Abstract
Class I myosins participate in various membrane-cytoskeletal interactions. Several class I myosins preferentially bind to acidic phospholipids such as phosphatidylserine and phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) through a tail homology 1 (TH1) domain. Here, we show that the second messenger lipid phosphatidylinositol 3,4,5-trisphosphate (PIP3) binds to the TH1 domain of a subset of Dictyostelium class I myosins (ID, IE, and IF) and recruits them to the plasma membrane. The PIP3-regulated membrane recruitment of myosin I was important for chemotaxis and induced chemoattractant-stimulated actin polymerization. Similarly, PIP3 recruited human myosin IF to the plasma membrane upon chemotactic stimulation in a neutrophil cell line. These data suggest a mechanism through which the PIP3 signal is transmitted through myosin I to the actin cytoskeleton.
Introduction
In eukaryotic cells, multiple pathways mediate intracellular signaling in chemotaxis, the process by which cells sense extracellular chemical gradients and migrate toward higher chemical concentrations (1-5). Phosphatidylinositol 3,4,5-trisphosphate (PIP3) functions as a key signaling molecule and is transiently produced upon chemotactic stimulation by phosphoinositide-3 kinases (PI3Ks) and the phosphatase and tensin homolog (PTEN). Downstream effectors of PIP3 are thought to stimulate actin polymerization and drive pseudopod extension at the leading edge of cells. In Dictyostelium cells, several pleckstrin homology (PH) domain-containing proteins have been identified as PIP3 effectors, including a homolog of AKT (PKBA), cytosolic regulator of adenylyl cyclase (CRAC), and three PIP3-binding proteins (PhdA, PhdB, and PhdG). However, the role of PIP3 signaling in actin polymerization at the leading edge of chemotaxing cells is still not well understood. We identified three Dictyostelium class I myosins (ID, IE, and IF) as PIP3-binding proteins in a proteomic study using PIP3-affinity purification and mass spectrometry (6).
Myosin I is a membrane-bound, actin-based motor protein that functions in membrane-cytoskeletal interactions involved in exocytosis, endocytosis, cell migration and plasma membrane tension (7-9). Several myosin I molecules preferentially bind to acidic phospholipids such as phosphatidylserine and phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) through a TH1 domain that contains a putative PH domain phosphatidylinositol-binding motif (7, 10). These phospholipids are relatively abundant in biological membranes, and their abundances change only slightly in response to intracellular signaling. In contrast, the abundance of PIP3 is regulated and changes in the abundance of PIP3 can trigger signaling events. If myosin I isoforms bind to PIP3, their localization may be regulated by PIP3 in cells. Here, we investigated the mechanisms and cellular function of PIP3-dependent membrane recruitment of class I myosins during chemotaxis and phagocytosis.
Results
To confirm our proteomic results, we used a lipid dot blot assay. All Dictyostelium class I myosins (IA, IB, IC, ID, IE, IF, and IK) were expressed as green fluorescent protein (GFP) fusions in Dictyostelium cells, and whole cell lysates were incubated with nitrocellulose membranes carrying different phosphatidylinositols (Fig. 1A–C). Consistent with our previous findings (6), myosins ID, E, and F bound to PIP3, but myosins IA, IB, IC, and IK did not (Fig. 1C). In addition, myosins ID, IE, and IF bound weakly to PI(3,4)P2. We used myosin IE to further characterize myosin I-PIP3 interactions. Immunopurified myosin IE-GFP directly bound to PIP3, but not phosphatidylinositol (Fig. S1). We confirmed the interactions between myosin IE and PIP3 using liposomes containing small amounts (5%) of PIP3 or PIP2 (Fig. S2A). Therefore, myosin IE specifically binds to PIP3 under physiological conditions.
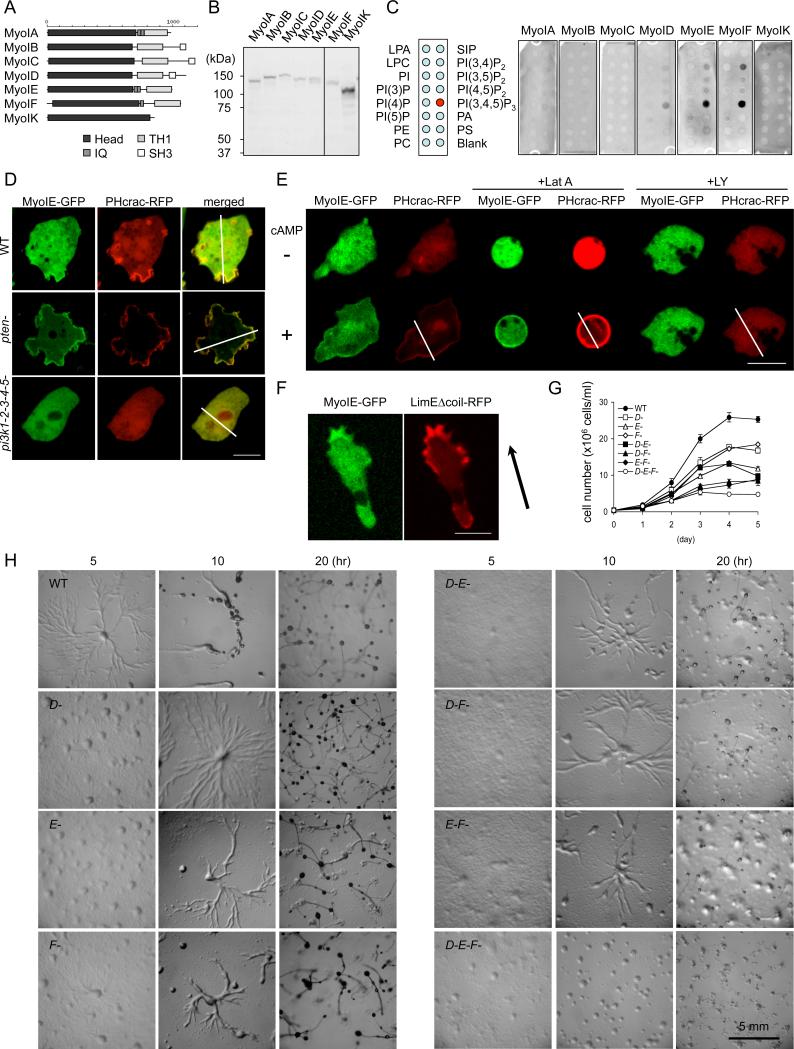
Figure 1. PIP3-binding class I myosins are required for normal cell growth and development.
(A) The domain structure of seven class I myosins in the Dictyostelium genome. (B) Immunoblotting of whole cell lysates prepared from Dictyostelium cells expressing myosin I-GFP fusions using anti-GFP antibodies. (C) Lipid dot blot assays show interactions of myosins ID, IE, and IF with PIP3. Images are representative of more than three independent experiments. (D) Localization of myosin IE-GFP and PHcrac-RFP in wild-type, pi3k-null, and pten-null cells (n ≧ 3, more than 10 cells analyzed in each experiment). Fluorescence intensity was quantified along the lines shown in Fig. S3. (E) Cells expressing myosin IE-GFP and PHcrac-RFP, a biomarker for PIP3, were observed before and after cAMP stimulation, in the presence or absence of latrunculin A or LY294002 (n ≧ 2, more than 25 cells analyzed in each experiment). (F) Cellular localization of myosin IE-GFP and LimEΔcoil-RFP, a biomarker for F-actin, in a cAMP gradient (n ≧ 6, more than 5 cells analyzed in each experiment). Arrow indicates the direction of cell migration. (G) Cell growth in wild-type and myosin I-null cells. Cells were counted daily. Values represent the mean ± SEM from more than four independent experiments. (H) Cells were plated on non-nutrient (DB) agar and examined for development. Images are representative of more than three independent experiments.
To determine whether myosin IE binds to PIP3 in cells, we observed the localization of myosin IE-GFP in both undifferentiated and differentiated cells by quantitative fluorescence microscopy (Fig. 1D and S3A). In undifferentiated cells, myosin IE-GFP colocalized with PHcrac-RFP, a fluorescent reporter for PIP3 (11), at macropinocytic cups and pseudopods at the plasma membrane. The membrane association of myosin IE-GFP depended on PIP3, because both myosin IE-GFP and PHcrac-RFP were not associated with the plasma membrane in pi3k-null cells, which contain decreased amounts of PIP3 and lack macropinocytic cups and pseudopods. Conversely, membrane association of myosin IE-GFP and PHcrac-RFP was enhanced in pten-null cells, which have higher amounts of PIP3 (12). Unlike undifferentiated cells, differentiated cells only transiently produce PIP3 in response to the chemoattractant cAMP, which promotes actin polymerization in chemotaxis (13). Myosin IE-GFP and PHcrac-RFP were observed primarily in the cytosol in the absence of cAMP, but were transiently recruited to the plasma membrane after cAMP stimulation (Fig. 1E, S3B and Movie S1-3). The PI3K inhibitor LY294002 blocked the transient recruitment of myosin IE-GFP and PHcrac-RFP, suggesting that myosin IE-GFP localization requires PIP3. In contrast, latrunculin A, which disrupts the actin cytoskeleton, did not affect cAMP-stimulated membrane recruitment of myosin IE-GFP and PHcrac-RFP. Finally, myosin IE-GFP was observed at the leading edge of chemotaxing cells (Fig. 1F), where PIP3 is concentrated, and colocalized with LimEΔcoil-RFP, a biomarker for F-actin (14). Consistent with our findings, a previous study reported myosin IE at the leading edge of randomly migrating cells (15).
To determine the function of myosins ID, IE, and IF, we generated single, double, and triple knockouts in Dictyostelium cells by homologous recombination (Fig. S4). Single knockouts exhibited only minor growth defects, whereas double and triple knockouts showed more severe growth defects (Fig. 1G). These results suggest that myosins ID, IE, and IF have overlapping functions. When starved, Dictyostelium cells display chemotactic migration toward aggregation centers, which secrete cAMP and differentiate into fruiting bodies (1, 4). Single knockouts normally developed, but double knockouts produced fewer, smaller fruiting bodies (Fig. 1H). Triple knockout cells were defective in chemotactic aggregation and fruiting body formation, developmental phenotypes that were rescued by myosin IE-GFP overexpression (Fig. S5). These developmental defects were not due simply to impaired expression of developmentally regulated genes, because cAMP receptor 1 and adenylate cyclase were present at normal abundances in triple knockout mutants during development (Fig. S6).
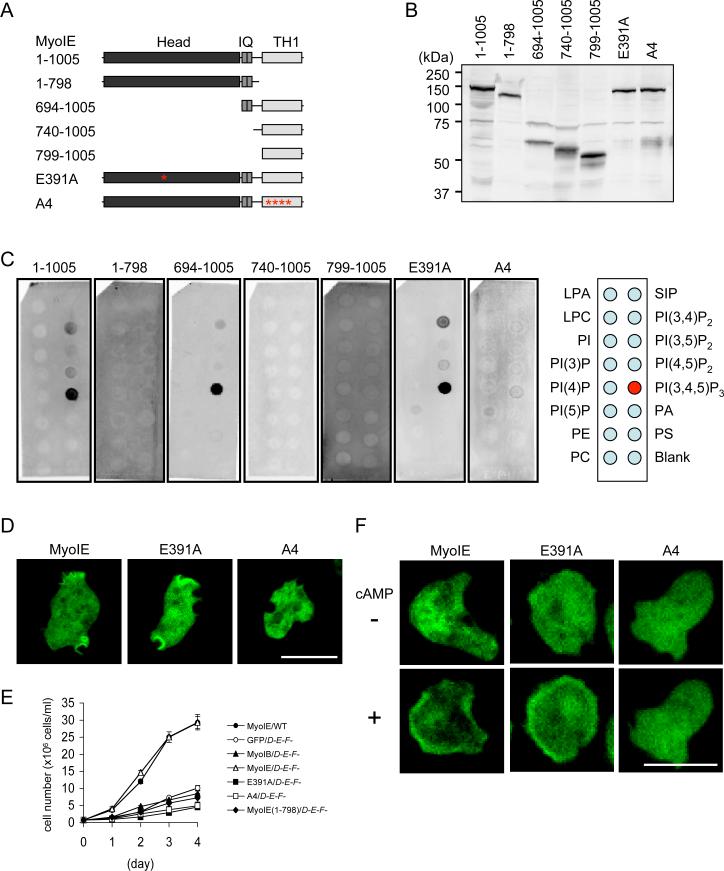
To map PIP3-binding regions in myosin IE, we performed lipid blot assays using truncated forms of this protein. Deletion of the TH1 domain blocked interactions with PIP3, showing that this region is necessary (1–798 in Fig. 2A–C). Furthermore, alanine substitutions of the four conserved positive residues (Lys859, Lys863, Arg868, and Arg869) in the second beta sheet of the PH domain of the TH1 domain (16) also inhibited the interaction (A4 in Fig. 2A–C). However, the TH1 domain was not sufficient for the interaction and the IQ domain was also required (694–1005, 740–1005, and 799–1005 in Fig 2A–C). We confirmed that the TH and IQ domains were sufficient for PIP3-interactions, which were reduced by A4 mutations in the liposome binding assay (Fig. S2B). Mutants that could not interact with PIP3 (A4) failed to localize to the plasma membrane in undifferentiated cells (Fig. 2D) and were unable to rescue the growth defect of triple knockout cells whereas wild-type myosin IE-GFP fully rescued the growth defects of triple knockouts (Fig. 2E). Similarly, these mutant proteins did not translocate to the plasma membrane in differentiated cells upon cAMP stimulation (Fig. 2F and Movie S4). A mutation in the conserved critical residue in the motor domain (Glu391→Ala; E391A) (17) did not interfere with PIP3-interactions in vitro or in vivo (Fig. 2C, D, F and Movie S4), but abolished the ability of myosin IE to rescue the growth defect of triple knockout cells (Fig. 2E).
Figure 2. The IQ and TH1 domains are required for PIP3-myosin IE interactions.
(A) Truncated and mutated forms of myosin IE used in the present study are shown. (B) Immunoblotting of whole cell lysates confirms expression of myosin I constructs in Dictyostelium cells. (C) Results of lipid dot blot assays show that the IQ and TH1 domains are necessary and sufficient for PIP3 interactions. Images are representative of more than two independent experiments. (D) Mutations in the TH1 domain (A4), but not in the motor domain (E391A), block myosin IE localization in undifferentiated triple knockout cells (n = 3, more than 50 cells analyzed in each experiment). (E) Wild-type and triple knockout cells expressing the indicated myosin IE constructs were examined for cell growth. Cells were counted daily with a hemocytometer. Values represent the mean ± SEM from more than four independent experiments. (F) cAMP-stimulated membrane recruitment in differentiated triple knockout cells expressing different myosin IE-GFP constructs (n ≧ 2 experiments, more than 50 cells analyzed in each experiment).
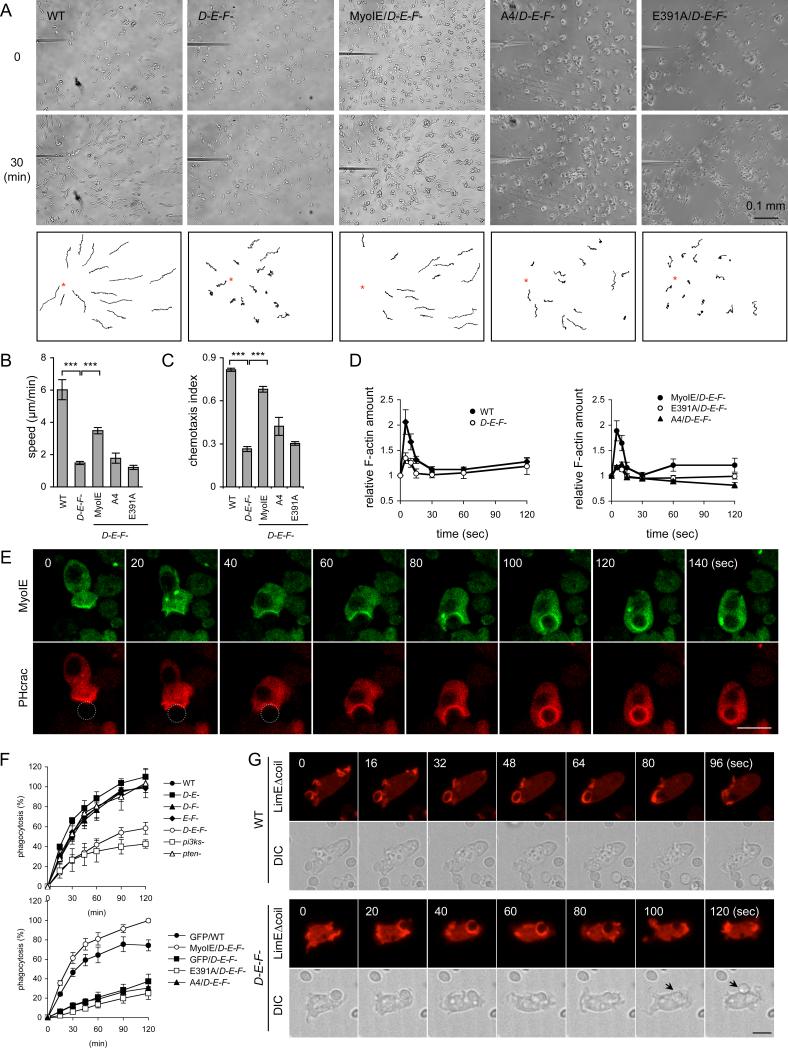
Next we directly tested whether myosins ID, IE, and IF are important for chemotaxis (Fig. 3A–C). When placed in a cAMP gradient, wild-type cells migrated toward the tip of the microneedle releasing cAMP. In contrast, chemotaxis speed was reduced in triple knockout cells by approximately 70%. The chemotaxis index was also significantly decreased. To examine actin polymerization, we measured filamentous actin (F-actin) in the Triton X-100-insoluble cytoskeletal fraction, which represents the amount of F-actin in the cell cortex (18). cAMP stimulated actin polymerization and transiently increased the amount of cortical F-actin in wild-type cells, as previously reported (Fig. 3D). In triple knockout cells, the basal amount of F-actin was not affected; however, cAMP-stimulated actin polymerization was decreased. The defects in chemotaxis (Fig. 3A–C) and cAMP-stimulated actin assembly (Fig. 3D) were partially rescued by wild-type myosin IE-GFP, but not by myosin IE(A4)-GFP or myosin IE(E391)-GFP. PIP3 production was not affected in triple knockout cells, because PHcrac-GFP was recruited to the plasma membrane in both wild-type and triple knockout cells upon cAMP stimulation (Fig. S7A and Movie S5). PIP3-simulated PKBA phosphorylation and PIP3-independent TORC2-mediated PKBR1 phosphorylation (19) also appeared normal in triple knockout cells (Fig. S7B).
Figure 3. Myosins ID, IE, and IF function in chemotaxis and phagocytosis.
(A) The movement of wild-type cells and triple knockout cells expressing the indicated myosin IE constructs toward a micropipette containing cAMP was tracked. (B) Chemotaxis speed was calculated as the distance travelled towards the micropipette divided by the elapsed time (20 min) (n ≥ 3 experiments). (C) Chemotaxis index was defined as the distance traveled in the direction of the gradient divided by the total distance traveled in 20 min (n ≥ 3 experiments). At least 15 cells were analyzed in each experiment in (B) and (C). (D) cAMP-stimulated actin polymerization was determined in wild-type and triple knockout cells expressing the indicated myosin IE constructs. At the indicated time points after stimulation, 5×106 cells were harvested and lysed, and amounts of F-actin were determined (36). Values represent the mean ± SEM from more than six independent experiments. (E) Localization of myosin IE-GFP and PHcrac-RFP was examined during phagocytosis. Circles indicate yeast cells in the first three time points. (F) Quantification of yeast uptake during phagocytosis. At the indicated time points, samples were collected and phagocytosed yeast cells were quantified. Values represent the mean ± SEM from more than four independent experiments. (G) Wild-type and triple knockout cells expressing LimEΔcoil-RFP were observed during phagocytosis of yeast cells. Arrows indicate yeast cells released from phagocytic cups.
We assessed phagocytosis in triple knockout cells, because this process also involves PIP3 signaling and actin polymerization. PIP3 is produced at phagocytic cups during phagocytosis, and PIP3-induced actin polymerization facilitates engulfment of yeast cells and bacteria (11). Myosin IE-GFP colocalized with PHcrac-RFP and LimEΔcoil-RFP at the phagocytic cups, and all three proteins dissociated from the membrane after engulfment was complete (Fig. 3E, S8 and Movie S6-7). Quantitative measurements of yeast uptake (20) revealed defective phagocytosis in triple knockout cells (Fig. 3F). Triple knockout cells formed phagocytic cups which contained F-actin as shown by LimEΔcoil-RFP, but failed to complete phagocytosis (Fig. 3G and Movie S8). Triple knockout cells lost yeast cells from the phagocytic cups (n = 123, 54%) more frequently than wild-type cells (n = 30, 10%) after apparently attempting to engulf them (Fig. 3G and Movie S8). This phenotype was statistically significant (p < 0.0001 in Fisher's exact test). These results suggest that myosins ID, IE and IF specifically control the completion of phagosome formation. Phagocytosis defects were rescued by wild-type myosin IE-GFP, but not GFP, myosin IE(A4)-GFP, or myosin IE(E391)-GFP (Fig. 3F). Myosin IE(A4)-GFP failed to associate with phagocytic cups in triple knockout cells (Fig. S9 and Movie S9). Myosin IE-GFP overexpression enhanced yeast uptake in wild-type cells (Fig. 3F) as reported previously (15). Similar to triple knockout cells, pi3k-null cells showed reduced rates of phagocytosis (Fig. 3F). In contrast, pten-null cells exhibited normal phagocytosis, suggesting that overproduction of PIP3 does not inhibit phagocytosis. These results suggest that PIP3-regulated recruitment of myosin I to the plasma membrane is critical for phagocytosis.
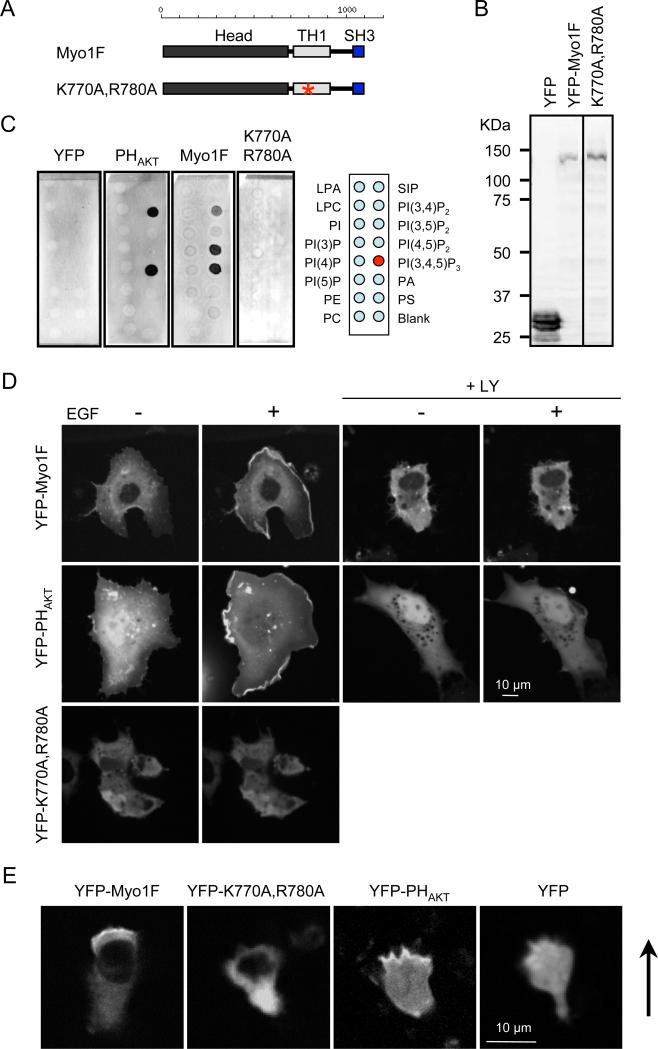
Similar to Dictyostelium myosin IE, human myosin IF also binds to PIP3 (21). Yellow fluorescent protein (YFP) fused to myosin IF (YFP-MyoIF) and a PIP3-binding PH domain from AKT (YFP-PHAKT), but not YFP alone, bound to PIP3 in lipid blot assays (Fig. 4A–C) and liposome assays (Fig. S2C). When expressed in COS-7 cells, YFP-myosin IF and YFP-PHAKT were recruited to the plasma membrane upon addition of epidermal growth factor (EGF) to the culture medium, which increases the amount of PIP3 in the plasma membrane (Fig. 4D and Movie S10) (22). The PI3K inhibitor LY294002 blocked the membrane recruitment of YFP-myosin IF and YFP-PHAKT (Fig. 4D and Movie S11). Like Dictyostelium myosin IE, mutations in the second beta sheet in the PH-like sequence in the TH1 domain (K770A, R780A) blocked membrane association in vitro and in vivo (Fig. 4A–D and Movie S11). When expressed in the human neutrophil cell line HL-60, YFP-myosin IF and YFP-PHAKT, but not YFP or YFP-myosin IF (K770A, R780A), were located at the leading edge of cells stimulated with a chemoattractant, N-formyl-methionine-leucine-phenylalanine (fMLP) (Fig. 4E). These results suggest that interactions with PIP3 are critical for the localization of human myosin IF in chemotaxis. Our data are consistent with a previous study showing that myosin IF regulates F-actin abundance, exocytosis of integrin-containing granules, and cell motility in neutrophils (23).
Figure 4. PIP3 regulates the intracellular localization of human myosin IF.
(A) The domain structure of human myosin IF. An asterisk indicates the location of the mutations introduced in the TH1 domain (K770A, R780A). (B) Immunoblotting of whole cell lysates prepared from COS-7 cells expressing YFP-human myosin IF using anti-GFP antibodies. (C) Lipid dot blot assays show that interactions between YFP-human myosin IF and PIP3 depend on the TH1 domain. YFP was used as a negative control and PHAKT-YFP was used as a positive control. Images are representative of >3 independent experiments. (D) Localization of YFP-human myosin IF, YFP-human myosin IF (K770A, R780A), and PHAKT-YFP was examined in COS-7 cells before and after EGF treatment (n = 3 experiments, more than 10 cells analyzed in each experiment). In the presence of LY294002, EGF-stimulated membrane recruitment of YFP-myosin IF and PHAKT-YFP was blocked (+LY). (E) Localization of YFP-human myosin IF, YFP-human myosin IF (K770A, R780A), PHAKT-YFP, and YFP was evaluated in HL-60 cells stimulated by fMLP (n = 3 experiments). Arrows indicate the direction of cell migration.
Discussion
Our results demonstrate that myosins ID, IE, and IF bind to PIP3 and function as downstream effecters of PIP3. The triple knockout of these three PIP3-binding class I myosins revealed their overlapping functions in chemotaxis and phagocytosis. In chemotaxis, recruitment of the PIP3-binding myosin I to the plasma membrane likely stimulates actin polymerization at the leading edge of migrating cells. Our data are consistent with previous studies showing that PIP3 is important for cAMP-stimulated actin polymerization. In phagocytosis, these myosin I molecules may control the actin cytoskeleton at the later stages, when the phagocytic membrane dissociates from the plasma membrane. These roles of myosin I in chemoattractant-stimulated actin polymerization and phagosome closure are consistent with results of previous studies (7-9). Dictyostelium myosins IA, IB, IC, and IK, which do not bind to PIP3, also regulate chemotaxis and phagocytosis (24-27). In mammals, several myosin I molecules have been reported to bind to phosphatidylserine and PI(4,5)P2 (7, 10). The findings of these previous studies and our current studies expand the repertoire of regulatory mechanisms and functions of myosin I in various membrane-cytoskeletal interactions.
The mechanism by which PIP3-myosin I interactions promote actin polymerization remains to be determined. One possible role for this interaction is to deliver the Arp2/3 complex which stimulates actin polymerization to the leading edge of chemotaxing cells. Consistent with this model, myosin I forms protein complexes with the Arp2/3 complex by associating with the scaffold protein CARMIL (28). In addition to its scaffolding role, CARMIL itself may promote actin polymerization by uncapping actin filaments and directly inhibiting capping protein (29, 30). Recruiting CARMIL to the leading edge may be a critical function for myosin I in generating free barbed ends of actin filaments (31). The interaction of myosin I with CARMIL is mediated by an SH3 domain that is present in myosin ID, but not in myosins IE or IF. Future studies to examine whether myosins ID, IE, and IF associate with CARMIL through SH3-dependent or -independent mechanisms would be interesting. Finally, myosin I has been suggested to facilitate transport of intracellular membrane vesicles along actin filaments (7). During chemotaxis, Golgi-derived vesicles carrying WASP, which stimulates actin polymerization through the Arp2/3 complex, are transported to the leading edge to reorganize the actin cytoskeleton (32). Myosin I may transport such vesicles to the leading edge using its motor activity and PIP3-binding as a targeting mechanism.
Materials and Methods
Plasmids
The PCR primers and plasmids used in this study are listed in Tables S1 and S2, respectively. Full-length myosins IB, IC, ID, IE, IF, and IK were amplified by PCR from a cDNA library (a gift from Dr. P. Devreotes, Johns Hopkins University). Myosin IA was amplified from cDNA (SHL 748) obtained from the Dictyostelium discoideum cDNA project (http://dictycdb.biol.tsukuba.ac.jp/cDNAproject.html). DNA fragments were cloned into the BglII and XhoI sites of pIS1, a plasmid carrying GFP (33) Truncated versions of myosin IE were amplified from pIS1-myosin IE. Human myosin IF was amplified from a cDNA containing full-length myosin IF (ID 5213035, Invitrogen) and cloned into pEYFP-C1 (Clontech). Mutations were created by overlap extension PCR as previously described (6). To produce pDM181-PHcrac-RFP (mCherry), a plasmid expressing the PH domain of CRAC fused to mCherry, DNA fragments containing PHcrac-RFP was generated by overlap extension PCR using primers for PHcracRFP1-4 and subcloned into pDM181. Plasmid pDM181-PHcrac-RFP-myosin IE-GFP was generated by subcloning myosin IE-GFP into pDM181-PHcrac-RFP. All constructs were confirmed by DNA sequencing.
Dictyostelium cell culture
All D. discoideum cell lines were cultured in HL5 medium at 22°C. Dictyostelium cells transformed with myosin I-GFP and PHcrac-GFP or PHcrac-RFP plasmids were selected in HL5 medium containing 20 μg/ml G418. To select transformants carrying the LimEΔcoil-RFP plasmid, 50 μg/ml hygromycin was used. To inhibit actin polymerization and PI3 kinase, 5 μM latrunculin A and 20 μM LY294002 were used.
Gene knockout
Myosin I-null cells were generated by homologous recombination. Vectors targeting genes encoding myosins ID, IE, and IF were constructed as follows. DNA fragments containing the 5' untranslated region (UTR) of each myosin I was amplified from genomic DNA using primers MyoDd1 and MyoDd2 for myosin ID, primers MyoE2 and MyoE3 for myosin IE, and primers MyoFd1 and MyoFd2 for myosin IF. A DNA fragment containing the 3' UTR region was amplified with primers MyoDd3 and MyoDd5 for myosin ID, primers MyoE4 and MyoE5 for myosin IE, and primers MyoFd3 and MyoFd4 for myosin IF. These DNA fragments and the blasticidin-S-resistance cassette from pLPBLPv2 (Dr. M. Landree, Johns Hopkins University) were cloned into pBluescript (Stratagene) to generate the targeting vectors. The targeting vectors were linearized by restriction enzymes and introduced into growing AX2 cells by electroporation (34). Transformants were selected on HL5 medium containing 5 μg/ml blasticidin (ICN Biomedicals Inc.). After 5 days, cells were harvested and plated on SM agar with Klebsiella aerogenes. Randomly selected clones were screened for gene disruption by PCR using primer L-A15P-1, which recognizes the actin 15 promoter of the blasticidin-S-resistance cassette, in combination with MyoDd5, MyoE1, or MyoFd5 for myosins ID, IE, or IF, respectively. Southern blot analysis confirmed gene disruption as follows. DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen). Then, the DNA was digested with enzymes (fig. S2), resolved by electrophoresis, and transferred to nitrocellulose membrane (Hybond-N+, Amersham). Hybridization was performed using the Amersham ECL Direct Nucleic Acid Labelling and Detection System according to the manufacturer's instructions. The probes were generated by PCR amplification of genomic DNA with the primers used to amplify the 3' regions of the targeting vectors. The pi3k-null cells which lack five PI3 kinases were obtained from the Dicty Stock Center (35).
Lipid-binding assays
A lipid dot blot assay was performed as described (6). Cells were cultured in HL5 medium to 3-5 ×106 cells/ml and starved for 2 hours in development buffer (DB; 10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2). Cells were then collected by centrifugation at 500 × g for 5 min at 4°C and washed twice in 20 ml ice-cold 10 mM sodium phosphate (pH 7.0). Then, cells were resuspended to 5×107 cells/ml in 10 mM sodium phosphate (pH 7.0) containing 1% protein inhibitor cocktail (P8340, Sigma), and filter-lysed by passing through polycarbonate membranes with 5-μm pores (110613, Whatman) three times on ice. Cell lysates were centrifuged at 10,000 × g for 10 min at 4°C, then at 110,000×g for 20 min at 4°C. The supernatants were mixed with equal volumes of 2× binding buffer (10 mM sodium phosphate [pH 7.0], 0.5% NP40, 300 mM NaCl). Membranes spotted with different phospholipids (PIP membrane P-6001; Echelon) were incubated in phosphate buffered saline with Tween 20 containing 3% fatty acid-free bovine serum albumin (Sigma A-7030) to block nonspecific binding and then mixed with the lysates for at least 3 hr. After extensive washing, the membranes were probed with anti-GFP antibodies followed by Cy5-labeled anti-rabbit IgG antibodies (816116, Invitrogen) as described (Zhang et al., 2010). The membranes were scanned with a PharosFX Plus molecular imager and analyzed with Quantity One software (Bio-Rad).
For liposome binding assays, Dictyostelium cell lysates produced by filter-lysis and clarified as described above. Cell lysates (400 μl) were mixed with liposomes (10 μl) containing PIP3 (5%) (PIP3 PolyPIPosomes Y-P039; Echelon) for 2 hours at 4°C. Liposomes were washed with PBS by centrifugation at 45,000 rpm (TLA55, Beckman) for 20 min at 4°C and resuspended in 20 μl of SDS-PAGE sample buffer. To examine interactions with a different PIP2, liposomes containing PI(3,4)P2 (Y-P034), PI(3,5)P2 (Y-P035), and PI(4,5)P2 (Y-P045) were also used. Liposomes containing no phosphoinositol (Y-000) were used as a control. For human myosin IF, HEK293T cells were transfected with plasmids containing YFP-myosin IF and YFP-PHAKT or YFP and sonicated for 10 s. Cell lysate were clarified and used for liposome binding assays as described for Dictyostelium cell lysate.
Cell growth and development
To examine cell growth, 4×105 cells/ml were cultured in 25 ml of HL5 in a 250-ml flask on a rotary shaker at 180 rpm at 22°C and cells were counted daily with a hemocytometer. To assess developmental phenotypes, cells growing exponentially were washed twice in DB (10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2) and plated on 1% non-nutrient DB agar (5×105 cells/cm2).
Dictyostelium chemotaxis
A chemotaxis assay using Dictyostelium cells was performed as described (6, 33, 36). Briefly, cells were cultured in HL5 medium to approximately 2×106 cells/ml, washed twice with DB, resuspended to 2×107 cells/ml, and shaken for 1 hour before being induced to differentiate with 100 nM cAMP pulses at 6-min intervals for 4 hours. Differentiated cells were plated on a chambered coverslip (Lab-Tek, Nalge Nunc) and allowed to adhere to the surface. A cAMP gradient was generated by a micropipette (Femtotips; Eppendorf) containing 1 μM cAMP and a microinjector with a compensation pressure of 100 hPa (FemtoJet; Eppendorf). Images of moving cells were recorded at 30-sec intervals for 30 min using an Olympus CKX41 inverted microscope equipped with a 10×/0.25 objective connected to a gray scale digital camera (CFW-1308M). ImageJ software was used to collect and process data. Chemotaxis speed was calculated as the distance towards the micropipette divided by the elapsed time (20 min). The chemotaxis index was defined as the distance moved in the direction of the gradient divided by the total distance moved for 30-sec intervals averaged over 20 min. Localization of myosin IE-GFP in chemotaxing cells was observed with a microscope consisting of a fully automated DMI6000 (Leica) and a Yokogawa CSU10 spinning disc confocal.
Actin polymerization assay
After cells were developed for 5 hours and pretreated with 3 mM caffeine for 20 min, cells were stimulated with 1 μM cAMP (33). At various time points after stimulation, 5×106 cells were harvested and lysed in Triton X-100 buffer (1% Triton X-100, 10 mM KCl, 10 mM imidazole, 10 mM EGTA, 50 μg/ml NaN3) (36). Samples were vortexed, held on ice for 10 min, and then incubated at room temperature for 10 min. The samples were spun at 8000 × g for 4 min. The pellet fractions were washed once with Triton X-100 buffer and resuspended in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were resolved by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. Actin was quantified by densitometric analysis using ImageJ software (National Institutes of Health).
Phagocytosis
A quantitative phagocytosis assay was performed as described with some modifications (20). Exponentially growing cells were collected, washed, and resuspended at 2×106 cells/ml in Sorenson's buffer (17 mM KH2PO4, 2 mM Na2HPO4, pH 6.0) and tetramethyl rhodamine isothiocyanate-labeled yeast cells were added (1.2×107 cells/ml). Samples (1 ml) were transferred to a 1.5-ml tube containing 0.1 ml 0.4% Trypan blue to quench background fluorescence from yeast cells that were not phagocytosed. Samples were vortexed and centrifuged at 800 × g for 2 min, and the pellets were washed and resuspended in 1 ml Sorenson's buffer. Fluorescence was measured at 544 nm (excitation) and 574 nm (emission). Fluorescence intensity was normalized to total protein. Uptake by wild-type cells at 120 min was set at 100%.
For microscopic observation of phagocytosis, exponentially growing Dictyostelium cells were transferred to an 8-well chambered-coverslip (Lab-Tek, Nalge Nunc) and allowed to adhere to the surface. HL5 was replaced with phosphate buffer (17 mM KH2PO4/Na2HPO4, 5 mM CaCl2, and 10 mM MgCl2, pH 6.0). After 30 min, yeast cells (YSC2, Sigma) that were hydrated by incubation in phosphate buffer at room temperature for 30 min were added to the chamber; excess yeast cells were removed within 10 min. Before observation, the chamber was covered with a coverslip and sealed with silicone grease. Confocal time-lapse images were captured using a microscope consisting of an automated AxioObserver Z1 (Zeiss), a Yokogawa CSU22 spinning disc confocal, and a Roper Cascade II 512b camera.
Mammalian cell culture and transfection
HL-60 cells were cultured in RPMI-1640 medium (Invitrogen) supplemented with 15% fetal bovine serum in an incubator with 5% CO2 at 37°C. Granulocytic differentiation was induced by treating 5×104 cells/ml with 1.3% dimethyl sulfoxide in RPMI-1640 medium for 5 days (37). For transient transfection, 5×106 cells in 100 μl nucleofector solution containing 1 μg DNA were electroporated using Amaxa program T-19, according to the manufacturer's instructions (Amaxa Nucleofection, Lonza). Cells were then transferred to a single-well chambered coverslip (Lab-Tek, Nalge Nunc) with 2 ml RPMI-1640 medium and incubated for 5 hr before observation. 4 μM fMLP was used to stimulate HL-60 cells.
COS-7 cells were maintained in Dulbecco's modified Eagle medium (Gibco, 11965) supplemented with 10% fetal bovine serum in an incubator with 5% CO2 at 37°C. Cells were plated in an 8-well chambered coverslip (Lab-Tek, Nalge Nunc) for 24 hours at low confluency to minimize cell–cell interactions. Cells were transiently transfected with Lipofectamine 2000 (Invitrogen) for 5 hours. After the culture medium was changed, cells were cultured for an additional 8 hours before observation. 100 ng/ml of EGF was used to stimulate cells. 20 μM LY294002 were used to block PI3 kinase.
Statistical analysis
All values are means ± SEM. Results were statistically analyzed using the t-test (*, p < 0.05; **, p < 0.01). Fisher's exact test was used for the phenotype of triple knockout cells losing yeast cells from the phagocytic cups.
Supplementary Material
Acknowledgements
We thank Dictyostelium cDNA project in Japan and T. Inoue for plasmids and members of the Iijima and Sesaki laboratories for helpful discussion and technical support. Funding: This work was supported by NIH grants (GM084015 to MI and GM089853 to HS). Author contributions: C.C, Y.W and M.I. performed experiments. C.C, H.S. and M.I. designed experiments, analyzed data and wrote the paper.
Footnotes
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Haastert PJ. Chemotaxis: insights from the extending pseudopod. J Cell Sci. 2010;123:3031. doi: 10.1242/jcs.071118. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Chen CL, Iijima M. Signaling mechanisms for chemotaxis. Dev Growth Differ. 2011;53:495. doi: 10.1111/j.1440-169X.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia GL, Parent CA. Signal relay during chemotaxis. J Microsc. 2008;231:529. doi: 10.1111/j.1365-2818.2008.02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SV, Flavell RA. Myosin I: from yeast to human. Cell Mol Life Sci. 2008;65:2128. doi: 10.1007/s00018-008-7435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Roche MA, Cote GP. Regulation of Dictyostelium myosin I and II. Biochim Biophys Acta. 2001;1525:245. doi: 10.1016/s0304-4165(01)00110-6. [DOI] [PubMed] [Google Scholar]
- 10.Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17:4856. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormann D, Weijer G, Parent CA, Devreotes PN, Weijer CJ. Visualizing PI3 kinase-mediated cell-cell signaling during Dictyostelium development. Curr Biol. 2002;12:1178. doi: 10.1016/s0960-9822(02)00950-8. [DOI] [PubMed] [Google Scholar]
- 12.Tang M, Iijima M, Kamimura Y, Chen L, Long Y, Devreotes P. Disruption of PKB signaling restores polarity to cells lacking tumor suppressor PTEN. Mol Biol Cell. 2011;22:437. doi: 10.1091/mbc.E10-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 14.Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Durrwang U, Fujita-Becker S, Erent M, Kull FJ, Tsiavaliaris G, Geeves MA, Manstein DJ. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J Cell Sci. 2006;119:550. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- 16.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans. 2004;32:707. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 17.Tang S, Liao JC, Dunn AR, Altman RB, Spudich JA, Schmidt JP. Predicting allosteric communication in myosin via a pathway of conserved residues. J Mol Biol. 2007;373:1361. doi: 10.1016/j.jmb.2007.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall AL, Warren V, Condeelis J. Transduction of the chemotactic signal to the actin cytoskeleton of Dictyostelium discoideum. Dev Biol. 1989;136:517. doi: 10.1016/0012-1606(89)90277-7. [DOI] [PubMed] [Google Scholar]
- 19.Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr Biol. 2008;18:1034. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivero F, Maniak M. Quantitative and microscopic methods for studying the endocytic pathway. Methods Mol Biol. 2006;346:423. doi: 10.1385/1-59745-144-4:423. [DOI] [PubMed] [Google Scholar]
- 21.Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, Davidson K, Eguinoa A, Ellson CD, Lipp P, Manifava M, Ktistakis N, Painter G, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Dove SK, Michell RH, Grewal A, Nazarian A, Erdjument-Bromage H, Tempst P, Stephens LR, Hawkins PT. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- 22.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SV, Mehal WZ, Dong X, Heinrich V, Pypaert M, Mellman I, Dembo M, Mooseker MS, Wu D, Flavell RA. Modulation of cell adhesion and motility in the immune system by Myo1f. Science. 2006;314:136. doi: 10.1126/science.1131920. [DOI] [PubMed] [Google Scholar]
- 24.Falk DL, Wessels D, Jenkins L, Pham T, Kuhl S, Titus MA, Soll DR. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J Cell Sci. 2003;116:3985. doi: 10.1242/jcs.00696. [DOI] [PubMed] [Google Scholar]
- 25.Bretschneider T, Anderson K, Ecke M, Muller-Taubenberger A, Schroth-Diez B, Ishikawa-Ankerhold HC, Gerisch G. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J. 2009;96:2888. doi: 10.1016/j.bpj.2008.12.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieckmann R, von Heyden Y, Kistler C, Gopaldass N, Hausherr S, Crawley SW, Schwarz EC, Diensthuber RP, Cote GP, Tsiavaliaris G, Soldati T. A myosin IK-Abp1-PakB circuit acts as a switch to regulate phagocytosis efficiency. Mol Biol Cell. 2011;21:1505. doi: 10.1091/mbc.E09-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung G, Wu X, Hammer JA., 3rd Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., 3rd The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. The Journal of cell biology. 2001;153:1479. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uruno T, Remmert K, Hammer JA., 3rd CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. The Journal of biological chemistry. 2006;281:10635. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Developmental cell. 2005;9:209. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. International review of cell and molecular biology. 2008;267:183. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, Han JW, Leeper L, Gruver JS, Chung CY. Regulation of the formation and trafficking of vesicles from Golgi by PCH family proteins during chemotaxis. Biochim Biophys Acta. 2009;1793:1199. doi: 10.1016/j.bbamcr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 34.Gaudet P, Pilcher KE, Fey P, Chisholm RL. Transformation of Dictyostelium discoideum with plasmid DNA. Nat Protoc. 2007;2:1317. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- 35.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, Sesaki H, Iijima M. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22:2270. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millius A, Weiner OD. Manipulation of neutrophil-like HL-60 cells for the study of directed cell migration. Methods Mol Biol. 2010;591:147. doi: 10.1007/978-1-60761-404-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.