined by polyacrylamide electrophoretic mobility shift (gel-shift) assay (Figure 7A). The KD value of 0.9 ± 0.1 μM for RRM1/2 binding to this 12-nt sequence (Figure 7B,C) is comparable to ITC-measured KD value of 0.65 ± 0.07 μM for RRM1/2 binding to GUUGUUUUGUUU (Figure 1B,E), as well as to those previously measured for full-length CUGBP1 binding to (UG)15 (0.25 ± 0.1 μM) and (UUG)10 (3.6 ± 0.1 μM) repetitive sequences determined by surface plasmon resonance (SPR) (Mori et al., 2008).
Figure 7. Binding of CUGBP1 RRM1/2 to UUGU-containing RNAs and Impact of Base Substitution Mutants within the GU Segment.
(A) Electrophoretic mobility gel shift data for binding of GUUGUUUUUGUU RNA (sequence 2) to RRM1/2, establishing 1:1 stoichiometry of the complex.
(B) RNA sequences used in binding experiments. GU motifs and their substitutions in each sequence are shown in red. Percentage of the bound RNA fraction and apparent equilibrium binding constants (KD) measured by nitrocellulose filter-binding assay are listed together with ± fitting error.
(C) Filter-binding assays for complex formation of RRM1/2 with GUUGUUUUUGUU RNA (black circles) and RNAs containing double substitutions of either G4 and G10 or U5 and U11 in this sequence (colored circles).
(D) Filter-binding assays for complex formation of RRM1/2 with GUUGUUUUUGUU (designated G4G10, black circles), GUUGUUUUGUUU (designated G4G9, green circles), UUGUU (red circles) RNAs, as well as mutants containing single substitutions of G4 to A (blue circles) and G10 to A (cyan circles).
RNA sequences used in panels C and D and measured binding constants are listed in panel B. The plots represent mean ± standard deviation (SD) for at least two independent measurements. Solid lines indicate non-linear least-squares fit according to equation 1 in the Methods section.
(E) Chemical structures of the bases guanine (G), inosine (I), 2-aminopurine (2AP) and adenine (A). Differences in chemical structures compared to guanine are highlighted in pink.
(F) Intermolecular contacts defining recognition of G4 and U5 in the complex. Hydrogen-bonding of the O6 and N7 of G4 with the backbone amide of Met114; N2 and N1 of G4 with U3 O2; and water-mediated hydrogen bonding of the N2 and N1 of G4 with U5. Lys109 side chain Nζ is hydrogen-bonded to U5 O4.
We observed undetectable binding for the A4A10, C4C10 and U4U10 dual mutants in which guanines in positions 4 and 10 are simultaneously replaced by either A, C or U (Figures 7B and 7C), whereas substitutions of single guanine at either position 4 or 10 for adenine, reduced binding efficiency of A4G10 and G4A10 mutants by a factor of 4.4 and 2.5, respectively, as well as a drop to 30–40% value of maximum bound RNA fraction (Figures 7B and 7D). Similar binding properties were observed for a 5-nt UUGUU sequence (Figures 7B and 7D), indicative that each of the two UGUU motifs within the 12-nt sequence must be involved in the interactions with the tandem RRM1/2 domain. The two 12-nt RNA sequences used in crystallization of the complex, G4G10 (complex 2) and G4G9 (complex 1) (Figure 7B), in which either a 2 or 1-nt spacer separates the two UGUU motifs, respectively, bind the CUGBP1 RRM1/2 with similar affinities (Figure 7D).
The O6 functional group of the bound G4/G9 base is involved in intermolecular hydrogen-bonding interactions in the crystal structures, while the N2 amino group is involved in water-mediated intramolecular hydrogen bond and is located within hydrogen-bonding distance to O2 of U3/U8 (Figure 3). To assess the contribution of these functional groups of G4/G10 to the CUGBP1 RRM1/2 binding affinity, we have monitored the binding of inosine (I) or 2-aminopurine (2-AP) (Figure 7E) substitutions for guanines in positions 4 and 10. Simultaneous replacement of G4 and G10 by inosines (I4I10) resulted in app. 3.5-fold loss in binding affinity, with an estimated maximum fraction of bound RNA of 56% (Figures 7B,C). A similar reduction in binding affinity of app. 3.4-fold, was measured for the guanine to 2-aminopurine substitutions at positions 4 and 10, (2AP)4(2AP)10 (Figures 7B,C). However, the fraction of RNA bound at saturating protein concentrations for this mutant was twice lower than for the unmodified sequence. These data are indicative of reduced binding efficiencies following substitution of guanine by analogs, with the bigger impact for the 2AP-substitution (loss of two hydrogen bonds per substitution) compared to I-substitution (loss of one hydrogen bond per substitution). Furthermore, the loss of all three functional groups following adenine for guanine substitution (A4A10) resulted in undetectable binding (Figures 7B,D), consistent with the loss of all hydrogen bonding interactions and the unfavorable close contact of the adenine (syn) 6-amino group with the protein backbone amide (Figure 7F).
Substitution of uracils at the U5 and U11 positions by cytosines reduced binding affinity by a factor of app. 5 (Figures 7B and 7C). However, the KD value for the C5C11 mutant was measured with high (40%) error, as only 16% of total RNA was bound at the highest protein concentration used. A previously reported 10-fold reduction in CUGBP1 binding affinity for (CUG)10, which contains GC rather than GU steps, with respect to the (UG)15 sequence (Mori et al., 2008), is consistent with our observations.
DISCUSSION
In this study, we report on the first crystal structures of single and tandem RRM domains of CUGBP1 bound to their UGUU- and UGUG-containing single-stranded RNA targets. The crystal structures reveal that RRM1 in the RRM1-RNA complex and RRM2 in the tandem RRM1/2-RNA complex target UGU(U/G)-containing sequences in which RRM1 and RRM2 use the same principles for sequence-specific RNA recognition. Specifically, both RRM domains recognize the UG sugar-phosphate backbone and the base edges in the GU step through direct and water-mediated intermolecular hydrogen-bond formation and stacking interactions, as well as by selecting the G(syn) conformation on the basis of shape complementarity. The ability of both RRM1 and RRM2 to recognize UGU(U/G) steps explains the preference of CUGBP1 for UGU and GU repeats found in many mRNAs (Charlet et al., 2002; Kalsotra et al., 2008; Paillard et al., 2002; Paillard et al., 1998; Paillard and Osborne, 2003; Philips et al., 1998; Savkur et al., 2001; Vlasova et al., 2008) and SELEX-based RNA targets (Faustino and Cooper, 2005; Marquis et al., 2006). In solution, in the absence of crystal packing effects, similar NMR chemical shift perturbations demonstrate that both RRM1 (Figure 6C) and RRM2 (Figure 6D) in RRM1/2 bind to a pair of UGUU segments via analogous sets of amino acids located on the surface of the β-sheet and adjacent loops of each RRM.
CUGBP1 RRM-bound U-G Step Adopts a Left-handed Z-helix
The sugar-phosphate backbone of the U3-G4 and U8-G9 steps in the GUUGUUUUGUUU sequence in complexes with bound RRM1 and tandem RRM1/2 domains share unanticipated structural features with that reported for the Z-RNA helix. Thus, superposition of U3-G4 in the RRM1/2-RNA complex with the C3-G4 step of the Z-RNA-ADAR1 Zα complex (Placido et al., 2007), highlights their striking structural similarity (stereo pair in Figure 5A). The sugar-phosphate backbone parameters closely follow those that have been observed for the Z-RNA structure (Table S1). In particular, the ribose of U3 adopts a C2′-endo pucker, as has been reported for all cytidine residues in Z-RNA, whereas the ribose of G4, similar to guanine residues in Z-RNA, adopts the typical C3′-endo RNA pucker (Table S1). Similarly, U8-G9 in the RRM1-RNA complex superpositions well with the C3-G4 step of the Z-RNA-ADAR1 Zα complex (stereo pair in Figure 5B). A more detailed comparison is outlined in the Supplemental Information.
It should be noted that only the left-handed Z-RNA conformation (but not its right-handed counterpart) for the U-G steps can be accommodated within its target pockets on RRM1 and RRM2 in the complex. Further, the Watson-Crick edges of the left-handed U-G steps are exposed outwards and available for further recognition. Thus, our study demonstrates that the Z-RNA conformation interacts with proteins not only at the duplex level as established previously (Placido et al., 2007), but also at the single strand level as shown for the first time in the present study.
Comparison of RNA-binding Surfaces of CUGBP1 RRM1 and RRM2
At the crystallographic level, a striking similarity is observed in the intermolecular contacts defining recognition between RRM1 and the U3-G4-U5-G6 step in the RRM1-RNA complex and between RRM2 and the U8-G9-U10-U11 step in the RRM1/2-RNA complex (comparison of Figures 2B with 2D, Figures 3A with 3B in stereo, and Figures 4A,B,C,D with Figures 4E,F,G,H).
The NMR chemical shift perturbation analysis demonstrated that RRM1 and RRM2 of the CUGBP1 RRM1/2 construct are both involved in binding the GUUGUUUUUGUU 12-nt RNA (complex 2; Figure 6). Notably, each of the two UGUU recognition motifs of this RNA are bound by individual RRM domains of RRM1/2, as substitutions of the central guanine and uracil bases in either of the two UGUU motifs caused reduction of binding affinity as monitored by filter-binding assays (Figures 7B,C,D). The striking structural similarity between RRM1 (Figure 6C) and RRM2 (Figure 6D) folds, as well as the high conservation of side chains recognizing the uracil and guanine bases (Figure 1A), reinforces the similar modes for recognition of RRM1 and RRM2 by UGUU in the crystal structures of the complexes.
The qualitative trend (1.5-fold higher binding affinity by ITC measurements) for complex formation between UUGUU 5-mer RNA and RRM1 compared to RRM2 (Figure 1E) could reflect formation of a direct intermolecular hydrogen bond between O4 of U8 and side chain of Q93 in RRM1 recognition of RNA (Figure 4E), that is absent in the corresponding RRM2 recognition of RNA (Figure 4A). In addition, a direct hydrogen bond between O6 of G9 and side chain of Q22 in RRM1 recognition of RNA (Figure 4F) is absent in the corresponding RRM2 recognition of RNA (Figure 4B).
Relative Alignments of RRM Domains and UGUU Sites in Models of the CUGBP1 RRM1/2-RNA Complex
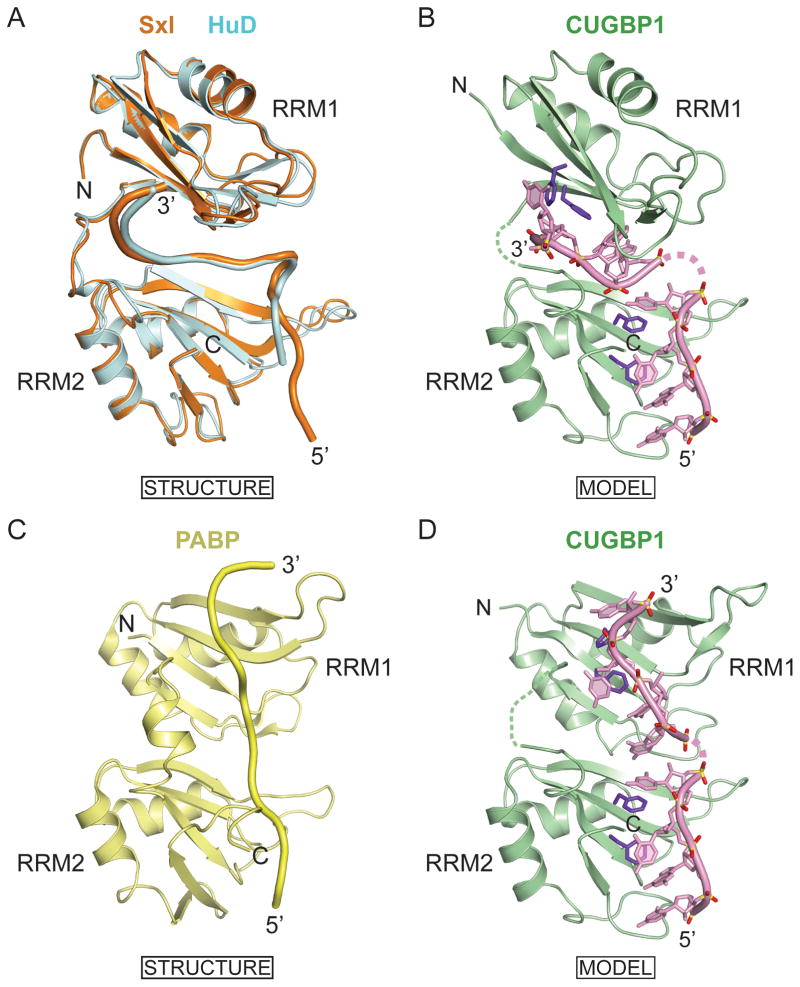
The relative arrangement of tandem RRM domains can generate an extended interface, thereby facilitating targeting of longer RNA sequences. This results in increased binding affinity and specificity, as shown previously for Sex-lethal (Sxl) (Handa et al., 1999), HuD (Wang and Tanaka Hall, 2001), PABP (Deo et al., 1999), nucleolin (Allain et al., 2000) and Hrp1 (Perez-Canadillas, 2006) RNA-binding proteins. A search of the Dali database (Holm and Sander, 1995) revealed that the folds of the first two RRM domains of CUGBP1 most closely match the folds of the RRM1 of the Sxl and the first two RRM domains of HuD protein. Crystal structures of the tandem RRM1 and RRM2 domains of Sxl bound to a pyrimidine-rich tract (Handa et al., 1999), as well as that of HuD bound to AU-rich elements (AREs) (Wang and Tanaka Hall, 2001), have shown that both RRM domains in these proteins engage in multiple base-specific contacts, thereby explaining their sequence preferences. The relative orientation of the two RRMs in the RNA bound complexes of Sxl and HuD complexes are very similar (Figure 8A), thereby facilitating recognition of a 8–10 nucleotide RNA sequence. In the structures of both complexes, the RNA backbones adopt very similar folds, including a sharp turn, when positioned in a deep cleft between the two RRM domains (Figure 8A).
Figure 8. Models of CUGBP1 RRM1/2 Binding to Adjacently Positioned UGUU Motifs.
(A) Superposition of structures of RRM1 and RRM2 domains of Sex-lethal (PDB code 1B7F) and HuD (PDB code 1FXL) in complexes with their respective RNA targets. RRMs and RNAs of Sex-lethal and HuD are shown in orange and cyan, respectively.
(B) Model of CUGBP1 RRM1/2-RNA complex generated by using coordinates of RRM1 from the RRM1-RNA complex and RRM2 from the RRM1/2-RNA complex. The relative orientation of CUGBP1 RRM1 and RRM2 is modeled based on alignments in the Sx1 and HuD RRM1/2-RNA complexes shown in panel A. Each CUGBP1 RRM interacts with a UGUU RNA element in the model.
(C) Structure of RRM1 and RRM2 domains of PABP (PDB code 1CVJ) in complex with its RNA target. RRMs and RNAs are shown in light green.
(D) Model of CUGBP1 RRM1/2-RNA complex generated by using coordinates of RRM1 from the RRM1-RNA complex and RRM2 from the RRM1/2-RNA complex. The relative orientation of CUGBP1 RRM1 and RRM2 is modeled based on alignments in the PABP RRM1/2-RNA complexes shown in panel C. Each CUGBP1 RRM interacts with a UGUU RNA element in the model.
Pair-wise superposition of the first two RRM domains of CUGBP1 (in green), each bound to a UGUU element (Figure 8B), with the two RRMs of Sxl (in orange) and HuD (in cyan) proteins bound to their RNA targets, indicates that the two UGUU segments can bind to RRM1 and RRM2 of CUGBP1 in the same orientation as was found for Sxl- and HuD-RNA complexes. Similar length linker segments (12–13 amino acids) separate RRM1 and RRM2 in all three proteins, thereby suggesting that RRM1 and RRM2 in CUGBP1 could adopt a similar orientation upon binding to the GUUGUUUUUGUU 12-nt sequence. The relative alignment of CUGBP1 RRM1 and RRM2 should allow binding of two UGUU motifs when separated by at least a single nucleotide linker (Figure 8B). This model is supported by the virtually identical measured binding affinities of CUGBP1 RRM1/2 for 12-nt GUUGUUUUGUUU and GUUGUUUUUGUU sequences (Figure 7B), in which a spacer of one or two uridines separates the two UGUU motifs, and by a 45–70-fold lower binding affinity of individual RRMs for a single UGUU motif (Figure 1E).
The complex of RRM1/2 domains of PABP bound to their RNA target (Deo et al., 1999) provides an alternate template for modeling alignment of the CUGBP1 RRM1/2 with its RNA target. The relative alignments of the tandem RRM domains of PABP in its RNA complex (Figure 8C) differ from their counterparts in Sxl and HuD in their RNA complexes (Figure 8A), resulting in a linear trajectory of the bound RNA as it spans the two RRM domains in the PABP complex (Figure 8C). Thus, an alternate model of CUGBP1 RRM1/2 bound to a pair of UGUU targets involving a linear trajectory of the bound RNA is shown in Figure 8D.
At this time, our data do not allow us to differentiate between models shown in Figure 8B and 8D for the solution structure of the complex of CUGBP1 RRM1/2 bound to a pair of UGUU motifs.
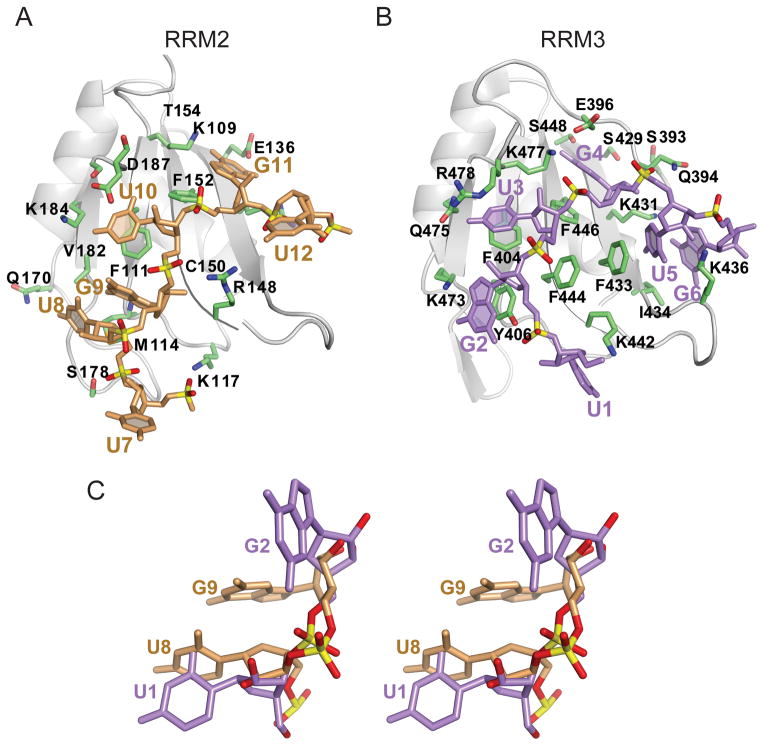
Comparison of RNA-binding Surfaces of CUGBP1 RRM2 and RRM3
While our manuscript on the structure-based analysis of the CUGBP1 RRM1-RNA and RRM1/2-RNA complexes was under preparation, a paper appeared on the solution NMR structure of the CUGBP1 RRM3 bound to the (UG)3 (Tsuda et al., 2009). RRM2 (109–187) and RRM3 (402–478) adopt similar folds, except that a conserved 7-residue N-terminal extension (394–400) of RRM3 interacts with the β-sheet surface forming a unique pocket that plays an important role in RNA recognition. The binding of UGU(U/G) to RRM2 (Figure 9A) exhibits both similarities and differences when compared to the UGUGUG binding by RRM3 (Figure 9B). The RRM3 RNA-binding surface accommodates six nucleotides (U1-G2-U3-G4-U5-G6), four of which (G2, U3, G4 and U5) are stacked with aromatic rings that are conserved among CELF RRM3 domains (Figure 9B). RRM2 binds four nucleotides (U8-G9-U10-G11) in complex 3, only two of which (U10 and G11) are stacked with canonical aromatic rings of RNP2 and RNP1 motifs in the crystal structure of the complex with UGUGUGUUGUGUG RNA target (Figure 9A).
Figure 9. Comparison of structures of CUGBP1 RRM2 bound to U7-U8-G9-U10-G11-U12 element in the crystal with that of CUGBP1 RRM3 bound to U1-G2-U3-G4-U5-G6 element in solution, with an emphasis on bound UG segment.
(A) Ribbon and stick representation of the CUGBP1 RRM2 in complex with U7-U8-G9-U10-G11-U12 (beige) in the crystal structure of the RRM1/2-RNA complex.
(B) Ribbon and stick representation of the CUGBP1 RRM3 in complex with U1-G2-U3-G4-U5-G6 (purple) (PDB code 2RQC) in the NMR solution structure of the RRM3-RNA complex. The U1-G2-U3-G4 segment bound by RRM3 is equivalent to U8-G9-U10-G11 bound by RRM2.
(C) Stereo-view of the superimposed structures of U8-G9 (beige) in complex with CUGBP1 RRM1/2 in the crystal and U1-G2 (purple) in complex with CUGBP1 RRM3 in solution (PDB code 2RQC, molecule 1). Though the guanines adopt a syn alignment in both structures, very different conformations are adopted by the bases and the sugar-phosphate backbones.
See also Figure S5.
The conformations of the first UG dinucleotide segments, U8-G9 bound to RRM2 and U1-G2 bound to RRM3, differ considerably in the two structures (stereo view of superpositioned UG steps, Figure 9C), although a syn conformation is adopted by G9 in RRM2-UG9U(U/G) and G2 in RRM3-UG2UGUG complexes. Noticeably, the U1-G2 step does not adopt a Z-RNA helical conformation for CUGBP1 RRM3 bound to the (UG)3 (Tsuda et al., 2009). A more detailed comparison of the interaction of U8-G9-U10-G11-U12 with RRM2 in the RRM1/2-RNA complex in the crystalline state (Figure 9A), with the interaction of U1-G2-U3-G4-U5-G6 with RRM3 in the RRM3-RNA complex in solution (Figure 9B) can be found in Supplemental Information and Figure S5.
Implications for Alternative Splicing Regulation
CELF proteins have been shown to regulate alternative splicing and to contribute as factors involved in the pathogenesis of myotonic dystrophy (Ho et al., 2004; Philips et al., 1998; Timchenko et al., 1996). CELF proteins bind to UG-rich motifs within muscle specific intronic elements (MSEs) downstream of the cTNT exon 5 to promote exon inclusion (Charlet et al., 2002; Ladd et al., 2001). CELF2 (also called ETR-3) binding sites were mapped to UGUU and UGUG motifs of MSE2 separated by 18 nucleotides, and to two UGUG motifs of MSE3 separated by a 3-nt UCC linker. The sequence specificity of CUGBP1 for UGUU and UGUG motifs revealed by the crystal structures of RRM1-RNA and RRM1/2-RNA complexes reported in this study could have biological relevance, because mutation of these binding sites within elements of cTNT, reduced the ability of CUGBP1 to cross-link with this RNA, as well as its ability to activate inclusion of cTNT exon 5 (Charlet et al., 2002). The 3-nt separation of the two UGUG sites in MSE3, makes it a potential candidate for targeting by the tandem RRM1 and RRM2 domains of CUGBP1.
A recent study (Kalsotra et al., 2008) demonstrated that postnatal changes in CELF and muscleblind-like (MBNL) protein expression determine a large subset of splicing transitions that occur during postnatal heart development. Splicing microarrays and computational screens identified conserved GUGUG CUGBP1-binding motifs among 8 mammalian species, located within the intronic region immediately downstream of the alternative exon, that promotes exon inclusion. Less conserved CGUGU and GUGUC motifs, enriched within the last 250 nucleotides of the downstream intron, were associated with decreased exon inclusion. Thus, CUGBP1 can have both positive or negative effects depending on where it is recruited to the transcript, as has also been shown for the brain-specific splicing regulator NOVA (Ule et al., 2006). The structures of CUGBP1 RRM1/2 bound to UGUGU (this study) and RRM3 bound to UGUGUG (Tsuda et al., 2009) explain the sequence preferences of the CUGBP1 for motifs identified by the splicing microarray analysis. Nevertheless, additional studies are required to clarify the mechanism by which CUGBP1 binding to its RNA targets regulates alternate splicing events, given that it could also involve conformational changes in the RNA structure or competition with the other splicing factors to promote or repress the spliceosome formation.
Implications for Translation Regulation and mRNA decay
In addition to modulating pre-mRNA splicing, CUGBP1 binding to GU-rich elements also plays a key role in the control of mRNA translation and stability. Highly conserved GU-rich elements composed of a consensus UGUUUGUUUGU 11-nt sequence (GRE), that can be targeted by CUGBP1, have been identified as sequence elements enriched in the 3′-UTR of a subset of short-lived transcripts, which encode important regulators of cell cycle and apoptosis (Vlasova et al., 2008). Thus, interaction between CUGBP1 and GRE elements appears to be required for promotion of rapid decay of these transcripts, thereby providing a mechanism to turn off their expression depending on the needs of the cell. A very similar GU-rich EDEN15 motif was found to be overrepresented in the 3′-UTR of numerous targets of embryo deadenylation element-binding protein (EDEN-BP), a Xenopus laevis homolog that is 88% identical to CUGBP1 (Graindorge et al., 2008). EDEN-BP binding to the EDEN element activates deadenylation and subsequent translational repression of EDEN-containing transcripts (Paillard et al., 2003; Paillard et al., 1998).
The similar sequence specificities of CUGBP1 RRM1 and RRM2 revealed by the crystal structures of their RNA complexes could explain the binding preferences of each RRM for UGUU motif found in two copies in 11-nt GRE (Vlasova et al., 2008) and in three copies in 15-nt EDEN15 (Graindorge et al., 2008) consensus sequences. Since the relative orientation of the RRM1 and RRM2 can change significantly as a function of linker flexibility, it is conceivable that the two consecutive UGUU tetranucleotides found in 11-nt GRE and 15-nt EDEN15 can be bound by two N-terminal RRM’s of CUGBP1 (models shown in Figures 8B,D). Further structural analysis of the two N-terminal RRMs bound to two UGUU motifs as a function of spacer length could shed light on the functional features of the CELF protein family.
EXPERIMENTAL PROCEDURES
Detailed procedures for protein and RNA preparation, ITC measurements, crystallization and data collection, structure determination and refinement, NMR sample preparation, NMR spectroscopy and chemical shift assignments, NMR relaxation measurements, gel electrophoretic mobility shift binding assays, and filter binding assays are listed under Experimental Procedures in the Supplemental Information.
Supplementary Material
Acknowledgments
This research was supported by NIH grant CA049882 to DJP. We thank Dr. Haitao Li for x-ray data collection on one of the complexes, and Dr. Vitaly Kuryavyi for identification and helpful discussion regarding the Z-RNA motif and advice on the Curves+ program. We would like to thank the staff of X29 beamline at the National Synchrotron Light Source, Brookhaven National Laboratory, and of NE-CAT beamline at the Advanced Photon Source, Argonne National Laboratory, supported by the US Department of Energy, for assistance with data collection.
Footnotes
Accession codes. Protein Data Bank: Coordinates of the structures of the complexes have been deposited in the Protein Data bank as follows:
CUGBP1 RRM1/2-GUUGUUUUGUUU 12-mer complex: accession code 3NMR.
CUGBP1 RRM1/2-GUUGUUUUUGUU 12-mer complex: accession code 3NNA.
CUGBP1 RRM1/2-:UGUGUGUUGUGUG 13-mer complex: accession code 3NNC.
CUGBP1 RRM1-UGUGUUUUGUUU 12-mer complex: accession code 3NNH.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19:6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraczniak R, Gunderson SI. The regulatory element in the 3′-untranslated region of human papillomavirus 16 inhibits expression by binding CUG-binding protein 1. J Biol Chem. 2008;283:2286–2296. doi: 10.1074/jbc.M708789200. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Jun KY, Xia Y, Han X, Zhang H, Timchenko L, Swanson MS, Gao X. (1)H, (15)N and (13)C chemical shift assignments of RNA repeats binding protein -- CUGBP1ab. J Biomol NMR. 2004;30:371–372. doi: 10.1007/s10858-005-2598-y. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem. 2008;143:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15(Spec No 2):R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Beverley Osborne H. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J Biol Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol Cell. 2003;95:211–219. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Placido D, Brown BA, 2nd, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.